Open Access Article
Open Access ArticleInhibition of SARS-CoV-2 spike protein entry using biologically modified polyacrylonitrile nanofibers: in vitro study towards specific antiviral masks†
Merna H. Emama,
Hassan Nageha,
Fedaa Alia,
Mohamed Tahac,
Hasnaa A. ElShehabyb,
Rehab Amincd,
Elbadawy A. Kamounae,
Samah A. Loutfyab and
Amal Kasry *a
*a
aNanotechnology Research Center (NTRC), The British University in Egypt, El-Shorouk City, Suez Desert Road, P.O. Box 43, Cairo 11837, Egypt. E-mail: Amal.Kasry@bue.edu.eg; a.kasry@unesco.org
bVirology and Immunology Unit, Cancer Biology Department, National Cancer Institute, Cairo University, Egypt
cNano Gate, 9254 Hodashaarawy, Al Abageyah, El Mukkatam, Cairo 43511, Egypt
dNational Institute of Laser Enhanced Science (NILES), Cairo University, Giza 12613, Egypt
ePolymeric Materials Research Dep., Advanced Technology and New Materials Research Institute (ATNMRI), City of Scientific Research and Technological Applications (SRTA-City), New Borg Al-Arab City 21934, Alexandria, Egypt
First published on 31st May 2022
Abstract
With the increase of the contagiousness rates of Coronavirus disease (COVID-19), new strategies are needed to halt virus spread. Blocking virus entry by capturing its spike (S) protein is one of the effective approaches that could help in eliminating or reducing transmission rate of viruses. Herein, we aim to develop a nanofiber-based filter for protective face masks, composed of polyacrylonitrile (PAN) nanofibers (NFs)-loaded with Angiotensin Converting Enzyme-2 (ACE-2) for capturing the spike protein of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and blocking its entry. Docking simulations were performed to evaluate interactions of PAN with target proteins of both SARS-CoV-2 and Human Adenovirus type 5 (ADV-5) which was used as an in vitro model of human respiratory viruses. Scanning electron microscopy (SEM) and Fourier transformed infrared (FT-IR) spectroscopy was employed to investigate the surface morphology and to analyze the functional groups of the NFs, respectively. The mechanical properties of the electrospun NFs were investigated, according to which the tensile strengths of PAN and modified PAN NFs were 4.9 ± 1.2 GPa and 4.5 GPa. Additionally, elongations at break were 25 ± 2.5% to 24 ± 1.48% for PAN and modified PAN NFs. The tensile strength test showed good mechanical characteristics of the NFs. The ACE-2-loaded NFs were shown to be safe, with promising antiviral activity towards ADV-5. Meanwhile, a binding affinity study between the spike protein and ACE-2 was performed and the dissociation constant (KD) was found to be 1.1 nM. Accordingly, the developed antiviral filters have a potential role to stand as a base for combating various human respiratory viruses.
1 Introduction
Viral outbreaks are considered as one of the primary causes of morbidity and mortality worldwide.1 Viral infections are associated with several complications, such as cardiovascular, respiratory, and thromboembolic diseases, which might lead to secondary bacterial infections alongside the viral infection.2 The emergence of Coronavirus disease-19 (COVID-19) caused by severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) in Wuhan, China, led to the call of a viral emergency worldwide.3,4 It was reported that SARS-CoV-2 virus was discovered to be similar to severe acute respiratory syndrome Coronavirus (SARS-CoV) that caused severe acute respiratory syndrome (SARS).4During the past two decades, the three pandemics caused by Coronavirus family (SARS-CoV-1, MERS-CoV and SARS-CoV-2) have affected about 29 million people around the world, according to the World Health Organization (WHO). In September 2020, it has been reported that SARS-CoV-2 virus had already taken more than 917![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 417 lives globally.5
417 lives globally.5
Coronavirus infection is mediated first through binding of spike (S) glycoprotein to angiotensin-converting enzyme-2 (ACE-2) receptors on the host cells.6 This receptor is considered as an active site for S protein binding and hence its entry into host cells.6 Since ACE-2 is expressed in various human organs such as lungs, small intestine, and liver, infection can be extended to multiple human organs. It has been reported that, SARS-CoV-2 demonstrated a stronger affinity to ACE-2.7 This can distinguish it from SARS-CoV-1, and play a crucial role in both viral entry and transmission.8
Adenoviruses (ADV-2 and ADV-5) are double-stranded DNA (dsDNA) non enveloped viruses of 90–100 nm size, with genomes of about 36 kb long.9 They are associated with variety of human diseases, such as respiratory infections, ocular and gastrointestinal disorders. They mainly affect children and young adults.10 No specific treatment regimen is used for adenovirus infection except only two antiviral drugs, cidofovir and ribavirin have been approved by the Food and Drug Administration (FDA).11
For these reasons, development of antiviral compounds and advanced protective tools are essential to reduce morbidity and mortality among patients especially those with immunocompromised diseases, as well as to reduce economic losses.
Molecular docking and computational simulations served as scientific advances in drug discovery of newer antimicrobial agents against various microbial infections.12 Two stages are utilized for docking computations, sampling of various ligand configurations and binding pockets in protein and, calculating the binding affinity between molecular systems. Auto Dock Vina is quite possibly the most utilized docking packages in protein–ligand interactions.13,14 Phytochemical compounds have been given a great attention, as many of them are used as antimicrobial agents,15 where they cause minimal adverse effects to patients.
Various nano-carriers types have been employed, to a great extent, in combating infectious diseases.15 After more than 70 years of intensive research and investigation, the field of antiviral materials, both therapeutics and anti-infective, remain relatively underdeveloped. While antiviral polymeric biomaterials showed to be a promising avenue for addressing these challenges.
Currently available face masks are protective tools that can play crucial role in avoiding viral infections, due to presence of filters that can reduce inhalation of air-borne particles. However, no antiviral masks are available to provide specific protection against respiratory viruses, like SARS-CoV-2.
Nanofibers (NFs) possess unique physical and chemical characteristics, they provide a large surface area and a good potential to incorporate active chemistry,16 they have a range of advantages in terms of diameter, permeability, and, most significantly, large surface area, therefore, they have recently attracted a lot of attentions.17 The small pore size of nanofibrous mats have a significant effect on air permeability and surface porosity. Electrospinning has become one of the most common methods for fabricating nanofibrous mats, owing to its simplicity and efficiency. Poly acrylonitrile (PAN) is a synthetic thermo-plastic polymer that has good mechanical and chemical stability,18 moreover it has a relatively low cost which allows incorporation in many fields including: protective clothing, filtration media, and biomedical textiles.19 PAN NFs were previously used to fabricate protective face masks incorporating an anti-bacterial agent.17 Previously, PAN NFs were loaded with silver nanoparticles to filter air from pollutants and microbes.20 Whereas, coating PAN NFs with bovine serum albumin (BSA) protein were utilized for wound dressing applications.18
The aim of the current study is to fabricate PAN NFs loaded with anti-spike protein of corona virus, to block viral entry, thereby, specifically protects highly susceptible people, e.g. doctors and medical staff, against SARS-CoV-2 infection. Accordingly, in silico study was performed followed by in vitro study evaluating antiviral activity against ADV-5 as a model of human respiratory virus and against SARS-CoV-2 using binding affinity assay.
2 Materials and methods
2.1 Materials
Poly acrylonitrile (PAN, average molecular weight = 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), (3-aminopropyl)triethoxysilane (APTES, ≥98%), EDC N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride, and N-hydroxy succinimide (NHS) were all purchased from Sigma-Aldrich, USA. N,N-Dimethyl formamide (DMF, ≥99%, Laboratory reagent grade) was purchased from Fisher Chemical, USA.
000 g mol−1), (3-aminopropyl)triethoxysilane (APTES, ≥98%), EDC N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride, and N-hydroxy succinimide (NHS) were all purchased from Sigma-Aldrich, USA. N,N-Dimethyl formamide (DMF, ≥99%, Laboratory reagent grade) was purchased from Fisher Chemical, USA.
Blank PAN NFs, amine-modified PAN NFs, and ester-chemistry activated PAN NFs loaded with ACE-2 materials were subjected for cytotoxicity test. Vero cell line was obtained from ATCC (American tissue culture collection). Dulbecco's Modified Eagle Medium (DMEM), sodium pyruvate (100 mM), and L-glutamine (200 mM), penicillin (100 IU mL−1), streptomycin (10 mg mL−1), and trypsin/versine (0.05%) were all purchased from Lonza, Belgium. Fetal Bovine Serum (FBS) was purchased from Gibco, South America. MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) dye was purchased from Serva Electrophoresis GmbH, Germany. Phosphate Buffer Saline tablets (PBS, pH 7.4) was purchased from Loba Chemie, India. Each tablet was dissolved in 200 mL distilled water. Dimethyl sulfoxide (DMSO) was purchased from Analysis Company, Egypt.
Blank PAN NFs, and amine-modified PAN NFs materials were subjected to anti-viral assay test. Adenovirus (VR-5) was obtained from ATCC (American tissue culture collection, USA). QIAamp viral RNA extraction kit was purchased from Qiagen, Valencia, USA. qRT-PCR kit (Power SYBR™ Green PCR Master Mix) was purchased from Thermo-Scientific, USA. cDNA synthesis kit was purchased from Roche Diagnostics, Germany.
Recombinant human coronavirus SARS-CoV-2 spike glycoprotein S1 (active), (Cat. No. ab273068), recombinant anti-SARS-CoV-2 spike glycoprotein antibody H6 (chimeric), (Cat. No. ab272854), recombinant human ACE-2 protein, (Cat. No. ab151852), Goat anti-Mouse IgG H&L (Alexa Flour 647), (Cat. No. ab150115), Anti-SARS-CoV-2 spike glycoprotein antibody Coronavirus, (Cat. No. ab272504), and Goat anti-rabbit IgG Fc (Alexa Flour 647), (Cat. No. ab150091), were all purchased from Abcam, Cambridge, United Kingdom.
2.2 Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) distilled water as 9
distilled water as 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).21,22 PAN-APTES NFs were washed several times using hot alcoholic-distilled water to remove the unreacted APTES.
1).21,22 PAN-APTES NFs were washed several times using hot alcoholic-distilled water to remove the unreacted APTES.
The mechanical strength of NFs was measured by uniaxial tensile test machine (Z050, ZwickRoell AG, Ulm, Germany). Tensile strength of NFs was measured by standard uniaxial tensile test. The NFs were cut into rectangular pieces with dimensions (40 × 10 mm) and incorporated into paper frames to protect them before the definite tensile testing. To obtain the stress–strain curves, specimens were elongated at a speed of 10 mm min−1 with an initial length of 20 mm and 50 N cell load. Measurements were taken for at least six specimens of each composition to obtain average values and standard deviations.
A modified method was performed utilizing MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) dye, that is based on the reduction of the dye by mitochondrial dehydrogenases of metabolically active cells into insoluble formazan crystals according to previously published protocol.24 Cell viability (%) was calculated by dividing average of 3 replicas of each material on average of the control and multiplied by 100.
Viral model of human adenovirus type 5 using Real-Time PCR assay: investigating antiviral activity: This was based on the following steps.
(A) Infectivity assay. Human ADV-5 (ATCC VR-5) was propagated into Vero cells and examined every day under an inverted microscope until 80–90% of CPE (cytopathic effect) was achieved. The virus culture was then frozen and thawed for three times before being tested for viral load using a quantitative real-time PCR assay.9 The standard curve was generated using 10-fold serial dilutions of standard VR-5 control (10–106 copies per mL).9
(B) Titration of adenovirus-5 DNA in cell culture. Adenoviral titration was carried out by seeding 2 × 104 cells per well into a 96-well tissue culture plate. The plate was incubated for 24 h at normal conditions of 37 °C and 5% CO2, after which the cells were infected with a two-fold serial dilution of an adenovirus stock with established viral load and incubated for 48 h until 80–90% cell lysis was observed in the undiluted viral stock, then quantitation of viral load was detected using real-time PCR assay.25
(C) Quantitation of adenovirus-5 DNA in cell culture using Real-time PCR assay.
(C.1) Nucleic acid extraction. QIAamp viral RNA extraction kit was used to extract adenovirus DNA from infected and uninfected cultures (Qiagen, Valencia, USA). The extraction was carried out in compliance with the manufacturer's guidelines. DNA extracts were put on ice and used for PCR right away, or they were kept at −20 °C before analysis (if needed). A Nano-Drop 2000 spectrophotometer (Thermo- Scientific/US, Canada) was used to determine the volume of viral DNA, and 100 ng of DNA template was used in the PCR assays.9
(C.2) Real-time PCR assay. Adenovirus DNA viral load was detected and quantified using a real-time detection system (Applied Bio systems 7500 Fast Real-time PCR). The ADV viruses were amplified using the common forward (ADV-F: ATG ACT TTT GAG GTG GAT CCC ATG GA) and the common reverse (ADV-R: GCC GAG AAG GGC GTG CGC AGG TA) primers targeting Hexon gene at nucleotide. The reaction mixture contained 12.5 μL of Maxima SYBR Green/ROX qPCR master mix (2×) (Fermentas, California, USA), 10 μM of each adeno primers, and 100 ng of extracted DNA in a final volume of 25 μL distilled water. The ADV-5 cycle protocol consisted of initial denaturation at 94.0 °C for 10 min followed by 94.0 °C for 30 s; 50.0 °C for 30 s; and at 72.0 °C for 30 s for 40 cycles then final extension at 72 °C for 7 min, finally melting curve; 95.0 °C for 15 s; 60 °C for 1 min, and 95 °C for 15 s. The PCR products were subjected to a temperature rise of up to 85 °C during the post-amplification melting temperature (Tm) study, resulting in a progressive decrease in fluorescence. It was possible to differentiate between non-specific PCR products from Adeno PCR related products in this manner.9
(D) Antiviral activity of the anti-spike protein-loaded modified PAN NFs against adenovirus (ADV-5). The safest materials were subjected for the three antiviral mechanisms as follows:
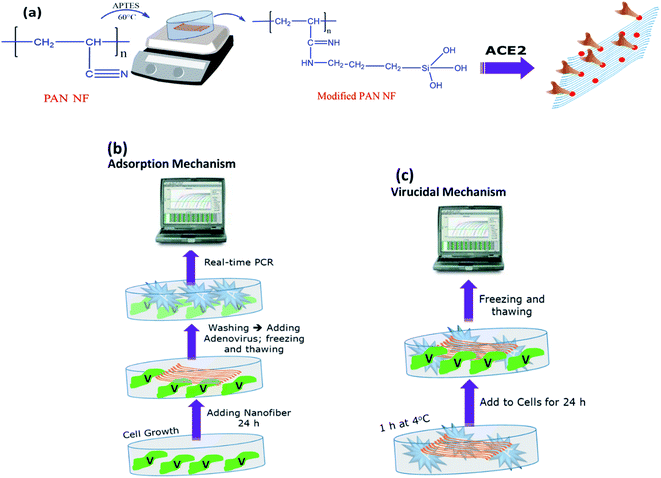
(D.1) Adsorption mechanism. As presented in (Fig. 1b), a six-well plate was seeded with 5 × 105 cells per mL and incubated at 37 °C and 5% CO2 as per normal procedure. The cells were treated with our materials after 24 h. The cells were infected with IC50 of adenovirus stock after 24 h and incubated for another 24 h under standard conditions.
(D.2) Viral replication mechanism. A six-well plate was seeded with 5 × 105 cells per milliliter and incubated at room temperature. After 24 h, the cells were infected with IC50 of adenovirus stock and incubated for another 24 h at normal conditions.
(D.3) Virucidal mechanism. This approach was used to investigate whether the tested materials that could neutralize the viral effect and then prevent it from entering the cells. As demonstrated in (Fig. 1c), a six-well plate was seeded with 5 × 105 cells per milliliter and incubated at room temperature. After incubation with IC50 of virus at 4 °C for 1 h, the cells were treated with our materials after 24 h, then incubated for another 24 h at standard condition.
In each antiviral assay, two controls were run: an untreated well for cell control and an untreated infected well for viral control. All plates were then subjected for quantitative real-time PCR assay to measure viral load.25
Ester chemistry was used to attach the ACE-2 protein to the electrospun NFs. The amine group was activated by a mixture of 1-ethyl-3-(30-dimethylaminopropyl)carbodiimide–N-hydroxysuccinimide (EDC–NHS) in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio.26 The NFs were rinsed after 30 min with distilled water to remove any excess of EDC-NHS. ACE-2 protein (100 nM) was immobilized onto the modified PAN NFs by incubation for 20 min. The NFs were then rinsed with PBS to remove excess unbound proteins.
1 ratio.26 The NFs were rinsed after 30 min with distilled water to remove any excess of EDC-NHS. ACE-2 protein (100 nM) was immobilized onto the modified PAN NFs by incubation for 20 min. The NFs were then rinsed with PBS to remove excess unbound proteins.
3 Results and discussion
3.1 Molecular docking calculation of PAN polymer against both spike protein in SARS-CoV-2 virus and Caspid protein Hexon in ADV-5
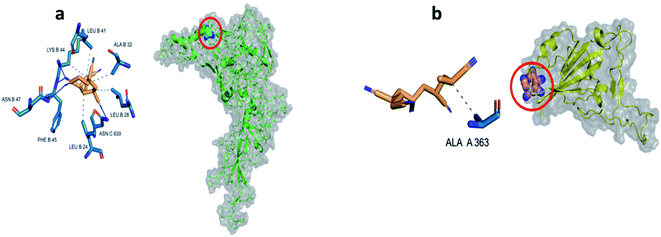
The Docking calculations usually use different scoring functions to study the likelihood of ligand to bind to the protein target, the more negative values indicate stronger binding of the ligand.27 The results demonstrate that the binding affinity of PAN against both of Hexon protein (PDB ID: 6CGV) and spike protein (PDB ID: 6M17) is about −6.3 and −5 kcal mol−1, respectively. The protein–ligand interaction profiler (PLIP) analysis was employed to determine different types of interaction within the predicted binding pockets (Fig. 2). The docking results suggest that stabilization of the interaction between PAN and Hexon protein in the predicted binding site is more likely due to due to hydrophobic interactions and hydrogen bonds with the nearby residues. Where that PAN forms hydrogen bonds with amino acid residues LYS-44B, PHE-45B, ASN-47B and ASN-639C at the predicted binding pocket in Hexon protein. Where that PHE 45B and ASN-639C are shown to form shorter (stronger) hydrogen bonds with PAN molecule with average distances 2.14 Å. On the other hands, PAN forms hydrophobic interactions with residues LEU-24, LEU-28, ALA-32, LEU-41 and LYS-44 in chain B. In contrary, Hexon protein, PAN possess only hydrophobic interactions through interacting with ALA-363A in the predicted site in SARS-CoV-2 protein (Table 1).| Index | PAN@Hexon | PAN@SARS-COV2 | ||
|---|---|---|---|---|
| Resn. | Distances [Å] | Resn. | Distances [Å] | |
| a —: not detected. | ||||
| Hydrophobic interactions | ||||
| 1 | 24B LEU | 3.84 | ALA 363 A | 3.74 |
| 2 | 28B LEU | 3.95 | — | — |
| 3 | 32B ALA | 3.93 | — | — |
| 4 | 41B LEU | 3.93 | — | — |
| 5 | 44B LYS | 3.75 | — | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Hydrogen bonds | ||||
| 1 | 44B LYS | 2.65 | — | — |
| 2 | 45B PHE | 2.17 | — | — |
| 3 | 47B ASN | 2.12 | — | — |
| 4 | 639C ASN | 2.26 | — | — |
3.2 NFs imaging results
SEM was intensively used to investigate the accepted nanofibrous mats, as shown in (Fig. 3), SEM images show that the surface morphology of NFs demonstrated accepted morphological structure; random, uniform, filamentous and continuous shape-structure of PAN NFs with no formed beads. Meanwhile, images of activated PAN/APTES/EDC-NHS/ACE-2 protein revealed major changes after incubation of protein, where loaded protein resulted in significant swollen NFs structure, compared to blank NFs. These changes might be a result of changing the functional groups after modification by APTES and incubation of proteins as confirmed by FT-IR spectra (Fig. 4). Moreover, NFs-loaded protein, preserved their entire interior and exterior morphological structure.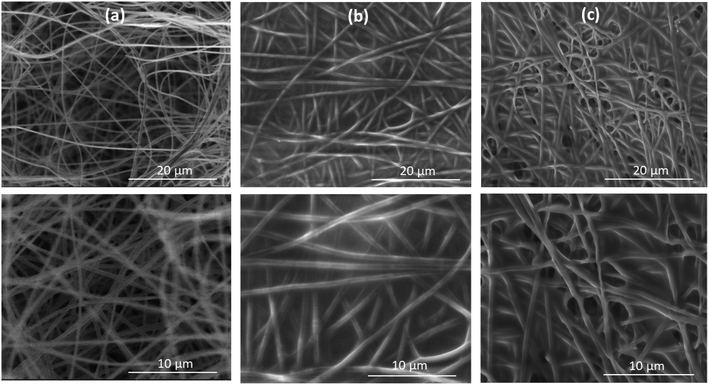 | ||
| Fig. 3 SEM micrographs of (a) blank PAN NFs, (b) PAN NFs/APTES, and (c) PAN NFs/APTES/EDC-NHS/ACE-2/S protein (with original magnification 4000× (up) and 8000× (down) at 7 kV). | ||
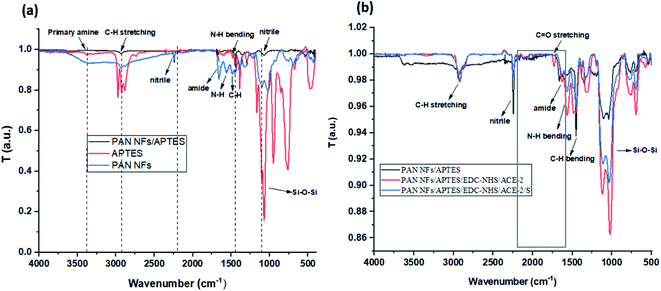 | ||
| Fig. 4 FT-IR spectra of (a) modified PAN NFs/APTES, and (b) loaded proteins onto amine-activated PAN NFs; PAN NFs/APTES, PAN NFs/APTES/EDC-NHS/ACE-2, and PAN NFs/APTES/EDC-NHS/ACE-2/S protein. | ||
3.3 NFs chemical modification
As illustrated in (Fig. 1a), PAN was modified by APTES through salinization process to modify nitrile group to amine group.22 The chemical structure of PAN NFs and modified PAN-APTES NFs were analyzed by FT-IR. As shown in (Fig. 4a), FT-IR spectra of PAN revealed appearance of amide (CO–NH) and amine (N–H) bands at ν 1657 and 1565 cm−1 respectively, upon the interaction between PAN and APTES. The main characteristic peaks of Si–O–Si that appear from ν 1020 cm−1 to 1200 cm−1, were detected in both APTES and modified PAN, which is verifying the modification of PAN by APTES. Results show that nitrile groups of PAN were successfully modified to amide and amine groups.28 The modified groups were then activated by EDC-NHS ester chemistry for protein attachment.The results presented in (Table 2) display the effect of modification of PAN NFs with APTES through measuring specific mechanical parameters e.g. Young's modulus, tensile strength, and elastic modulus. Obtained data revealed that modification of PAN NFs with APTES decreases their mechanical stability insignificantly. This indicates that APTES modification affects PAN NFs surfaces. These results are consistent with Wang et al., that demonstrated minor decrease in modified NFs' strength.22
3.4 Standard uniaxial tensile test
| Type of nanofiber | PAN NFs | |
|---|---|---|
| PAN | Modified PAN | |
| Elongation at break (%) | 25 ± 2.5 | 24 ± 1.48 |
| Max. displacement (mm) | 4.2 | 4.0 |
| Young's modulus (GPa) | 260 ± 5.52 | 258 ± 1.25 |
| Tensile strength (GPa) | 4.9 ± 1.2 | 4.5 |
| Elastic modulus (GPa) | 4.5 ± 0.85 | 4.1 ± 0.25 |
3.5 Cytotoxicity assay
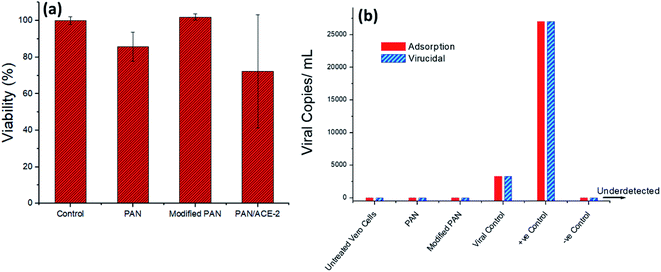
The cytotoxic effect of tested material was performed on Vero cells was determined using MTT colorimetric assay after 48 h of cell exposure. Morphological examination (Fig. S1†) confirmed MTT results, where no changes in cell morphology were revealed upon treatment of Vero cells with PAN NFs, modified PAN/APTES NFs and activated PAN NFs loaded with ACE-2 compared to untreated cells (cell control), as shown in (Fig. 5a), where the cell viability was ranged between 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100% of all tested NFs materials. The results indicate that our tested materials are safe on cells and hence can be tested for their antiviral activity.
100% of all tested NFs materials. The results indicate that our tested materials are safe on cells and hence can be tested for their antiviral activity.
3.6 Antiviral assay against ADV-5 using real-time PCR assay
Viral titration was performed using quantitative PCR assay, showed that IC50 (viral concentration can kill 50% of cells) determined at 105 copies per mL (Fig. 5b), that was used in the antiviral assay. Results showed that PAN and modified PAN/APTES NFs exerted antiviral activity via the two tested antiviral mechanisms, adsorption and virucidal/replication. They did not only prevent viral entry into cells, but also viral infectivity when they are incubated with the virus for 1 h. The virus was unable to replicate inside cells, as evidenced by undetected levels of viral copies using quantitative real-time PCR assay (Fig. 5b). This might be due to the affinity between the nanofiber and the viral receptors, which might lead to blocking viral path and preventing viral entry into the cells.29 The second reason might be due to the negative charges on the surface of PAN,30 which when bound to viral proteins, could form a complex with the virus and hence neutralize its ability to infect the cells.31Several approaches have been proposed to explain antiviral activity employed by anionic polymers depending on their complexation with virus proteins.31 This was shown to occur via formation of salt linkages or ion pairs between oppositely charged groups on PAN polymer and virus proteins, respectively, as previously explained in properties of anti-HIV-1 compounds.32 These results were supported by in silico study, where binding of PAN to Hexon protein is taking place through hydrogen bonds and hydrophobic interactions (Table 1).
3.7 Loading of anti-spike protein onto modified NFs
As shown in (Fig. 4b), FT-IR spectra of modified PAN revealed the appearance of N–H and C–H bending bands at ν 1621 and 1564 cm−1 respectively. Interestingly, both bands were shifted after incubation of proteins. The FT-IR spectra of (PAN NFs/APTES/EDC-NHS/ACE-2) revealed appearance of a peak at ν 1729 cm−1 after the incubation of ACE-2 protein, corresponding to –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group.33,34 The carbonyl band disappeared after incubation of S protein, owing to the interaction between S protein and its antibody, while the linkage between –COOH group of ACE-2, S proteins and –NH2 group of modified PAN NFs was confirmed by the appearance of the amide (CO–NH) stretching peak at ν 1631 cm−1, that was detected in both (PAN NFs/APTES/EDC-NHS/ACE-2) and (PAN NFs/APTES/EDC-NHS/ACE-2/S protein). This is besides, the characteristic peaks of N–H and C–H bending bands and their appearance at characteristic peaks at ν 1564 cm−1 and 1456 cm−1 respectively. The results verify the binding of ACE-2 and S proteins to modified PAN NFs.33,34
O group.33,34 The carbonyl band disappeared after incubation of S protein, owing to the interaction between S protein and its antibody, while the linkage between –COOH group of ACE-2, S proteins and –NH2 group of modified PAN NFs was confirmed by the appearance of the amide (CO–NH) stretching peak at ν 1631 cm−1, that was detected in both (PAN NFs/APTES/EDC-NHS/ACE-2) and (PAN NFs/APTES/EDC-NHS/ACE-2/S protein). This is besides, the characteristic peaks of N–H and C–H bending bands and their appearance at characteristic peaks at ν 1564 cm−1 and 1456 cm−1 respectively. The results verify the binding of ACE-2 and S proteins to modified PAN NFs.33,34
Regarding the stability of ACE-2-loaded PAN NFs, the samples are shown to be stable until up to 8 months at room temperature, stored under dry nitrogen. SEM images presented in (Fig. S2†) show that no visual cracks nor surface degradation. This might be due to the hydrophobic nature of PAN polymer and the storing conditions of the modified and loaded nanofibers. Moreover, the chemical structure of the protein-loaded PAN NFs, as prepared and after 8 months, doesn't indicate any structure change, as shown in the FT-IR spectra (Fig. S3†). It is expected that the samples can be stable up to a longer period if kept in refrigerator (4 °C).
3.8 Affinity test results
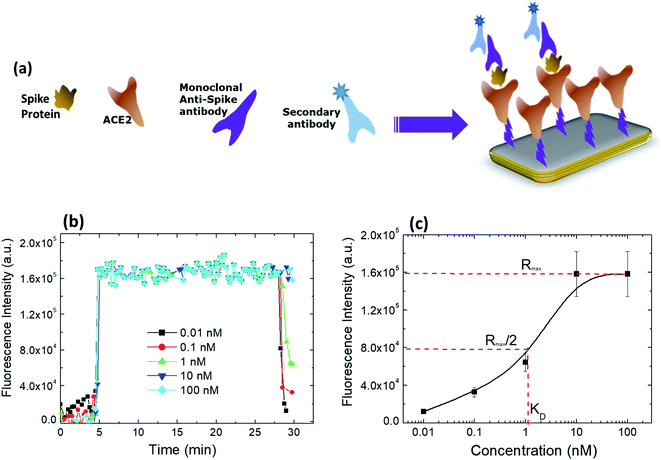
(Fig. 6a) is a schematic of the system used to study the binding interaction between ACE-2 and S protein, whereas (Fig. 6b) demonstrates a rapid increase and saturation of the fluorescence signal occurring after adding the labelled antibody. The samples were left for 20 min to ensure the binding of the secondary antibody. A decrease in the fluorescence signal is observed after the rinsing step. Despite the decrease in the signal, it is hard to observe the dissociation owing to the fact that it is a static type of measurement. However, after rinsing, the signals show a variation linearly dependent on the concentration of S protein, which is a strong proof that the binding is specific. Specificity is further proved by results of the two controls revealing a negligible signal demonstrating that there is no nonspecific binding.35 As illustrated in (Fig. 6c), the affinity constant between S protein of SARS-CoV-2 and its antibody equals 0.9 × 109 M−1 and KD equals 1.1 nM. These results indicate a high affinity and strong specific interactions between the S protein and ACE-2. The current findings are consistent with the reported results of Saponaro et al., where they demonstrated KD values of several binding affinity studies between S protein of SARS-CoV-2 and ACE-2.364 Conclusions
Developing broad-spectrum antiviral polymers holds enormous potential for two main reasons; to treat current viral diseases and to control transmission of viral pathogens, especially respiratory pathogens. ACE-2 protein loading onto NFs is a promising approach to develop a potential stable antiviral mask. It demonstrates a remarkable advancement to capture the virus on the nanofibrous surface and block its entry to the host cells. The loaded protein was specifically chosen considering viral pathologic features to enhance antiviral activity as evidenced by binding affinity. Molecular docking as well as physical, chemical, and biological investigations showed a promising antiviral activity against ADV-5 via adsorption and virucidal, and replication mechanisms. Testing the modified nanofibers over a period of 8 months showed significant stability. However, many queries are not yet answered such as the nature of this interaction, whether the virus can be reactivated or not, and the time it takes the virus to degrade. They can also serve as microbicides or antiviral drug-delivery vehicles. Further studies are still needed to explore the exact antiviral mechanism exerted by anionic polymers.A prototype was designed for the proposed anti-viral mask, in which ACE-2-loaded modified PAN NFs were placed on a commercial filter. For future prospective, PAN NFs should be investigated for their antiviral activity against other viruses as well. Besides, they can be loaded with many other proteins for suppressing infectious diseases. The proposed mask is in-progress to be tested as a potential commercial, specific antiviral mask.
It is worth mentioning that the fabrication of 1 mask is not expensive, taking in account the cost of the chemicals, antibody, labor, and overhead; the total cost of 1 filter with dimensions 2.5 × 2 cm2 would be 0.79 $ and the price of the mask that will be used to implant the filter is 0.5 $, so the total cost will be 1.28 $. The filer can be just used and changed in the same mask.
This cost was calculated for pilot scale production; therefore, it is expected to significantly decrease by about 60% in case of mass production, the price of this mask including the modified PAN filter is expected to be: 0.768 $. This low value makes the one-time use of PAN filter economically effective.
Author contributions
MHE performed the experimental work, conducted the graphical analysis for all the results, and initiated writing the original manuscript, under supervision of AK, EAK and SAL. HN prepared some NFs and helped in the chemical modification interpretations. FA performed the theoretical study. HE performed two antiviral experiments under the supervision and guidance of SAL. MT and RA supervised the embedding of NFs in the commercial masks. MHE and AK wrote the manuscript. All authors revised the manuscript and agreed with all its contents.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the Egyptian Academy of Scientific Research (ASRT) under the Covid 19 Emergency Call; Ideation, Project ID 6842, and was partially supported by the British University in Egypt through the NTRC facilities.References
- A. Habibi-Yangjeh, S. Asadzadeh-Khaneghah, S. Feizpoor and A. Rouhi, Review on heterogeneous photocatalytic disinfection of waterborne, airborne, and foodborne viruses: Can we win against pathogenic viruses?, J. Colloid Interface Sci., 2020, 580, 503–514 CrossRef CAS PubMed.
- R. He, S. He, X. Li, Y. Shi and Q. Liu, How can heart disease patients prevent complications from viral infections?, Eur. J. Prev. Cardiol., 2018, 25(7), 758 CrossRef PubMed.
- J. T. Ortega, M. L. Serrano, F. H. Pujol and H. R. Rangel, Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis, EXCLI J., 2020, 19, 410 Search PubMed.
- J. Yang, S. J. Petitjean, M. Koehler, Q. Zhang, A. C. Dumitru, W. Chen, S. Derclaye, S. P. Vincent, P. Soumillion and D. Alsteens, Molecular interaction and inhibition of SARS-CoV-2 binding to the ACE2 receptor, Nat. Commun., 2020, 11(1), 1–10 CrossRef PubMed.
- S. Basak and G. Packirisamy, Nano-based antiviral coatings to combat viral infections, Nano-Struct. Nano-Objects, 2020, 24, 100620 CrossRef CAS.
- R. Bayarri-Olmos, A. Rosbjerg, L. B. Johnsen, C. Helgstrand, T. Bak-Thomsen, P. Garred and M.-O. Skjoedt, The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization, J. Biol. Chem., 2021, 100536 CrossRef CAS PubMed.
- F. Ali, A. Kasry and M. Amin, The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant, Med. Drug Discovery, 2021, 10, 100086 CrossRef CAS PubMed.
- S. K. Saxena, Coronavirus disease 2019 (COVID-19): Epidemiology, pathogenesis, diagnosis, and therapeutics, Springer Nature, 2020 Search PubMed.
- E. M. Elmahdy, N. I. Ahmed, M. N. Shaheen, E.-C. B. Mohamed and S. A. Loutfy, Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis, J. Water Health, 2019, 17(2), 287–294 CrossRef PubMed.
- P. R. Kinchington, E. G. Romanowski and Y. Jerold Gordon, Prospects for adenovirus antivirals, J. Antimicrob. Chemother., 2005, 55(4), 424–429 CrossRef CAS PubMed.
- T. Melby and M. Westby, Inhibitors of viral entry, Antiviral Strategies, 2009, pp. 177–202 Search PubMed.
- L. Chen and J. Liang, An overview of functional nanoparticles as novel emerging antiviral therapeutic agents, Mater. Sci. Eng., C, 2020, 112, 110924 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31(2), 455–461 CAS.
- B. J. McConkey, V. Sobolev and M. Edelman, The performance of current methods in ligand–protein docking, Curr. Sci., 2002, 845–856 CAS.
- A. D. Fuzimoto and C. Isidoro, The antiviral and the coronavirus-host protein pathways inhibiting properties of herbs and natural compounds-Additional weapons in the fight against the COVID-19 pandemic?, Afr. J. Tradit., Complementary Altern. Med., 2020, 10, 405–419 CrossRef PubMed.
- C. Akduman and E. A. Kumbasar, Nanofibers in face masks and respirators to provide better protection, in IOP Conference Series: Materials Science and Engineering, IOP Publishing, 2018, p. 012013 Search PubMed.
- C. Huang, Y. Liu, Z. Li, R. Li, X. Ren and T.-S. Huang, N-halamine antibacterial nanofibrous mats based on polyacrylonitrile and N-halamine for protective face masks, J. Eng. Fibers Fabr., 2019, 14, 1558925019843222 CAS.
- S. Homaeigohar, T.-Y. Tsai, E. S. Zarie, M. Elbahri, T.-H. Young and A. R. Boccaccini, Bovine Serum Albumin (BSA)/polyacrylonitrile (PAN) biohybrid nanofibers coated with a biomineralized calcium deficient hydroxyapatite (HA) shell for wound dressing, Mater. Sci. Eng., C, 2020, 116, 111248 CrossRef CAS PubMed.
- N. Ucar, N. Kizildag, A. Onen, I. Karacan and O. Eren, Polyacrylonitrile-polyaniline composite nanofiber webs: Effects of solvents, redoping process and dispersion technique, Fibers Polym., 2015, 16(10), 2223–2236 CrossRef CAS.
- A. Sharma, S. R. Kumar, V. Katiyar and P. Gopinath, Graphene oxide/silver nanoparticle (GO/AgNP) impregnated polyacrylonitrile nanofibers for potential application in air filtration, Nano-Struct. Nano-Objects, 2021, 26, 100708 CrossRef CAS.
- R. Zhang, Y. Li, Y. Cai, Q. Han, T. Zhang, Y. Liu, K. Zeng and C. Zhao, Photocatalytic Poly(vinylidene fluoride) membrane of Ag3PO4/GO/APTES for water treatment, Colloids Surf., A, 2020, 597, 124779 CrossRef CAS.
- W. Wang, W. Li, C. Gao, W. Tian, B. Sun and D. Yu, A novel preparation of silver-plated polyacrylonitrile fibers functionalized with antibacterial and electromagnetic shielding properties, Appl. Surf. Sci., 2015, 342, 120–126 CrossRef CAS.
- T. Mosmann, Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays, J. Immunol. Methods, 1983, 65(1–2), 55–63 CrossRef CAS PubMed.
- M. B. Hansen, S. E. Nielsen and K. Berg, Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill, J. Immunol. Methods, 1989, 119(2), 203–210 CrossRef CAS PubMed.
- L. Betancur-Galvis, G. Morales, J. Forero and J. Roldan, Cytotoxic and antiviral activities of Colombian medicinal plant extracts of the Euphorbia genus, Mem. Inst. Oswaldo Cruz, 2002, 97(4), 541–546 CrossRef CAS PubMed.
- M. J. Fischer, Amine coupling through EDC/NHS: a practical approach, in Surface plasmon resonance, Springer, 2010, pp. 55–73 Search PubMed.
- D. Afriza, W. Suriyah and S. Ichwan, In In silico analysis of molecular interactions between the anti-apoptotic protein survivin and dentatin, nordentatin, and quercetin, J. Phys.: Conf. Ser., 2018, 032001 CrossRef.
- A. Dastbaz and A. R. Keshtkar, Adsorption of Th4+, U6+, Cd2+, and Ni2+ from aqueous solution by a novel modified polyacrylonitrile composite nanofiber adsorbent prepared by electrospinning, Appl. Surf. Sci., 2014, 293, 336–344 CrossRef CAS.
- R. H. Bianculli, J. D. Mase and M. D. Schulz, Antiviral Polymers: Past Approaches and Future Possibilities, Macromolecules, 2020, 53(21), 9158–9186 CrossRef CAS.
- J. K. Aronson, Meyler's side effects of drugs: the international encyclopedia of adverse drug reactions and interactions, Elsevier, 16th edn, 2016 Search PubMed.
- N. Jarach, H. Dodiuk and S. Kenig, Polymers in the Medical Antiviral Front-Line, Polymers, 2020, 12(8), 1727 CrossRef CAS PubMed.
- A. R. Neurath, N. Strick and Y.-Y. Li, Anti-HIV-1 activity of anionic polymers: a comparative study of candidate microbicides, BMC Infect. Dis., 2002, 2(1), 1–11 CrossRef PubMed.
- H. A. Alhazmi, FT-IR spectroscopy for the identification of binding sites and measurements of the binding interactions of important metal ions with bovine serum albumin, Sci. Pharm., 2019, 87(1), 5 CrossRef.
- R. P. Singh, G. Sharma, P. Agrawal, B. L. Pandey, B. Koch and M. S. Muthu, Transferrin receptor targeted PLA-TPGS micelles improved efficacy and safety in docetaxel delivery, Int. J. Biol. Macromol., 2016, 83, 335–344 CrossRef CAS PubMed.
- A. Bozdogan, R. F. El-Kased, V. Jungbluth, W. Knoll, J. Dostalek and A. Kasry, Development of a specific troponin I detection system with enhanced immune sensitivity using a single monoclonal antibody, R. Soc. Open Sci., 2020, 7(10), 200871 CrossRef CAS PubMed.
- F. Saponaro, G. Rutigliano, S. Sestito, L. Bandini, B. Storti, R. Bizzarri and R. Zucchi, ACE2 in the era of SARS-CoV-2: Controversies and novel perspectives, Front. Mol. Biosci., 2020, 7, 1–25 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra01321e |
| This journal is © The Royal Society of Chemistry 2022 |