Open Access Article
Open Access ArticleTetraphenylethene derivative that discriminates parallel G-quadruplexes†
Lei Liu‡
a,
Wei Zhang‡b,
Ming-Qing Zhonga,
Meng-Hao Jiaa,
Fei Jianga,
Yan Zhangd,
Chao-Da Xiao *ac,
Xin Xiao
*ac,
Xin Xiao *b and
Xiang-Chun Shen
*b and
Xiang-Chun Shen *ac
*ac
aState Key Laboratory of Functions and Applications of Medicinal Plants, Guizhou Medical University, University Town, Guian New District, Guizhou 550025, P. R. China. E-mail: shenxiangchun@126.com; xcd@gmc.edu.cn
bKey Laboratory of Macrocyclic and Supramolecular Chemistry of Guizhou Province, Guizhou University, Guiyang 550025, P. R. China. E-mail: gyhxxiaoxin@163.com
cThe Key Laboratory of Optimal Utilization of Natural Medicine Resources, Guizhou Medical University, University Town, Guian New District, Guizhou 550025, P. R. China
dDepartment of Radiology, Affiliated Hospital of Guizhou Medical University, Guiyang, Guizhou 550001, P. R. China
First published on 16th May 2022
Abstract
G-Quadruplex (G4), as a non-canonical nucleic acid secondary structure, has been proved to be prevalent in genomes and plays important roles in many biological processes. Ligands targeting G4, especially small-molecular fluorescent light-up probes with selectivity for special conformations, are essential for studying the relationship between G4 folding and the cellular response. However, their development still remains challenging but is attracting massive attention. Here, we synthesized a new tetraphenylethene derivative, namely TPE-B, as a parallel G4 probe. Fluorescence experiments showed that TPE-B could give out a strong fluorescence response to the G4 structure. Moreover, it gave a much higher fluorescence intensity response to parallel G4s than anti-parallel ones, which indicated that TPE-B could serve as a special tool for probing parallel G4s. The circular dichroism (CD) spectra and melting curves showed that TPE-B could selectively bind and stabilize parallel G4s without changing their topology. ESI-MS studies showed that TPE-B could bind to parallel G4 with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. The gel staining results showed that TPE-B was a good candidate for probing parallel G4s. Altogether, the TPE-B molecule may serve as a promising new probe that can discriminate parallel G4s.
1 stoichiometry. The gel staining results showed that TPE-B was a good candidate for probing parallel G4s. Altogether, the TPE-B molecule may serve as a promising new probe that can discriminate parallel G4s.
Introduction
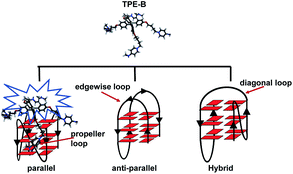
G-Quadruplexes (G4s) are non-canonical DNA and RNA structures found in nucleic acid sequences that are rich in guanine (G) residues. X-Ray1,2 and NMR3–8 studies have elucidated that such G-rich sequences can form G4s with different topologies, which usually can be categorized as parallel, hybrid and anti-parallel. Parallel G4s contain four strands oriented in the same direction, and propeller loops that surround the G-stem forming grooves. The anti-parallel G4s consist of two strands in opposition to the others and edgewise loops or diagonal loops above the external G-tract. However, hybrid G4s have three strands oriented in one direction and the fourth strand in the opposite direction with both the propeller loop and edgewise loop.9,10 Bioinformatics analysis has shown that G4s are prevalent in the human genome.11–14 After a decade of research effort, evidence shows that G4s exert important biological functions.15–18 For example, G4s in the oncogene promoter region can regulate the protein expression of VEGF19 and NRAS.20 The G4s formed by the TTAGGG repeat sequence in telomere end play an essential role in telomere homeostasis.21–24 Furthermore, G4s in the mRNA play different roles in regulating the transcription.19,25,26 Thus, developing molecules targeting G4s for studying their function has attracted massive attention.To analyze the existence and function of G4s in cells, probe molecules are highly required.27,28 Recently, antibodies for G4 have been developed and could successfully visualize both DNA and RNA G4s in human cells.29–31 However, this raises some issues related to the antibodies; for example, the artefacts caused by induced G4 formation, and the fixing and permeabilizing procedure of the immunodetection, which may impair the morphological integrity of the G4s in cells, which limit the application of antibodies.32,33 Thus, fluorescent light-up probes that directly detect the G4s are a good alternative. Many fluorescent probes have been reported; for example, the organic molecules, including cyanine,34 thiazole,35,36 triphenylmethane,37 or carbazole,38 and metal complexes.39,40 However, poor selectivity and ligand-induced conformation changes limit their application.32 Different topologies may endow the G4s with distinct functions. Numerous proteins can recognize G4s with a special topology and play different biofunctional roles. For example, transcription factor ICP4 has been shown to bind preferentially to parallel G4s, which is important for the transcription of HSV-1 in herpes virus-infected human cells.41 Heterochromatin protein 1α42 and insulin-like growth factor type I43 have both been shown to have selectivity for G4s in different topologies, which are essential for heterochromatin and cell growth, respectively. Therefore, the molecules with selectivity toward special G4s, which can analyze the relevant gene, are worth exploring.
Here, we tried to find and develop a small molecule that could discriminate particular G4s. We tried to find the molecule with the following principles: first, it should give out a fluorescence response to the G4 structure, which can be used for a G4 probing study; second, it should be able to distinguish different G4s; third, it should not induce conformation changes while binding to the G4s. Tetraphenylethene (TPE) as a typical aggregation-induced emission (AIE) dye has attracted attention as a light-up probe for G4s, but most probes developed with TPE did not show good selectivity towards the special G4 topology.44–46 Thus, to develop a G4 fluorescent probe with selectivity towards the special G4 topology, we modified the TPE with four side arms containing pyridinium rings. Pyridinium has shown good affinity towards nucleobase or phosphate oxygen atoms of DNA through π–π stacking, hydrogen bonds and electrostatic interactions.47–49 Thus, it may help the arms be accommodated within the loop and enhance the selectivity. We synthesized the tetraphenylethene derivative TPE-B, and through fluorescence, CD, ESI-MS, and docking analyses, we found that it had selectivity towards parallel G4s. The gel-staining experiments and cell-staining results demonstrated the good possibility of TPE-B for selectively imaging the parallel G4. Altogether, the TPE-B was demonstrated to have promise as a parallel G4-selective probe.
Experimental
Materials and reagents
All DNAs and RNAs were synthesized on a 1 μmol scale on an automatic DNA/RNA synthesizer (Nihon Techno Service Co., Ltd., NTS M-2). After the automated synthesis, the oligonucleotides were detached from the support and deprotected. All the oligonucleotides were purified by HPLC (Shimadzu (China) Co., Ltd., LC-16). The RNA and DNA sequences used in this study are listed in Table 1.| Oligo | Sequence (from 5′ to 3′) | Structure |
|---|---|---|
| NRAS | UGUGGGAGGGGCGGGUCUGGG | Parallel52 |
| TERRA | UUAGGGUUAGGG | Parallel53 |
| C-Myc | TGAGGGTGGGTAGGGTGGGTAA | Parallel54 |
| VEGF RNA | GGAGGAGGGGGAGGAGG | Parallel15 |
| TBA 15 | GGTTGGTGTGGTTGG | Anti-parallel54 |
| hRAS1 | TCGGGTTGCGGGCGCAGGGCACGGGCG | Anti-parallel55 |
| Bom 17 | GGTTAGGTTAGGTTAGG | Anti-parallel56 |
| Telomere DNA | AGGGTTAGGGTTAGGGTTAGGG | Anti-parallel57 |
| ODN-51 | TAGGGTTAGGGTTAGGGTTAGGG | Hybrid58 |
| AT | ATATATATATAT | Duplex |
| AU | AUAUAUAUAUAUAUAUAUAU | Duplex |
| i-motif | CCCTAACCCTAACCCTAACCC | i-motif59 |
| mut NRAS | UGUGUGAGUGGCGUGUCUGUG | Single-stranded |
| mut VEGF RNA | UGAUGAGUGUGAUGAUG | Single-stranded |
| mut TERRA | UUAGUGUUAGUG | Single-stranded |
| mut TBA 15 | TGTTGTTGTTGTTGT | Single-stranded |
TPPE was the final compound obtained from tetra-(4-pyridylphenyl) ethylene purchased by Aladdin (ML3771904) and modified through subsequent operations. First, 3 M hydrochloric acid was added for protonation, followed by heating to reflux, and finally the product was concentrated and dried to obtain TPPE. Details of the synthesis and characterization of TPE-B are available in the ESI.† TPE-B and TPPE were water soluble, and aqueous solutions were prepared for subsequent studies.
Cell lines and culture conditions
HeLa and HEK293 cells were obtained from the Kunming cell bank of the Chinese Academy of Sciences, MCF-7 cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA). The HeLa cells were kept in Dulbecco's modified eagle's medium (DMEM; Gibco, USA, Cat: C11995500BT) with 10% heat-inactivated fetal bovine serum (FBS; Si Ji Qing, Hangzhou, China, Cat: 11011-8611). The HEK293 cells were kept in Dulbecco's modified eagle's medium (DMEM; Gibco, USA, Cat: C11995500BT) with 15% heat-inactivated fetal bovine serum (FBS; Si Ji Qing, Hangzhou, China, Cat: 11011-8611). The MCF-7 cells were kept in Dulbecco's modified eagle's medium (DMEM; Gibco, USA, Cat: C11995500BT) with 5% heat-inactivated fetal bovine serum (FBS; Si Ji Qing, Hangzhou, China, Cat: 11011-8611). All the cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C.Fluorescence spectroscopy
Fluorescence spectra were recorded on a VARIANCARYE-CLIPSE fluorescence spectrophotometer (Varian, USA) with a 10 mm-path-length micro quartz cell. Oligo solutions were prepared as 3 mL DNA/RNA samples at different concentration gradients in the presence of 100 mM KCl/NaCl and 10 mM Tris–HCl buffer (pH 7.4). Each solution was heated to 95 °C for 5 min and then gradually cooled to room temperature. TPE-B/TPPE solutions (final concentration was 5 μM) were added to the oligo samples prior to the spectral measurements. During the emission spectral measurements, both the excitation and emission spectral slits were set to 10 nm and the excitation wavelength for TPE-B was set at 339 nm, while both the excitation and emission spectral slits were set to 5 nm and the excitation wavelength was set at 356 nm for TPPE.For the fluorescence quantum yield measurements, coumarin 6H (C6H) (ΦF = 0.58) was used as a standard. The fluorescence quantum yield was calculated by the formula (1):
| Yu = (Ys × Fu × As)/(Fs × Au) | (1) |
For the fluorescence titration experiments, the TPE-B concentration was fixed at 5 μM, and then the TPE-B was titrated with increasing concentrations of different oligos. The oligo solutions were prepared as described above, and the fluorescence spectra of TPE-B in the range of 300–650 nm (TPPE in the range of 350–800 nm) were recorded. The dissociation constant (Kd) from the fluorescence titration experiments was analyzed by GraphPad Prism 9.2.0. Data was fitted using the saturation binding eqn (2) and one site specific binding mode.
| Y = Bmax × X/(Kd + X) | (2) |
CD spectroscopy
CD experiments were carried out on a Chirascan spectrophotometer (Applied Photophysics, Surry, UK). The quartz cuvette with a 2 mm path length was employed for the spectra recorded over a wavelength range of 220–320 nm at 1 nm bandwidth, 100 nm min−1, and 0.25 s per point. For each experiment, an average of three scans was taken, while the corresponding buffer blanks were used to correct the background. G4 solutions for the CD spectra were prepared as 0.4 mL samples at 5 μM (VEGF RNA, NRAS, TERRA) or 15 μM (hRAS1, ODN-51, telomere DNA) concentrations individually in the presence of 100 mM KCl/NaCl, 10 mM Tris buffer (pH 7.4). The i-motif solutions (15 μM) for the CD measurements (400 μL) were prepared in 10 mM sodium phosphate buffer (pH 5.0) containing 100 mM NaCl. The G4 samples were prepared by heating the oligonucleotides at 95 °C for 5 min and gradually cooling them to room temperature. TPE-B solution (1–10 or 1.5−30 μM as final concentrations) was added to the G4 solutions prior to measurement. The melting curves were obtained by monitoring the 265, 285, 290 and 295 nm CD band.Mass spectrometry
The ESI-MS spectra was measured under the condition modified from the previous report using an Exactive Orbitrap mass spectrometer (Thermo Scientific, USA) in the negative ion mode.50 Data was acquired using Xcalibur software (Thermo Scientific). A final strand concentration of 20 μM G4 oligomers was mixed with TPE-B at different concentrations in the presence of 50% methanol. The trimethylammonium acetate (TMAA) (final concentration: 100 mM) was mixed with G4/TPE-B solution prior to the injection. The ESI spray voltage and capillary voltage were 2.75 kV and −20 V, respectively, with a heated capillary temperature of 150 °C. The ion accumulation time was 100 ms. The sample solutions were infused into the ESI source at a flow rate of 20 μL min−1 by using the instrument's syringe pump. The tube lens and skimmer voltage were fixed to −180 V and −10 V. In order to help the de-solvation process to occur, the HCD cell voltage was set to 10 eV and the cell pressure was 2.5 × 10−5 mbar.Molecular docking
The 3D coordinates of the DNA G4 structures were retrieved from the RCSB Protein Data Bank. The G-quadruplex structure for docking was prepared as described.33 With the MMFF force field, the molecular structures of TPE-B and TPPE were respectively optimized using the Discovery Studio 4.5 (Accelrys Software Inc., San Diego, USA). The molecular docking study was carried out using Autodock 4.2 with the Lamarckian genetic algorithm following the protocols developed for DNA G-quadruplex and ligand docking.33 The figures were presented using Discovery Studio 4.5.Native PAGE experiments
Here, 20% (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) polyacrylamide gels were used for the gel electrophoreses. Various oligonucleotides (5 μM) were run in 1 × TBE buffer in the presence of 20 mM KCl, at a temperature of 4 °C for 3 h. The gel was post-stained with TPE-B (10 μM) in 1 × TBE and another gel was post-stained with GelStar™.
1) polyacrylamide gels were used for the gel electrophoreses. Various oligonucleotides (5 μM) were run in 1 × TBE buffer in the presence of 20 mM KCl, at a temperature of 4 °C for 3 h. The gel was post-stained with TPE-B (10 μM) in 1 × TBE and another gel was post-stained with GelStar™.
Cell staining
Prior to the staining experiments, the cultured cells were fixed by 99% methanol (4 °C) for 1 min and washed twice with PBS for 5 min. Then, the cell membrane was permeabilized by immersing the cells in 1% Triton X-100 (2 min), and then the well was washed twice with PBS. The pre-treated HeLa cells were stained with TPE-B (5 μM) for 15 min at room temperature and subsequently washed twice with PBS for 5 min. The cells were then also stained with AO dye (1 μg mL−1) and EB dye (1 μg mL−1) for 5 min and washed by PBS twice for 5 min each time. A total of 1000 μL PBS (as control), DNase I (70 μg mL−1, Sigma), or DNase-Free RNase A (50 μg mL−1, GE) was added into the three adjacent wells and incubated at 37 °C for 2 h. The cells were rinsed by PBS twice before imaging. For each dye used in the tests, equal parameters and exposure time were used. Channel filter cube DAPI (emission wavelength: 460 nm), filter cube FITC (emission wavelength: 527 nm), and filter cube Y3 (emission wavelength: 605 nm) were used for the TPE-B, AO, and EB staining respectively.MTT assays
The in vitro cytotoxicity of TPE-B was tested by using standard methyl thiazolyl tetrazolium (MTT) assays. HeLa, HEK293 and MCF-7 cells were cultured in DMEM containing 10%, 15%, and 5% fetal bovine serum, respectively (37 °C in a humidified 5% CO2/95% air atmosphere). The cells were seeded in 96-well plates and cultured with TPE-B at different concentrations (0, 2.5, 5, 10, 20, 40, 80 μM) for 24 h. MTT solution (20 μL, 5 mg mL−1 in PBS buffer) was added to each well of the 96-well plates. After incubation for 4 h under moist conditions, followed by removal of the culture medium, 150 μL of dimethyl sulphoxide (Sigma) was added to the well and the solution was gently shaken for 10 min. The absorbance was measured at 570 nm using a microplate reader (Bio-Rad). The experiment was repeated three times. The IC50 values from the MTT experiments were analyzed by GraphPad Prism. Data were fitted using the dose–response–inhibition equation and the log[inhibition] vs. normalized response–variable slope.Absorption spectra
UV-Vis absorption spectra were measured using a micro quartz cell with a 10 mm path-length by a UV-Vis spectrophotometer (SHIMADZU UV-2700). For the study of the absorption spectra of the TPE-B and G-quadruplexes with different structures, oligo solutions were prepared as 3 mL DNA/RNA samples at different concentration gradients in the presence of 100 mM KCl, 10 mM Tris–HCl buffer (pH 7.4). Each solution was heated to 95 °C for 5 min and then gradually cooled to room temperature. The TPE-B solutions (final concentration was 5 μM) were added to the oligo samples prior to measurement.NMR experiments
DNA/RNA sequences were analyzed on a BRUKER (AV-600M) magnetic resonance spectrometer. The spectra were recorded at 25 °C. DNA/RNA samples (0.6–5.2 mM) were dissolved in 90% H2O/10% D2O (total volume: 0.15 mL) containing 10 mM potassium phosphate buffer (pH 7.0) and 100 mM KCl for TERRA, NRAS, C-Myc, ODN-51, DS RNA, DS DNA or 10 mM sodium phosphate buffer (pH 7.0) and 100 mM NaCl for telomere DNA, i-motif.Results and discussion
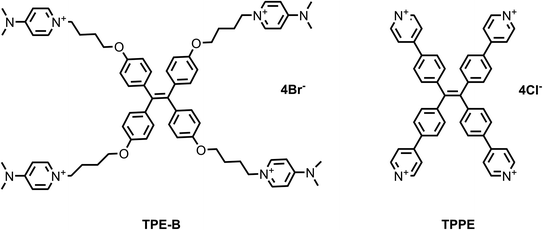
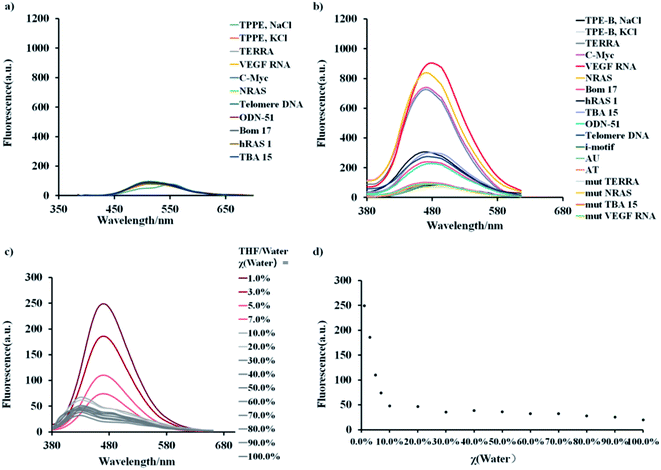
First, we synthesized two TPE derivatives: the 1,1′,1′′,1′′′-(((ethene-1,1,2,2-tetrayltetrakis(benzene-4,1-diyl)) tetrakis(oxy)) tetrakis(butane-4,1-diyl)) tetrakis(4-(dimethylamino) pyridin-1-ium) bromide (TPE-B) (Scheme S1 and Fig. S1–S6†) and 1,1,2,2-tetrakis(4-(pyridin-4-yl) phenyl) ethane tetrakis(1-chloropyridin-1-ium) chloride (TPPE) (Scheme S2†). The synthesis of TPE-B started from 4,4′-dihydroxybenzophenone. Through three steps, four flexible arms were added to the TPE core. The structural formulas of TPPE and TPE-B are shown in Scheme 1. Next, their fluorescence responses to different G4s were compared, including four parallel G4s, four anti-parallel G4s, and one hybrid G4 (Table 1).As shown in Fig. 1, both the free TPPE and TPE-B emitted a weak broad emission band ranging from 400 to 600 nm when excited at 356 nm/339 nm (Fig. 1a and b). Next, we measured the emission spectra of TPE-B. As shown in Fig. 1c and d, TPE-B displayed a strong emission band around 480 nm only with the poorer solvent THF at a high content (above 95%). While, with the addition of water content (above 5%), the emission band disappeared, which was ascribed to the typical AIE feature. Notably, TPE-B gave a strong fluorescence enhancement in response to the G4 structure, while TPPE gave barely no response, and the fluorescence enhancement of TPE-B towards parallel G4s was near 40-fold compared to the double-stranded DNA/RNA or i-motif, which indicated that TPE-B can act as a light-up probe specific for the G4 structure. That encouraged us to further investigate the TPE-B molecule. To verify that the fluorescence response of TPE-B was dependent on the G4 structure, we mutated the G4 sequences and checked the fluorescence of TPE-B mixed with the mutant sequences. As shown in Fig. 1b, TPE-B gave out no fluorescence response to the mutant sequences. Furthermore, the fluorescence enhancements of TPE-B mixed with parallel G4s were much higher than those of the anti- or hybrid G4s. This indicated that TPE-B may have a preference for parallel conformations of the G4 structure.
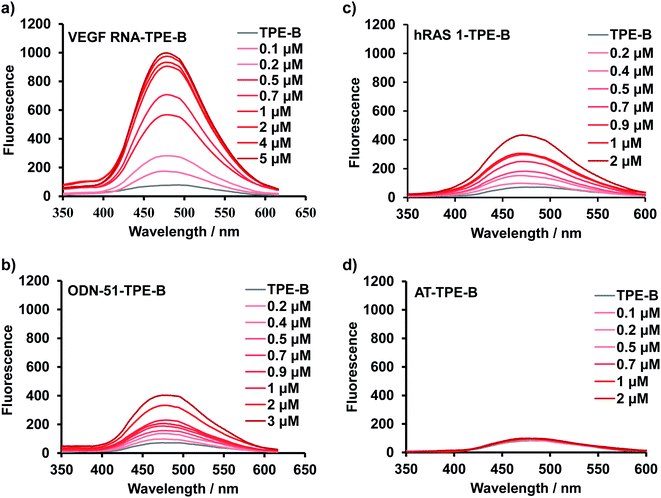
Next, fluorescence titration experiments of TPE-B were carried out with different kinds of oligonucleotides to further study the binding affinity of TPE-B towards the G4s. Fluorescence titrations were performed at a fixed concentration (5 μM) of TPE-B, and G4 sequences were continuously added until the fluorescence intensity did not significantly change. As shown in Fig. 2a, and S7–S9,† with the addition of G4s, the fluorescence was significantly enhanced, while no enhancement was observed with the addition of double-stranded DNA/RNA, i-motif or the mutant sequences, which again suggested the fluorescence response of TPE-B depended on the G4 structure (Fig. 2d and S9†). Also, the anti- or hybrid G4s induced a much lower fluorescence enhancement compared to the parallel G4s, which suggested that TPE-B can be developed as a parallel G4 selective probe (Fig. 2 and S7–S9†). The dissociation constants (Kd) between the TPE-B and G4s were analyzed through fluorescence titration experiments. The results showed that TPE-B had a higher affinity towards parallel G4s than anti- or hybrid ones. For example, the Kd values of TPE-B towards parallel G4s, such as NRAS and VEGF RNA, were more than 10-fold lower than those towards anti- or hybrid G4s, such as Bom 17 and ODN-51 (Fig. 2 and Table S1†). Also, measurements of the fluorescent quantum yields showed that the quantum yields upon binding with parallel G4s were higher than the anti- or hybrid G4s, which was in agreement with the results of the titration experiment and showed that fluorescence enhancement was always more pronounced for parallel G-quadruplexes (Table S2†). Such results again indicated that TPE-B can be used as a probe for selectively detecting parallel G4s. We investigated the structure of such sequences by nuclear magnetic resonance (NMR). The 1H-NMR spectra indicated such sequences indeed formed the G4, double strands, and i-motif topologies (Fig. S10†).
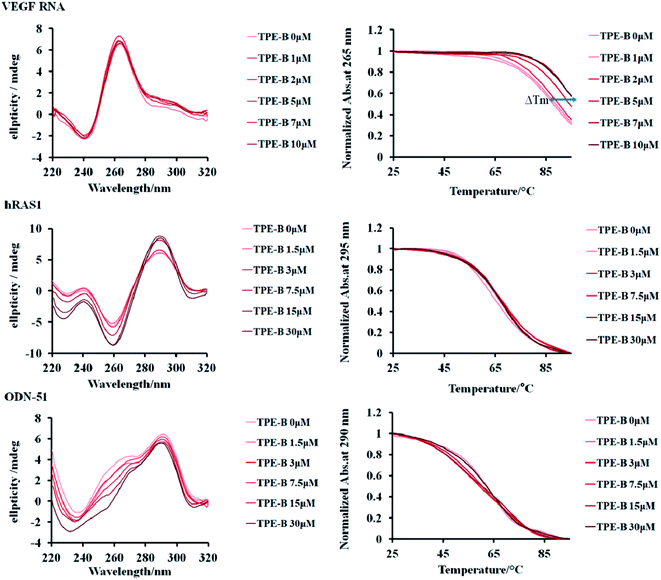
CD is particularly useful for monitoring the conformational change of macromolecules; therefore, we performed CD spectral measurements to analyze the influence of TPE-B on the conformation of G4s.51 The results of the CD experiments showed that the parallel G4s (VEGF RNA, TERRA, NRAS in the presence of K+) had a positive peak at 265 nm and a negative peak at 240 nm, while anti-parallel G4s (hRAS1 in the presence of K+; Telomere DNA in the presence of Na+) had a positive peak at 295 nm and a negative peak at 260 nm, and hybrid G4s (ODN-51 in the presence of K+) had a positive peak at 290 nm, a shoulder peak near 265 nm and a negative peak at 235 nm. Also as shown in Fig. 3 and S11,† with the addition of TPE-B, the CD spectral features of the G4s (including parallel, anti- and hybrid G4s) and i-motif did not change significantly. That result indicated that TPE-B would not induce the conformational change of G4. Further, the CD melting curves showed that TPE-B significantly increased the melting temperature (Tm) value of parallel G4s in a dose-dependent manner. The ΔTm (the Tm values of G4-TPE complex minus the Tm value of G4) for parallel G4s in the presence of TPE-B (ratio of TPE![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G4 was 1
G4 was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were above 3 °C. The ΔTm value for NRAS was 13.51 °C. However, no significant Tm change for i-motif, or the anti-, or hybrid G4s were observed (ΔTm for hRAS1 and ODN-51 were 0.07 °C and −1.50 °C, respectively), which suggested TPE-B could selectively stabilize parallel G4s (Fig. 3, S11 and Table S3†).
1) were above 3 °C. The ΔTm value for NRAS was 13.51 °C. However, no significant Tm change for i-motif, or the anti-, or hybrid G4s were observed (ΔTm for hRAS1 and ODN-51 were 0.07 °C and −1.50 °C, respectively), which suggested TPE-B could selectively stabilize parallel G4s (Fig. 3, S11 and Table S3†).
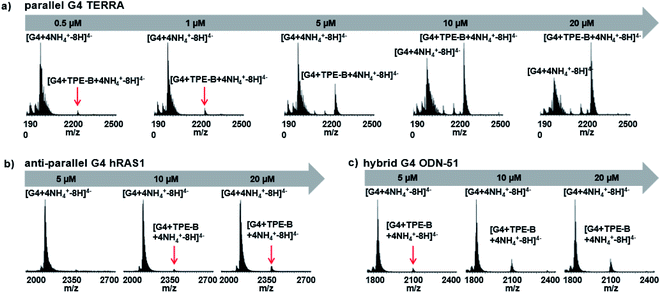
Due to the soft ionization, the ESI technique was used to determine both the mass of the G4 and the mass of the G4-ligand complex with minimal fragmentation, as it is a very straightforward way to measure the stoichiometry of the complex.60 Next, we used ESI-MS to further characterize the binding of TPE-B with G4s. As shown in Fig. 4a, for the parallel G4s, both the peaks of G4 and G4-TPE-B were directly observed, whereby the ions near m/z [1962.2] and [2228.5] corresponded to [G4 + 4NH4+–8H]4− and [G4 + TPE-B + 4NH4+–8H]4−, respectively. Such results showed the stoichiometry of the G4–TPE complex was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Also, the G4-ligand peak was significantly increased depending on the concentration of TPE-B, which confirmed the binding of the TPE-B and parallel G4. While, for the anti- and hybrid G4s, although the G4-ligand peaks were observed (for anti-parallel G4, the ions near m/z [2402] corresponded to [G4 + TPE-B + 4NH4+–8H]4−; for hybrid G4, the ions near m/z [2109] corresponded to [G4 + TPE-B + 4NH4+–8H]4−), and they were very weak even with TPE-B at a high concentration, which further confirmed the poor binding of TPE-B towards anti-parallel and hybrid G4s (Fig. 4b and c).
1. Also, the G4-ligand peak was significantly increased depending on the concentration of TPE-B, which confirmed the binding of the TPE-B and parallel G4. While, for the anti- and hybrid G4s, although the G4-ligand peaks were observed (for anti-parallel G4, the ions near m/z [2402] corresponded to [G4 + TPE-B + 4NH4+–8H]4−; for hybrid G4, the ions near m/z [2109] corresponded to [G4 + TPE-B + 4NH4+–8H]4−), and they were very weak even with TPE-B at a high concentration, which further confirmed the poor binding of TPE-B towards anti-parallel and hybrid G4s (Fig. 4b and c).
To gain structural insights into the TPE-B/G4 complexes, we performed TPE-B docking binding analysis on parallel G4s, whose structures in solution had been elucidated by NMR studies. The molecular structure of TPE-B was first optimized using BIOVIA Discovery Studio 4.5 and then docked into the G4 by Autodock 4.2. As shown in Fig. 5e, a remarkable edge to the face π–π stacking between the TPE core and G-tetrad was observed at the end of the quadruplex. Meanwhile, the proton in the 4-(dimethylamino)pyridinium group at the end of the arms could form a hydrogen bond with the “O” atom of the phosphate backbone located in the groove and the N+ may form electrostatic interactions with the phosphate backbone, which may help the four arms of TPE-B to be stretched and accommodated in the external grooves around the G-stem (Fig. 5a, b and d). The absorption spectrum of TPE-B treated with different G4s also confirmed this. As shown in Fig. S12,† TPE-B showed a peak around 287 nm. With the addition of parallel G4s, the 259 nm absorption band was greatly enhanced. Such a blue-shift is always ascribed to the n → π* transitions induced by binding between the side chain of TPE-B to the grooves of the parallel G4s (Fig. S13†). This may increase the selectivity of TPE-B for parallel G4s. Due to that the loops are located near or above the external G-tetrad in the anti- or hybrid G4s, it may induce huge steric hindrance and prevent TPE-B from stacking with the terminal G-tetrad, which may result in poor binding of TPE-B toward anti- or hybrid G4 (Scheme 2). Also, the blue-shift was not observed in the absorption spectrum of TPE-B treated with anti- or hybrid G4s, which further confirmed the selectivity of TPE-B. Comparing TPE-B with the TPPE molecule, they both have the TPE core. For TPPE, pyridinium rings are directly linked to TPE core, which may induce steric hindrance and prevent the TPPE stacking on to the terminal G-tetrad. While, for TPE-B, the flexible oxybutyl groups can be easily stretched and accommodated in groove regions (Fig. 5c). We compared the docking results of TPE-B with a larger number of literature-known G4s (Fig. S13†). Those results also showed that the TPE-B preferably binds with parallel topologies over non-parallel ones.
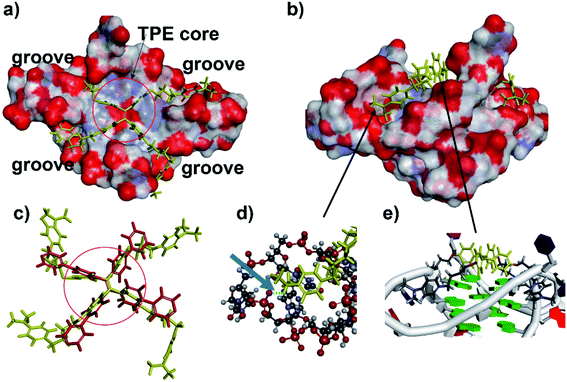 | ||
| Fig. 5 Model of the TPE-B complex with parallel G4 (PDB ID: 3IBK). Surfaces coloured by atom type: (a) TPE-B presented stacks to the G-tetrad core at the end of the quadruplex (TPE-B is in yellow). Red circle indicated the TPE core. (b) Side view of the complex showing the flexible arms of TPE-B extended into the grooves of G4. (c) Comparison of the TPE-B with TPPE (red) from top. Red circle indicated the TPE core. (d) Zoom from the (b) blue arrow, indicating hydrogen bonds (green dash), while the G4 is presented as a stick and ball, and the TPE-B molecule is presented as sticks in yellow. (e) Zoom from (b) showing the edge to face π–π stacking between the TPE core and G-tetrad. | ||
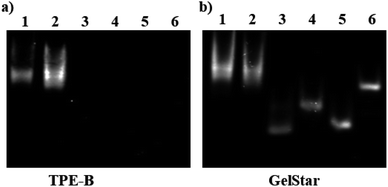
Next, we investigated the possibility of using the TPE-B as a fluorescent stain in non-denaturing polyacrylamide gel electrophoresis (PAGE). Many dyes are commercially available for staining nucleic acid in native gel but there is still no dye that can selectively stain specific G4s. We tried to stain PAGE gels with TPE-B (10 μM) for a number of nuclei acids. This showed that TPE-B could be selectively stained with parallel G4 and showed intense bands. In contrary to parallel G4s, almost no fluorescence was observed for non-parallel G4s, i-motif and mutant sequence (Fig. 6). GelStar™ (a universal nucleic acid gel stain) was also used to stain the gel conducted under the same conditions to demonstrate the specificity of the dye. Such results highlighted the potential application of TPE-B as a parallel G4 selective fluorescent staining agent.
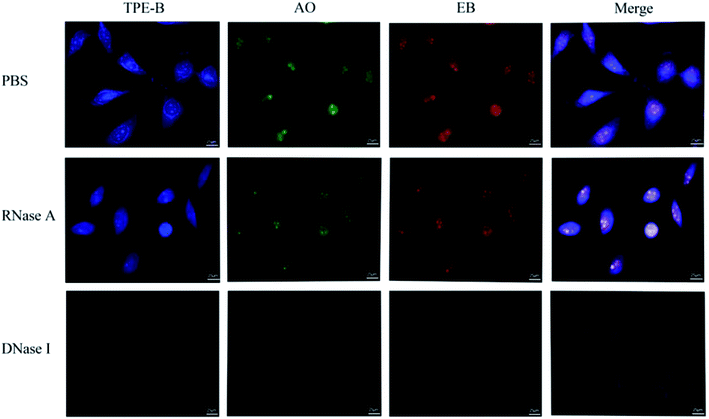
Cell staining experiments were performed to analyze the TPE-B in cells. As shown in Fig. 7, co-staining with the EB and AO dyes showed that TPE-B could be an efficient fluorescent probe for labelling foci in both the cytoplasmic and nucleolar regions. Next, to gain insights into the actual TPE-B cellular targets, fixed HeLa cells were subjected to both deoxyribouclease (DNase I) and ribonuclease (RNase A) treatments before TPE-B labelling.61 After the DNase I treatment, the enhanced fluorescence of TPE-B evidently disappeared, while the RNase A treatment only reduced the difference between the cytoplasmic and nuclear staining. These results indicated that TPE-B mainly interacted with the DNA G4s in cells.47,62 We also checked the cytotoxicity on different cell lines, including HeLa cells, MCF-7 cells and HEK293 cells. The results showed that the IC50 values of TPE-B towards HeLa cells, MCF-7 cells and HEK293 cells were 35.6 μM, 30.1 μM, and 110.7 μM respectively. Compared with other kinds of G4 fluorescent probes, such as platinum(II) Complex, which showed the IC50 (15.4 ± 0.5 μM) value against HeLa cells63 and acridine orange (AO) derivatives, which showed IC50 values on HeLa cells of 5.7, 2.4 and 0.9 μM, respectively,64 while TPE-B exhibited good safety for staining nucleic acids (Fig. S14†).
Conclusions
G4 structures have attracted massive attention as they have important biofunctions. Developing fluorescent probes that can discriminate different G4s is desirable for the detailed investigation of the features and functions of special G4s in the genome.65 In this study, we developed two tetraphenylethene derivatives and found TPE-B to be a new G-quadruplex fluorescent ligand. Compared with anti- or hybrid G4s, it showed a strong selective binding affinity for parallel G4s. Also, TPE-B showed a preferential stabilization effect on parallel G4s without inducing conformation changes. Docking analysis indicated that TPE-B can stack external G-quartets with the long side positioned in the G4 grooves. The binding between side chain TPE-B and the grooves of the parallel G4s endowed the TPE-B with selectivity of and led to a fluorescence enhancement, which may provide crucial factors for the design of AIE molecules for probing special G4s. Gel- and cell-staining experiments showed the good possibility for the use of TPE-B as a parallel G4 selective staining material. In conclusion, this study provides a new G4 probe for targeting parallel G4s.Author contributions
L. L. performed the fluorescence experiments, CD spectrum measurements and gel staining. TPE-B and TPPE were synthesized by W. Z. Molecular docking and ESI-MS experiments were performed by C. X. Data and statistical analysis were performed by M. Z and M. J, prepared all figures. F. J. helped some experiments. Y. Z. provided valuable conceptual advice. C. X, X. X and X. S. wrote the manuscript. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (32100981; U1812403-4-4; 82060772), High level Innovation Talents of Guizhou Province (2015-4029), Science and Technology Plan Projects of Guizhou Province (20191438), The Start-up Foundation for Doctors of Guizhou Medical University (2018007; GYFYBSKY-2021-49; GYFYNSFC-2021-32) and The base of International Scientific and Technological Cooperation of Guizhou Province (20175802). C.-D. X was supported by Ministry of Human Resources and Social Security of the People's Republic of China (201901) and Human resources and social security department of Guizhou province (202006083).Notes and references
- G. W. Collie, S. M. Haider, S. Neidle and G. N. Parkinson, Nucleic Acids Res., 2010, 38, 5569–5580 CrossRef CAS PubMed.
- H. Liu, R. Wang, X. Yu, F. Shen, W. Lan, P. Haruehanroengra, Q. Yao, J. Zhang, Y. Chen, S. Li, B. Wu, L. Zheng, J. Ma, J. Lin, C. Cao, J. Li, J. Sheng and J. Gan, Nucleic Acids Res., 2018, 46, 11627–11638 CrossRef CAS.
- C. D. Xiao, T. Ishizuka, X. Q. Zhu, Y. Li, H. Sugiyama and Y. Xu, J. Am. Chem. Soc., 2017, 139, 2565–2568 CrossRef CAS PubMed.
- C. D. Xiao, T. Ishizuka and Y. Xu, Sci. Rep., 2017, 7, 6695 CrossRef PubMed.
- C. D. Xiao, T. Shibata, Y. Yamamoto and Y. Xu, Chem. Commun., 2018, 54, 3944–3946 RSC.
- H. Martadinata and A. T. Phan, J. Am. Chem. Soc., 2009, 131, 2570–2578 CrossRef CAS PubMed.
- W. J. Chung, B. Heddi, M. Tera, K. Iida, K. Nagasawa and A. T. Phan, J. Am. Chem. Soc., 2013, 135, 13495–13501 CrossRef CAS PubMed.
- H. L. Bao, H. S. Liu and Y. Xu, Nucleic Acids Res., 2019, 47, 4940–4947 CrossRef CAS PubMed.
- A. T. Phan, FEBS J., 2010, 277, 1107–1117 CrossRef CAS.
- D. Yang, Methods Mol. Biol., 2019, 2035, 1–24 CrossRef CAS PubMed.
- A. K. Todd, M. Johnston and S. Neidle, Nucleic Acids Res., 2005, 33, 2901–2907 CrossRef CAS PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908–2916 CrossRef CAS PubMed.
- A. Bedrat, L. Lacroix and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1746–1759 CrossRef PubMed.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. Di Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef PubMed.
- M. J. Morris, Y. Negishi, C. Pazsint, J. D. Schonhoft and S. Basu, J. Am. Chem. Soc., 2010, 132, 17831–17839 CrossRef CAS.
- A. Bugaut and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 4727–4741 CrossRef CAS PubMed.
- M. M. Fay, S. M. Lyons and P. Ivanov, J. Mol. Biol., 2017, 429, 2127–2147 CrossRef CAS PubMed.
- J. T. Davis, Angew. Chem. Int. Ed., 2004, 43, 668–698 CrossRef CAS PubMed.
- D. Sun, K. Guo and Y. J. Shin, Nucleic Acids Res., 2011, 39, 1256–1265 CrossRef CAS PubMed.
- Y. Katsuda, S. Sato, L. Asano, Y. Morimura, T. Furuta, H. Sugiyama, M. Hagihara and M. Uesugi, J. Am. Chem. Soc., 2016, 138, 9037–9040 CrossRef CAS PubMed.
- K. Rippe and B. Luke, Nat. Struct. Mol. Biol., 2015, 22, 853–858 CrossRef CAS PubMed.
- J. J. Montero, I. López de Silanes, O. Graña and M. A. Blasco, Nat. Commun., 2016, 7, 12534 CrossRef CAS PubMed.
- L. Koch, Nat. Rev. Genet., 2017, 18, 453 CrossRef CAS PubMed.
- J. Abraham Punnoose, Y. Ma, M. E. Hoque, Y. Cui, S. Sasaki, A. H. Guo, K. Nagasawa and H. Mao, Biochemistry, 2018, 57, 6946–6955 CrossRef CAS PubMed.
- K. Guo, V. Gokhale, L. H. Hurley and D. Sun, Nucleic Acids Res., 2008, 36, 4598–4608 CrossRef CAS PubMed.
- R. C. Monsen, L. DeLeeuw, W. L. Dean, R. D. Gray, T. M. Sabo, S. Chakravarthy, J. B. Chaires and J. O. Trent, Nucleic Acids Res., 2020, 48, 5720–5734 CrossRef CAS PubMed.
- J. Mohanty, N. Barooah, V. Dhamodharan, S. Harikrishna, P. I. Pradeepkumar and A. C. Bhasikuttan, J. Am. Chem. Soc., 2013, 135, 367–376 CrossRef CAS PubMed.
- V. Dhamodharan, S. Harikrishna, A. C. Bhasikuttan and P. I. Pradeepkumar, ACS Chem. Biol., 2015, 10, 821–833 CrossRef CAS PubMed.
- G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS PubMed.
- A. Henderson, Y. Wu, Y. C. Huang, E. A. Chavez, J. Platt, F. B. Johnson, R. M. Brosh Jr, D. Sen and P. M. Lansdorp, Nucleic Acids Res., 2017, 45, 6252 CrossRef CAS.
- H. Y. Liu, Q. Zhao, T. P. Zhang, Y. Wu, Y. X. Xiong, S. K. Wang, Y. L. Ge, J. H. He, P. Lv, T. M. Ou, J. H. Tan, D. Li, L. Q. Gu, J. Ren, Y. Zhao and Z. S. Huang, Cell Chem. Biol., 2016, 23, 1261–1270 CrossRef CAS PubMed.
- V. Grande, F. Doria, M. Freccero and F. Würthner, Angew. Chem. Int. Ed., 2017, 56, 7520–7524 CrossRef CAS PubMed.
- S. Haider and S. Neidle, Methods Mol. Biol., 2010, 608, 17–37 CrossRef CAS.
- L.-J. Yu, W. Gai, Q.-F. Yang, J.-F. Xiang, H.-X. Sun, Q. Li, L.-X. Wang, A.-J. Guan and Y.-L. Tang, Chin. Chem. Lett., 2015, 26, 705–708 CrossRef CAS.
- J. W. Yan, W. J. Ye, S. B. Chen, W. B. Wu, J. Q. Hou, T. M. Ou, J. H. Tan, D. Li, L. Q. Gu and Z. S. Huang, Anal. Chem., 2012, 84, 6288–6292 CrossRef CAS PubMed.
- X. Xie, B. Choi, E. Largy, R. Guillot, A. Granzhan and M. P. Teulade-Fichou, Chem, 2013, 19, 1214–1226 CrossRef CAS PubMed.
- A. C. Bhasikuttan, J. Mohanty and H. Pal, Angew. Chem. Int. Ed., 2007, 46, 9305–9307 CrossRef CAS PubMed.
- T. Y. Tseng, Z. F. Wang, C. H. Chien and T. C. Chang, Nucleic Acids Res., 2013, 41, 10605–10618 CrossRef CAS PubMed.
- J. Weynand, A. Diman, M. Abraham, L. Marcélis, H. Jamet, A. Decottignies, J. Dejeu, E. Defrancq and B. Elias, Chem, 2018, 24, 19216–19227 CrossRef CAS PubMed.
- X. W. Liu, N. Y. Liu, Y. Q. Deng, S. Wang, T. Liu, Y. C. Tang, Y. D. Chen and J. L. Lu, J. Biomol. Struct. Dyn., 2021, 39, 5953–5962 CrossRef CAS.
- I. Frasson, P. Soldà, M. Nadai, S. Lago and S. N. Richter, Commun. Biol., 2021, 4, 510 CrossRef CAS.
- R. J. Roach, M. Garavís, C. González, G. B. Jameson, V. V. Filichev and T. K. Hale, Nucleic Acids Res., 2020, 48, 682–693 CrossRef CAS PubMed.
- H. Chen, H. Sun, Y. Chai, S. Zhang, A. Guan, Q. Li, L. Yao and Y. Tang, Biochim. Biophys. Acta, Gen. Subj., 2019, 1863, 31–38 CrossRef CAS.
- C. Kotras, M. Fossépré, M. Roger, V. Gervais, S. Richeter, P. Gerbier, S. Ulrich, M. Surin and S. Clément, Front. Chem., 2019, 7, 493 CrossRef CAS PubMed.
- Y. Hong, M. Häussler, J. W. Lam, Z. Li, K. K. Sin, Y. Dong, H. Tong, J. Liu, A. Qin, R. Renneberg and B. Z. Tang, Chem, 2008, 14, 6428–6437 CrossRef CAS PubMed.
- Q. Zhang, Y. C. Liu, D. M. Kong and D. S. Guo, Chem, 2015, 21, 13253–13260 CrossRef CAS PubMed.
- Y. J. Lu, D. P. Hu, K. Zhang, W. L. Wong and C. F. Chow, Biosens. Bioelectron., 2016, 81, 373–381 CrossRef CAS PubMed.
- A. Pipier, A. De Rache, C. Modeste, S. Amrane, E. Mothes-Martin, J. L. Stigliani, P. Calsou, J. L. Mergny, G. Pratviel and D. Gomez, Dalton Trans., 2019, 48, 6091–6099 RSC.
- V. H. Le, N. Nagesh and E. A. Lewis, PLoS One, 2013, 8, e72462 CrossRef CAS.
- A. Marchand and V. Gabelica, J. Am. Soc. Mass Spectrom., 2014, 25, 1146–1154 CrossRef CAS PubMed.
- R. Del Villar-Guerra, R. D. Gray and J. B. Chaires, Curr. Protoc. Nucleic Acid Chem., 2017, 68, 17.18.11–17.18.16 Search PubMed.
- M. Faudale, S. Cogoi and L. E. Xodo, Chem. Commun., 2012, 48, 874–876 RSC.
- G. W. Collie, G. N. Parkinson, S. Neidle, F. Rosu, E. De Pauw and V. Gabelica, J. Am. Chem. Soc., 2010, 132, 9328–9334 CrossRef CAS PubMed.
- J. M. Nicoludis, S. P. Barrett, J. L. Mergny and L. A. Yatsunyk, Nucleic Acids Res., 2012, 40, 5432–5447 CrossRef CAS PubMed.
- N. Kumar and S. Maiti, Nucleic Acids Res., 2008, 36, 5610–5622 CrossRef CAS PubMed.
- A. De Rache and J. L. Mergny, Biochimie, 2015, 115, 194–202 CrossRef CAS PubMed.
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263–282 CrossRef CAS PubMed.
- T. Frelih, B. Wang, J. Plavec and P. Šket, Nucleic Acids Res., 2020, 48, 2189–2197 CrossRef CAS PubMed.
- H. A. Assi, R. W. t. Harkness, N. Martin-Pintado, C. J. Wilds, R. Campos-Olivas, A. K. Mittermaier, C. González and M. J. Damha, Nucleic Acids Res., 2016, 44, 4998–5009 CrossRef PubMed.
- J. Jaumot and R. Gargallo, Curr. Pharm. Des., 2012, 18, 1900–1916 CrossRef CAS.
- N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027–1030 CrossRef CAS PubMed.
- A. Laguerre, K. Hukezalie, P. Winckler, F. Katranji, G. Chanteloup, M. Pirrotta, J. M. Perrier-Cornet, J. M. Wong and D. Monchaud, J. Am. Chem. Soc., 2015, 137, 8521–8525 CrossRef CAS PubMed.
- P. Wang, C. H. Leung, D. L. Ma, S. C. Yan and C. M. Che, Chem, 2010, 16, 6900–6911 CrossRef CAS PubMed.
- J. Carvalho, E. Pereira, J. Marquevielle, M. P. C. Campello, J. L. Mergny, A. Paulo, G. F. Salgado, J. A. Queiroz and C. Cruz, Biochimie, 2018, 144, 144–152 CrossRef CAS PubMed.
- S. Asamitsu, T. Bando and H. Sugiyama, Chem, 2019, 25, 417–430 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The synthetic route of TPE-B and TPPE, the results of fluorescence titration, CD, 1H-NMR spectra, molecular docking, absorption spectra and MTT assay. See https://doi.org/10.1039/d2ra01433e |
| ‡ These authors contributed equally to this work and should be considered co-first authors. |
| This journal is © The Royal Society of Chemistry 2022 |