Open Access Article
Open Access ArticleA novel construct of an electrochemical acetylcholinesterase biosensor for the investigation of malathion sensitivity to three different insect species using a NiCr2O4/g-C3N4 composite integrated pencil graphite electrode
Sehrish Bilal ab,
Muhammad Nasir
ab,
Muhammad Nasir b,
M. Mudassir Hassanb,
Muhammad Fayyaz ur Rehman
b,
M. Mudassir Hassanb,
Muhammad Fayyaz ur Rehman d,
Amtul Jamil Sami*ac and
Akhtar Hayat*b
d,
Amtul Jamil Sami*ac and
Akhtar Hayat*b
aSchool of Biochemistry and Biotechnology, University of the Punjab, Lahore 54000, Pakistan. E-mail: sehrishbilal33@gmail.com; Tel: +92 3364318117
bInterdisciplinary Research Center in Biomedical Materials (IRCBM), COMSATS University Islamabad, Lahore Campus, Lahore 54000, Pakistan. E-mail: muhammadnasir@cuilahore.edu.pk; mudassirhassan.27@gmail.com
cCenter for Biosensor Research and Development (CBRD), University of the Punjab, Lahore 54000, Pakistan. E-mail: amtuljamilsami@yahoo.com; Tel: +92 3317648259
dInstitute of Chemistry, University of Sargodha, Sargodha, Sargodha 40100, Pakistan. E-mail: muhammad.fayyaz@uos.edu.pk
First published on 7th June 2022
Abstract
Herein, an electrochemical biosensor has been prepared to assess the sensitivity of an organophosphate insecticide, malathion, to acetylcholinesterase (AChE) enzyme of three insects including Apis mellifera (honeybee), Tribolium castaneum (red flour beetle), and Zootermopsis nevadensis (dampwood termite). A composite of nickel chromite (NiCr2O4) and graphitic carbon nitride (g-C3N4) was prepared and characterized for its morphological, chemical and electrical properties. The NiCr2O4/g-C3N4 composite integrated pencil graphite electrodes were used to covalently immobilize insect AChE enzymes and amperometric response of bioelectrodes was determined through cyclic voltammetry. The prepared bioelectrodes exhibited high enzyme immobilization efficiency and electro-catalytic performance. The integrated bioelectrodes could efficiently detect malathion induced inhibition of insects' AChEs. The linear ranges for malathion were found to be 0.1–1.6 μM, 1–40 nM and 2–100 nM, and LODs were 2 nM, 0.86 nM and 2.3 nM for A. mellifera, T. castaneum, and Z. nevadensis, respectively. Additionally, the biosensing platform developed using A. mellifera AChE was found highly sensitive and effective for malathion recoveries from spiked wheat flour samples with high recovery rates. Moreover, the proposed method was adequately reproducible and selective. The results revealed that A. mellifera AChE is less sensitive to inhibition by malathion as compared to T. castaneum, and Z. nevadensis AChE. The experimental results were validated through computational docking of malathion with insect AChEs and the results were in correspondence to experimental outcomes. The proposed method can be a plausible alternate to conventional analytical methods to assess the pesticide sensitivity and toxicity of various compounds against insect enzymes.
1. Introduction
The control of insect infestation in agriculture is largely dependent on chemical pesticides. However, the unrestrained use of these pesticides is greatly disturbing the environment, human health, and survival of beneficial insects.1–3 The available toxicity data of various pesticides suggest that insect and pest enzymes behave disparately in the presence of different pesticides; for instance, honeybees have been found more susceptible to some insecticides and resistant towards other chemical or bio-pesticides, whereas red flour beetles possess reduced sensitivity against chemical and organic pesticides.4 A principle mechanism manipulated by insects or pests to resist pesticide effects is metabolic resistance. More than 500 species of insects are resistant to various classes of pesticides all over the world.5Among other widely used insecticides, malathion is a wide spectrum insecticide used for more than 50 years in agriculture, industrial and outdoor house applications to control insects or ectoparasites. Malathion is a phosphate ester having low persistence in the environment and readily converts to its metabolic forms such as malaoxon, which is a potent acetylcholinesterase (AChE) inhibitor. Malathion toxicity varies from moderate to high in target and non-targeted organisms. It has been found potentially toxic to birds, fish, aquatic larvae of terrestrial insects, and bees. It has been reported that malathion residues are highly toxic to honey bees and cause serious losses in other beneficial insect populations6,7 reported a huge loss of honeybees in apiaries due to malathion bait that was used to control mosquitos. Further this insecticide is commonly used on stored products considering its low mammalian toxicity and acute toxicity against insects such as Tribolium castaneum and other beetles which are highly resilient against many other insecticides that are considered suitable for food control.8 About half of the total applications of malathion in the US are on stored crops such as wheat, rice, sorghum, cotton and alfalfa. Many insects possess enzyme systems which are responsible for the detoxification of insecticides, explaining the selective toxicity of insects towards different insecticides. Several studies have reported the role of malathion carboxylesterases in conferring resistance to some insects including T. castaneum.9–12
Most commonly, the content of pesticides is indirectly determined by monitoring the degree of inhibition of AChE activity through methods based on enzyme-inhibition. To date a wide variety of analytical methods have been devised for both qualitative and quantitative determination of AChE activity and its inhibition, including bioassays, colorimetric Ellman's assay, radioactive methods, thin layer chromatography, gas chromatography or HPLC, etc. Although these methods are widely used and considered quite sensitive, they offer limitations such as low specificity, being labour-intensive and hold certain ethical limitations.13 From the past decade the use of electrochemical biosensors has emerged as sensitive and reliable tools to bridge the gap between traditional assays and newly developed techniques.14,15 These biosensors integrate AChE as a recognition element with an electrochemical signalling platform and work by amperometric detection of electron loss associated with irreversible oxidation of thiocholine (Tch) after AChE catalysis. These biosensors can be used to investigate pesticide toxicity to enzymes and detection of pesticide residues in environmental and agricultural samples.16–20
The enzyme-based biosensors provide fast response and high sensitivity and when combined with an efficient transducer material offer new ways in biosensor engineering. The key factor to develop a high performance biosensor is to retain the catalytic activity of the enzyme after immobilization on the electrode surface. Moreover, the strategy for transmission of electrochemical signals should be well defined because biological elements are naturally non-conductive. An appropriate sensing electrode fabricated with a suitable modifier material can fulfil the demands of modern electrochemical technology.21–25
A new class of binary transition metals known as spinals, having the chemical formula AB2O4, have emerged as attractive materials with promising applications in physics, materials science, geophysics, etc. They have also been used as advanced electrode materials for efficient energy storage devices.26 Chromites belong to normal type spinal structures, among which nickel chromite (NiCr2O4) is of significant importance due to its role in industry as an efficient catalytic material.27–29 The spinals offer high electrochemical activity and capacity than mono metal oxides and can be used to develop potential composite materials with enhanced catalytic selectivity and efficiency.30,31 Furthermore, the carbon based materials such as graphene have greatly improved the biosensor industry and its 2D analogue graphitic carbon nitride has replaced graphene as a new type of functional material with remarkable physiochemical properties.32 The NiCr2O4 particles when combined with graphitic carbon nitride (g-C3N4) form a novel NiCr2O4/g-C3N4 composite material combining the catalytic, electrical, electron rich and surface functionalities of both components.
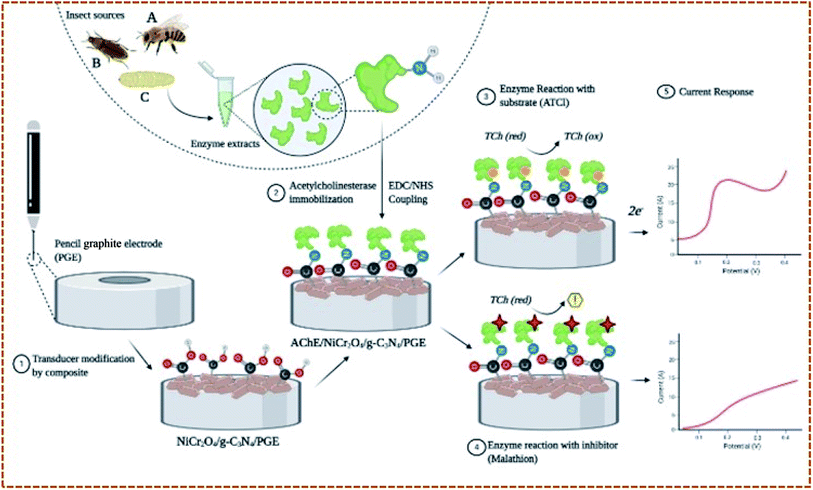
In this work, a pencil graphite electrode (PGE) modified with a NiCr2O4/g-C3N4 composite (referred further to as Ni/GCN) was exploited to immobilize AChE enzymes from two pests T. castaneum (red flour beetle) and Z. nevadensis (dampwood termite), and a non-target social insect A. mellifera (honeybee). The prepared composite was expected to possess high surface area with enhanced electrical conductivity and immobilization efficiency. Under optimized conditions, the fabricated bioelectrodes with the structure AChE/Ni/GCN/PGE were used to test the sensitivity of malathion against insect AChEs by electrochemical assay using cyclic voltammetry. The proposed structure was also applied for real time detection of malathion residues in wheat flour samples. Further, to evaluate the efficacy of the proposed method, in silico studies were performed to interpret the binding interaction of malathion with insect AChEs.
2. Results and discussion
2.1. Choice of material
For the proposed study a composite of graphitic carbon nitride and nickel chromite was employed to modify the surface of PGE before enzyme immobilization in order to improve the immobilization efficiency, electrical conductivity and catalytic efficiency of bioelectrodes. NiCr2O4 is a mixed metal oxide that has emerged as a novel material owing to its superior magnetic, electrical, optical and catalytic properties. The transition metal chromite has found potential in a wide range of applications including electrochemical sensors,33,34 gas sensors,35 electronic devices,36 catalytic materials37 and industrial processes.38 The incorporation of transition metal oxides on the surface of a potential support material is known to improve their electrocatalytic activity and surface area to volume ratio. The potential of g-C3N4 to develop novel nanocomposites with exceptional electrocatalytic and physiochemical properties has been reported by various researchers.39 The integration of NiCr2O4 to g-C3N4 would cause a unique surface contact between g-C3N4 and chromite, resulting in a large electrochemical surface area and accessibility of active sites that would lead to increased electrocatalytic efficiency of the modified electrodes.40 In addition, considering the role of nickel in specific enzyme immobilization,41 the composite was expected to possess effective biocatalyst immobilization ability. The NiCr2O4/g-C3N4 composite modified PGE will provide a suitable structure, reactive functional groups and appropriate binding sites for enzyme immobilization in addition to retaining its original activity and stability. Therefore, it will act as a conductive amplifier for enzymes. The principle lies in the EDC/NHS treatment of the material which activates the carboxyl groups on the surface of the composite, accelerating the formation of amide linkage between the enzyme and modified electrode. Therefore, the composite has dual importance in terms of increasing electrocatalytic efficiency of the biosensor and oriented, site specific immobilization of the enzyme on the modified electrode surface.2.2. Characterization of the NiCr2O4/g-C3N4 composite
Fig. 2A shows the FTIR spectra of bulk g-C3N4, NiCr2O4 and NiCr2O4/g-C3N4 composite. As shown in Fig. 2A(a) and (b) the FTIR spectra of g-C3N4 and NiCr2O4 exhibit their characteristic absorption peaks as reported earlier.42,43 For the bulk g-C3N4, the peak at 803 cm−1 is attributed to out of plane CN heterocyclic bending modes. Besides, the peak at 812 cm−1 is attributed to the breathing mode of triazine units. The set of peaks from 1200 to 1400 cm−1 are associated with aromatic C–N stretching.44 The peaks at 1537 and 1627 are attributed to cyano terminal groups C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group of the triple bond.45 The broad peak at the right corner at 3156 cm−1 has originated due to amine N–H stretching; this is because some nitrogen groups have partially hydrogenated during the calcination process.46 The absorption band at 663 cm−1 is typical of NiCr2O4 attributed to Ni–O stretching and bending vibrations. The absorption bands at 899 cm−1 arise due to Cr–O vibrational modes. The bands at 1631 and 3391 cm−1 are ascribed to stretching and bending vibrations of O–H due to adsorption of water molecules to the NiCr2O4 product. The FTIR spectra of NiCr2O4/g-C3N4 contain characteristic peaks of both NiCr2O4 and g-C3N4, suggesting that NiCr2O4 particles were embedded on the surface of g-C3N4 (Fig. 2A(c)).
N group of the triple bond.45 The broad peak at the right corner at 3156 cm−1 has originated due to amine N–H stretching; this is because some nitrogen groups have partially hydrogenated during the calcination process.46 The absorption band at 663 cm−1 is typical of NiCr2O4 attributed to Ni–O stretching and bending vibrations. The absorption bands at 899 cm−1 arise due to Cr–O vibrational modes. The bands at 1631 and 3391 cm−1 are ascribed to stretching and bending vibrations of O–H due to adsorption of water molecules to the NiCr2O4 product. The FTIR spectra of NiCr2O4/g-C3N4 contain characteristic peaks of both NiCr2O4 and g-C3N4, suggesting that NiCr2O4 particles were embedded on the surface of g-C3N4 (Fig. 2A(c)).
To study the crystal structure of the prepared composite material, X-ray diffraction (XRD) patterns of NiCr2O4/g-C3N4 were obtained and compared with those of their individual components, i.e. NiCr2O4 and g-C3N4. Typical XRD patterns of the prepared samples are shown in Fig. 2B(a)–(c). All diffraction peaks in NiCr2O4 can be indexed to a common orthorhombic phase space group Cmcm (no. 63) of NiCr2O4 with lattice constants a = 5.48 Å, b = 8.23 Å, c = 6.14 Å and β = 90° (JCPDS no. 072-1605). The diffraction peaks situated at 34.5°, 37.5°, 43.0°, 60.3°, 64.0°, and 71.1° and are indexed to (112), (130), (040), (151), (043) and (204) planes. These results demonstrate a typical XRD pattern of NiCr2O4. Moreover, g-C3N4 forms in the hexagonal shape and is in good agreement with the characteristic peaks (JCPDS no. 87-1526) of g-C3N4.36 Subsequently, the XRD pattern for NiCr2O4/g-C3N4 compared with NiCr2O4 and g-C3N4 showed two diffraction peaks at 27.6° and 13.0° that are indexed to (002) and (100) planes, confirming the presence of g-C3N4 with NiCr2O4. The two sample peaks have been marked with different symbols (+) and (*) to confirm their presence. Fig. 2B demonstrated less intense peaks of NiCr2O4 because the g-C3N4 peak is much higher and sharper, and it is a common observation that the peak intensities become weaker at larger angles and besides that micro strain broadening is also large at larger angles; therefore, some peaks of NiCr2O4 become broader at higher angles and are less noticeable in the pattern.
The PL spectra of g-C3N4 and NiCr2O4/g-C3N4 composite are shown in Fig. 2C. It can be seen that g-C3N4 had a broad and strong peak at around 833 nm, while NiCr2O4 at around 827 nm. It can be clearly observed that g-C3N4 produced higher intensity spectra than the NiCr2O4/g-C3N4 composite, which clearly indicates the increased efficiency of separation of charge carriers. This relates to the participation of more electrons and holes in oxidation and reduction reactions.47 The higher intense peak exhibits faster recombination of electrons and holes, thus lower redox reactions, while compared to the above a weaker and lower NiCr2O4/g-C3N4 composite depicts slow recombination of electrons and holes, indicating higher redox reactions.48
Further, the morphological nature and microstructure of the as-prepared NiCr2O4/g-C3N4 composite were inspected by SEM analysis and compared with those of its individual components, as shown in Fig. 3. Furthermore, the elemental composition of the as-prepared materials was also investigated by EDS analysis. It can be seen in Fig. 3A and B that the bulk g-C3N4 is in the form of irregular, rough, aggregated sub-blocks and stacked layers. Fig. 3A shows a rough and fissured morphology with lots of agglomerated particles of around 1.7 μm size. Some flaked stacks can also be seen which may have formed during the heating process of melamine, where HCL and NH3 were released which may hinder the thermal condensation into g-C3N4 sheets.49,50 Interestingly, some tubular stacked flake shaped structures also appeared on the surface with holes alongside, which can be seen in Fig. 3B. As discussed earlier the holes may be formed due to the release of ammonia and HCL gasses during the heating of melamine.50 The average size of these tubular stacked flakes is 0.81 μm. Fig. 3D shows the SEM image of NiCr2O4 and it can be clearly seen that spherical pumice stone shaped particles formed with diameters up to 0.96 μm. The particle agglomeration also happened which may be due to incomplete synthesis. The spongy surface of the particles, which can be seen in Fig. 3E, is linked to high reactivity of the catalyst due to the presence of numerous reaction sites. The SEM images of the NiCr2O4/g-C3N4 composite are shown in Fig. 3G and H. It can be seen that NiCr2O4 particles are evenly distributed on the surface of g-C3N4 flakes. In Fig. 3G the bright particles also confirmed the presence of NiCr2O4 which was previously undermined in the XRD spectra of the composite. This indicates that g-C3N4 acts like a sheet to support NiCr2O4 particles, which are attached and formed interconnects to behave like a single catalyst.47 Thus it can be conferred that the NiCr2O4/g-C3N4 composite with excellent nanojunctions has been successfully synthesized that would result in high electrocatalytic activity of the NiCr2O4/g-C3N4 composite due to efficient transfer of electrons during the electrochemical reaction.36,51,52 Further, energy-dispersive spectra of the composite and its components (Fig. 3C, F and I) revealed the presence of respective elements in the respective materials and confirmed the purity and successful synthesis of the as-prepared composite.
2.3. Electrochemical characterization of electrodes
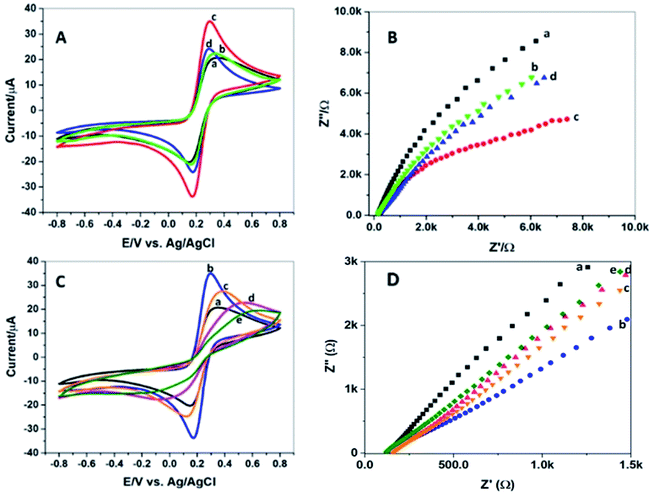
Electrochemical characterization of bioelectrodes at each fabrication step was done by cyclic voltammetry using a 5 mM equimolar solution of Fe(CN)63−/4− while potential swept across −0.8 to +0.8 V. The resistance to electron flow was measured by EIS at amplitude 234. It can be seen in Fig. 4A by the CV response that the current passing through bare PGE, GCN/PGE and Ni/GCN/PGE was subsequently increased, and the conductivity of the Ni/GCN modified pencil electrode was highest; similarly, the Nyquist plots (Fig. 4B) showed that resistance decreased in the same manner as the current was increased, whereas when the composite was functionalized with carboxyl groups, the resistance to electron flow enhanced, resulting in reduced current which ascertains successful introduction of carboxyl functional groups on the composite surface.After coating AChE on Ni/GCN/PGE, the value of peak current of AChE/Ni/GCN/PGE was decreased as shown in Fig. 4C, which verifies the attachment of the enzyme on the electrode surface. The attachment causes an increase in interface thickness, causing an interruption in electron flow and thus high impedance as shown by the EIS spectra of bioelectrodes in Fig. 4D. The results confirm the inertness of the enzyme molecule as it is not itself oxidized or reduced during the voltage cycling. As the source of AChE was three different insects, the electrochemical response of all three fabricated interfaces, i.e. Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE, and Zn-AChE/Ni/GCN/PGE was determined, and the peak oxidation current of thiocholine was found to be different among them. It was noticed that current was decreased maximally in the case of Z. nevadensis AChE (Zn-AChE) compared to T. castaneum AChE (Tc-AChE), and least in the case of A. mellifera AChE (Am-AChE). The impedance increased in the same reverse order (Fig. 4D). The results confirmed good electrical conductivity of the composite and successful immobilization of each component of the biosensor.
2.4. Analytical behavior of integrated bioelectrodes
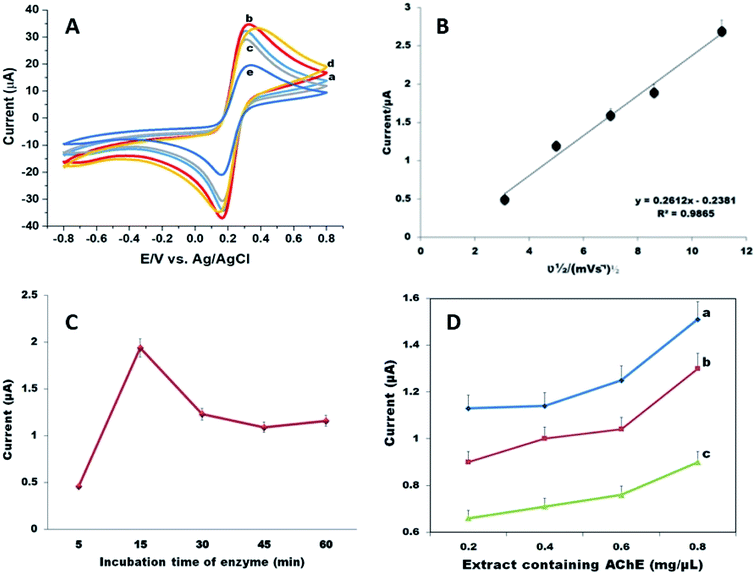
It is important to measure the electro-catalytic behaviour of the prepared bioelectrodes in response to thiocholine, which is the electroactive hydrolysis product of the AChE reaction and is responsible for electrochemical activity. For this purpose, CV was run for the AChE/Ni/GCN/PGE biosensor in the presence and absence of the substrate, ATCl. The response of three types of bioelectrodes, i.e. Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE and Zn-AChE/Ni/GCN/PGE, was compared in the presence of ATCl. As shown in Fig. 5, the current response of the bare electrode and negative controls did not produce any obvious oxidation peaks; however, in the case of Zn-AChE/Ni/GCN/PGE, a reversible oxidation peak arises at 0.7 V, whereas in Tc-AChE/Ni/GCN/PGE, a peak at 0.74 V can be observed. An obvious oxidation peak at 0.79 V was observed in Am-AChE/Ni/GCN/PGE with the highest peak signal compared to Tc-AChE and Zn-AChE. The absence of the oxidation peak in negative controls confirmed that the anodic peak was coming from the hydrolysis product of AChE, i.e. TCh. It can be inferred that AChE activity is considerably high in A. mellifera, and it provides suitable attachment sites, structural and surface features that stabilize protein conformation and orientation, providing efficient immobilization on the surface of the Ni/GCN modified electrode.2.5. Experimental optimization
The optimum experimental parameters were set in order to ensure the best performance of fabricated biosensors. In order to improve the analytical efficiency of the biosensor, the incubation time of the material was validated by incubating the bioelectrodes in composite suspension for a time period of 60 minutes. Fig. 6A shows that 15 minutes is a suitable time to achieve a better current response.Moreover, from Fig. 6B we could find that by increasing scan rate from 10 to 125 mV s−1, the peak current increased subsequently and the plots of square root of scan rate and current showed a linear response with the regression equation y = 0.2612x + 0.2381 (R2 = 0.9865), which indicates a surface controlled electrochemical process on the electrode, which means that the composite provides a conductive pathway and a large surface area for the effective immobilization of the enzyme on the electrode surface. However, 50 mV s−1 was used for all electrochemical measurements.
Similarly, the optimum incubation time of enzyme deposition was also determined to be 15 minutes (Fig. 6C). It was observed that under the given conditions, the current for the oxidation of acetylcholine increased with the increase in immobilization time of the enzyme; however, after 15 minutes a decrease in oxidation peak current was noticed probably due to interruption in current flow after a saturation point for the enzyme was achieved.
Further, the appropriate amount of extract that can be used for AChE was determined, because the amount of enzyme plays an important role in biosensor fabrication due to its insulating nature and possibility of increased film thickness. The varying amount of extracts of A. mellifera, T. castaneum and Z. nevadensis was deposited on the electrode surface, and 0.8 mg μL−1 was found suitable to obtain sufficient oxidation peak current (Fig. 6D).
The pH of phosphate buffer was adjusted at 7.4 for all electrochemical measurements. To ensure the maximum degree of enzyme inhibition, the effect of inhibition time was determined and it was observed that after 15 minutes of biosensor incubation with malathion, the degree of inhibition increased, after which it was stabilized by virtue of saturated binding between the active sites of the enzyme and pesticides (Fig. 8A). Thus, 15 min was employed for malathion inhibition assay.
2.6. Calibration curve of ATCl and enzyme kinetics
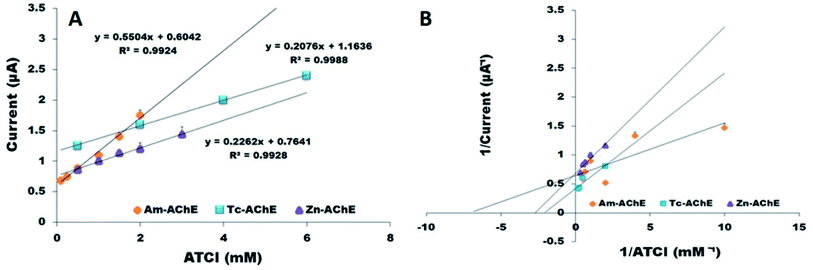
The influence of substrate concentration on the analytical performance of biosensors is an important parameter to determine the performance and sensitivity of the biosensor. A plot of current versus ATCl concentration is useful to determine the kinetics of the enzymatic reactions. The current response produced by the product of enzymatic catalysis corresponds to the velocity of the enzymatic reaction, which is proportional to the activity of the immobilized enzyme, according to the Michaelis–Menten equation. The effect of ATCl concentration was investigated with the already standardized parameters by varying its amount from 0.5 mM to 8 mM in 0.1 M pH 7.4 PBS. The substrate concentration at which the maximum current response obtained was different for each insect; therefore, detection limits and sensitivities for each biosensor were also different (Fig. 7A). For Am-AChE/Ni/GCN/PGE, the maximum current response was obtained at 2 mM, whereas in the cases of Zn-AChE/Ni/GCN/PGE and Tc-AChE/Ni/GCN/PGE, the maximum current response was attained at 6 mM and 8 mM, respectively. All readings were taken in triplicate with RSD < 4% (n = 3). The sensitivities of Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE and Zn-AChE/Ni/GCN/PGE were calculated to be 62.27, 22.29 and 22.41 μA mM−1 cm−2, respectively. To determine the affinity of the enzyme for the substrate, the apparent Michaelis–Menten constant (Kmapp) was calculated for each biosensor from the slope and intercept for the reciprocal plots of the steady-state current vs. ATCl concentration (Fig. 7B). The Kmapp for Am-AChE/Ni/GCN/PGE, Zn-AChE/Ni/GCN/PGE and Tc-AChE/Ni/GCN/PGE was 140 μM, 0.41 mM and 0.46 mM, respectively. It can be inferred from the results that honeybee AChE has higher affinity and sensitivity for the ATCl than red flour beetle and termite AChE, which have almost equal affinity for the substrate. However, the values are quite low and comparable to those of many other reported biosensors as shown in Table 1, which ensures good analytical behaviour and sensitivity of proposed biosensor platforms on account of the exceptional catalytic efficiency of the Ni/GCN composite using insect enzyme sources.| Biosensor platform | Kmapp | Sensitivity | Reference |
|---|---|---|---|
| AChE/Ti3C2Tx-CS/GR/GCE | 4.89 mM | NR | 53 |
| AChE/3D tree-like Ag/GCE | 137.5 μM | 0.056 μA mM−1 | 54 |
| AChE-Chit/Pd@Au NWsN/GCE | 190 μM | NR | 55 |
| f-MWCNT/poly(SNS-NH2)/AChE | 1.038 mM | 24.16 μA mM−1 cm−2 | 56 |
| Am-AChE/Ni/GCN/PGE | 140 μM | 62.27 μA μM−1 cm−2 | This work |
| Tc-AChE/Ni/GCN/PGE | 0.46 mM | 22.29 μA mM−1 cm−2 | This work |
| Zn-AChE/Ni/GCN/PGE | 0.41 mM | 22.41 μA mM−1 cm−2 | This work |
2.7. Electrochemical test for malathion sensitivity
To investigate the applicability of proposed biosensors for organophosphorus pesticide detection, the modified bioelectrodes were incubated with different concentrations of malathion under optimized conditions and CV responses were recorded (Fig. 8). The linear ranges for three types of biosensors were different. The linear range for Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE, and Zn-AChE/Ni/GCN/PGE was 0.1–1.6 μM, 1–40 nM, and 2–100 nM respectively. The LOD and LOQ values for each biosensor were determined by the method based on the standard deviation of the current response and the slope of the calibration line. The LODs were found to be 2 nM, 0.86 nM and 2.3 nM and LOQs were 6 nM, 2.6 nM, and 7 nM for Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE, and Zn-AChE/Ni/GCN/PGE, respectively. Similarly, the detection sensitivities were 1607, 58.09 and 21.4 μA nM−1 cm−2 for Am-AChE/Ni/GCN/PGE, Tc-AChE/Ni/GCN/PGE, and Zn-AChE/Ni/GCN/PGE, respectively. A high detection limit for Am-AChE/Ni/GCN/PGE may result from many factors such as population genetics or insecticide resistance among these insects. Previous reports have shown unique behaviours of honeybee enzymes towards certain insecticides and it has been found that many organic and chemical pesticides pose limited toxicity to honeybees, while others are highly toxic towards them, whereas the T. castaneum AChE is more susceptible to organophosphates and the genes and structure of AChE are widely studied in these insects.57–60 The insects damage a large mass of stored grains each year and multiply in large amounts in a very short time. In addition, they develop insecticide resistance, resulting in reduced sensitivity against insecticides. Based on its potential, the insect has been used in our previous study for the construction of biosensors for OP detection.21 Further, the damp wood termite species used in this study have not been significantly studied for their enzymes and the impact of pesticides on them. Even though the results indicate a similar inhibition pattern to T. castaneum AChE, the linear range for detection is wider than that for Tc-AChE. It can be recommended from the results that the platform is highly suitable to study the behaviour of insects' AChE towards OPs and can be utilized for all forms of AChEs. The result for Am-AChE suggests that the sensing design exhibits properties useful for organophosphate determination for commercial applications in honeybee populations contaminated by pesticides. The analytical performance of the proposed platforms was even better than that of various previously reported biosensor platforms for malathion detection (Table 2). Moreover, the presented design is tested on stored grains such as wheat for its practicality, which has not been done before.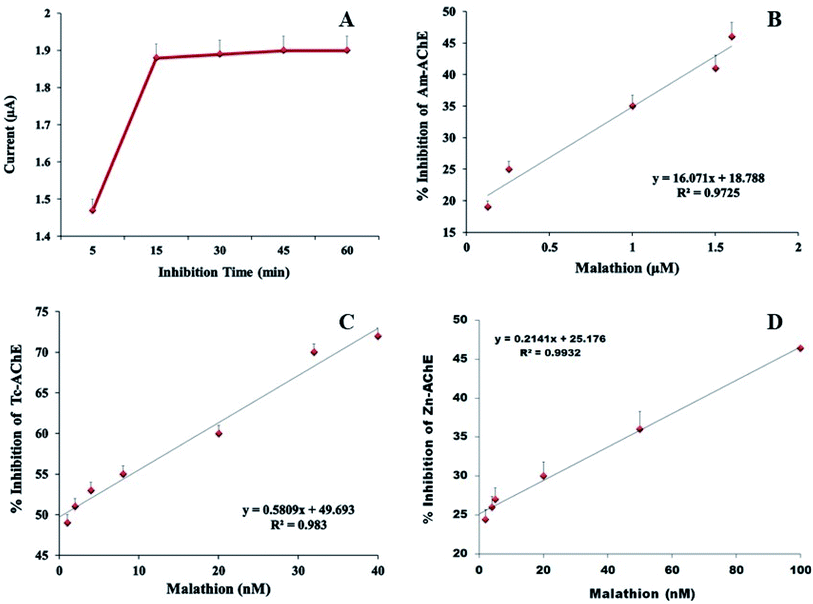 | ||
| Fig. 8 (A) Incubation time optimization for malathion, and (B) the calibration curve for malathion detection by (C) Am-AChE/Ni/GCN/PGE, (D) Tc-AChE/Ni/GCN/PGE, and (E) Zn-AChE/Ni/GCN/PGE. | ||
| Malathion biosensor | Linear range | LOD | References |
|---|---|---|---|
| AChE-PANI/ZnO/RGO/GCE | 10–40 nmol L−1 | 11 nmol L−1 | 61 |
| CHIT-g-PANI based electrode | 2–62.5 μM | 3.8 μM | 62 |
| AChE/HCS@PANI/GCEAChE/HCS@PANI/GCE | 1.0–10 μg mL−1 | 0.16 ng mL−1 | 47 |
| poly(TTP)/AChE/GCE | 9.99–99.01 nM | 4.08 nM | 63 |
| PANI-ES SWCNTS/GE | 0.2–1.4 μM | 0.2 μM | 64 |
| Am-AChE/Ni/GCN/PGE | 0.1–1.6 μM | 2 nM | This work |
| Tc-AChE/Ni/GCN/PGE | 1–40 nM | 0.86 nM | This work |
| Zn-AChE/Ni/GCN/PGE | 2–100 nM | 2.3 nM | This work |
2.8. Real time detection of malathion from wheat flour
The potential application of the proposed biosensor was demonstrated by detecting malathion in real samples of whole-wheat flour, which was not contaminated with pesticides. A standard addition method was employed to detect wheat samples for different concentrations of malathion. The Am-AChE/Ni/GCN/PGE biosensor exhibited good practical application showing good reliability with the recovery ranging from 99 to 100.9%, as well as RSDs less than 5% (Table 3). The other two interfaces did not provide satisfactory recoveries of malathion from wheat samples and therefore are not included here. The liability of the proposed method was assessed by comparing the obtained results with HPLC as a reference method, and the results were comparable as shown in Table 3. The results showed that the Am-AChE/Ni/GCN/PGE biosensor is most effective in recovery for malathion in real agricultural samples.| Source | Added (μM) | Found (μM) | % Recovery | RSD% |
|---|---|---|---|---|
| Biosensor method | ||||
| Apis mellifera (honeybee) | (i) 0.128 | 0.127 | 99.2 | 3.2 |
| (ii) 1 | 1.009 | 100.9 | 2.1 | |
| (iii) 1.6 | 1.594 | 99.6 | 4.1 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| HPLC method | ||||
| Apis mellifera (honeybee) | (i) 0.128 | 0.128 | 100 | 3.9 |
| (ii) 1 | 0.99 | 99 | 3.0 | |
| (iii) 1.6 | 1.61 | 100.6 | 3.6 | |
2.9. Selectivity and reproducibility
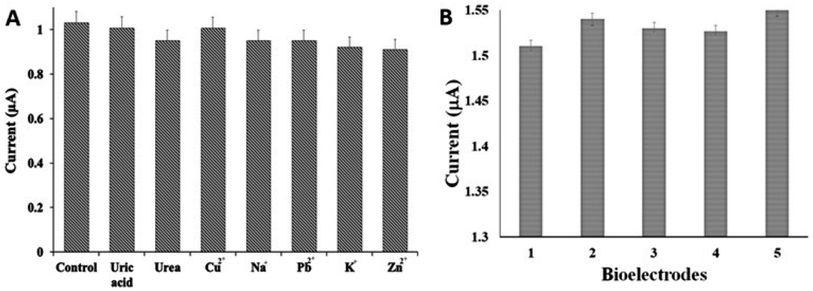
The selectivity of the biosensor was assessed by recording the current responses of Am-AChE/Ni/GCN/PGE in the presence of possible interfering analytes in the environment, like urea, uric acid, metal ions and transition metal ions. It can be seen in Fig. 9A that the interfering substances did not display any considerable disturbance in the current signals in the presence of 2 mM ATCl. Moreover, the current response of malathion in the presence of interfering substances was also investigated which did not get affected significantly; therefore, the results were not added in the manuscript. The obtained results propose an outstanding anti-interference capability of the biosensing interface.The precision was evaluated by evaluating the current response of five different electrodes using the same concentration of malathion and the RSD was acquired <5% (n = 4), showing good operational reproducibility of the biosensor (Fig. 9B). The above results validated a good reproducibility of the devised method.
2.10. Docking studies for AChE–malathion interaction
The study of the physicochemical properties of an enzyme and its structural and surface features is also very important to predict the immobilization efficiency of an enzyme on the transducer surface. AChE is a comparatively hard protein with a high degree of hydrophobicity that helps its binding on the hydrophobic substrates, prevents denaturation, and improves its thermal stability. Such immobilizations are known to improve organophosphate detection more efficiently. Furthermore, amino acids can form coordination metal complexes through their amino and carboxylate functional groups, while side chains of some amino acids possess metal-binding groups such as the imidazole ring of His or phenol ring of Tyr. For this study, the AChE has been immobilized through EDC/NHS chemistry, causing covalent binding of amine groups of AChE amino acids to the carboxylate-modified composite material. Moreover, the complexation of Ni with amino acids possibly improves the immobilization and catalytic efficiency of enzymes with the transducer surface. The imidazole ring of His and thiol sulfur group of Cys coordinates with Ni and is crucial for the interactions between peptides and Ni through amino or amide nitrogen atoms.65–67 Therefore, the offered transducer surface not only helps in catalysis and improved conductivity but also provides a suitable binding site for efficient enzyme immobilization with disturbing the structure of active sites, making them available to bind with the analyte molecules.Further, to gain an insight into the binding interaction of the model pesticide, malathion, with the amino acid residues of AChE enzymes of source insects, molecular docking simulations were performed. The molecular docking analyses show that malathion interacts with all three acetylcholinesterase enzymes, i.e., Tc-AChE, Am-AChE1, and Zn-TChE. The binding energies of malathion for Tc-AChE, Am-AChE1, and Zn-TChE were found to be −5.73, −5.95, and −5.54 kcal mol−1, while the dissociation constants were 62.6, 43.5, and 87.1 μM, respectively.
Malathion interactions with Am-AChE1 show that ASN117 and TYR385 are involved in the formation of two H-bonds (3.47 and 3.17 Å) with oxygen atoms of malathion, while other surrounding residues, including GLY168, GLY167, GLY112, GLU113, TRP116, PHE386, TYR389, TYR179, GLU252, and TYR104, are involved in stabilizing the interactions (Fig. 10A). TYR185 and TRP344 are present at the entrance gorge. The same amino acids have been previously reported to form the Am-AChE1 active site.68
In the case of Tc-AChE, O4 in malathion forms a comparatively strong H-bond (2.95 Å) with the backbone amine of TYR189, present at the opening of the active site gorge of Tc-AChE, therefore blocking the ATC entry site. Moreover, GLY187, GLY186, ASN153, ALA149, ASP140, ILE139, ALA508, TRP152, GLU266, SER267, TYR396, PHE397, and HIS507 were involved in the stabilization of the ligand–enzyme complex (Fig. 10B). Previously, Phosmet was also found to make H-bonding with Tyr189 and Ser190 of Tc-AChE.21 Distal methyl groups in malathion were found to bind deep in the active site gorge and show hydrophobic interactions with GLY187, SER256 and TRP300. Previously, azadirachtin, a saponin from Azadirachta indica, was found to interact with Tc-AChE with lower binding affinities and at different binding sites.4,69 In our study, malathion was also found to bind acetylthiocholine active site residues mentioned by ref. 21, with comparatively less affinity (−5.54 kcal mol−1 binding energy and 84.8 μM dissociation constant), including ILE138, ASP140, ILE139, ALA149, TRP152, ASN153, GLY185, GLY186, GLY187, TYR189, SER190, GLU266, SER267, ALA268, TYR396, PHE397, TYR400, TRP449, HIS507, and ALA508. Surprisingly, when 300 ppm (∼1 mM) malathion was used against T. castaneum, an 11-fold increase in Tc-AchE activity was found.70 Our molecular docking results may explain the reason for this increased Tc-AChE activity; the dissociation constant of malathion–Tc-AChE was found to be 62.6 μM, while at high concentrations, malathion binds nonspecifically at various sites in Tc-AChE that may bring conformational changes in enzyme structure and augment its activity. Malathion also interacts with Zn-AChE, while binding on the surface interactions is stabilized by GLU80, TRP83, GLU295, SER211, PHE343, ARG346, VAL347, ASN449, MET458, HIS459, GLY460, and ASP461 residues (Fig. 10C).
3. Materials and methods
3.1. Chemicals
Analytical grade acetylthiocholine chloride (ATCl), potassium ferricyanide (K3[Fe(CN)6]), potassium ferrocyanide (K4[Fe(CN)6]), malathion, Na2Cr2O7 and Ni2SO4 were purchased from Sigma-Aldrich. Co. Ltd (USA). EDC (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide) and MES (4-morpholinoethanesulfonic acid) were obtained from the Tokyo Chemical Industry Co Ltd (JP), and NHS (N-hydroxysuccinimide) was attained from CovaChem LLC Company (US). Deionized water (18 MΩ cm) was acquired from an ultra-pure purification unit.3.2. Instruments
The surface morphology of the as-prepared composite material was studied through Scanning Electron Microscopy (SEM) and elemental composition analysis using a Nova NanoSEM 450 field emission scanning electron microscope (FESEM) equipped with EDX. The crystallinity and phase structure of the composite were determined by X-ray diffraction (XRD) using a Rigaku-D/max-2500PC diffractometer at 40 kV with a graphite type monochromator working at 40 mA. The Fourier Transform Infrared (FTIR) spectroscopy measurements were recorded using a Thermo Fischer Scientific (Nicolet 6700, USA) spectrometer in the 600–4000 cm−1 scan range. The photoluminescence (PL) measurements of materials were obtained from a Xe lamp source F7000 Hitachi Spectrometer. Cyclic voltammetry was performed on an AMEL Electrochemistry Potentiostat (Model 2551) with a three-electrode system using an Ag/AgCl reference and platinum wire auxiliary electrode. The pencil graphite electrode (PGE) (diameter: 0.5 mm; length: 60 mm) was used as a working electrode acquired from Staedtler, Germany. Electrochemical impedance spectroscopy (EIS) was done on a 2700 Z-Pulse by AMEL Electrochemistry in the 0.1 Hz to 100 kHz range of frequency.3.3. Synthesis of the NiCr2O4/g-C3N4 composite
Graphitic carbon nitride (g-C3N4) was synthesized in bulk using a previously described method.21 The NiCr2O4/g-C3N4 composite was prepared by an in situ chemical reaction at ambient temperature. Typically, 1 g of NaCr2O7 and 2.5 g of Ni2SO4 were immersed in 100 mL of de-ionized water and ultra-sonicated for 30 minutes. Then 5 g of g-C3N4 was added and stirred for 1 hour at room temperature. The prepared suspension was then stirred for another 2 hours and the precipitates were collected by centrifugation and washed with ethanol and water three times. The collected sample was dried at 60 °C for 24 hours and stored at room temperature.3.4. Insects
Cultures of T. castaneum (Herbst) were reared in the laboratory under standard conditions. A. mellifera was collected from the honey bee collection centre of University of the Punjab and stored at −20 °C. Z. nevadensis was obtained from fallen logs and tree stumps from north Pakistan.3.5. Preparation of AChE/Ni/GCN/PGE bioelectrodes
Three types of AChE/Ni/GCN/PGE biosensors were prepared differing in the source of AChE. The crude extracts were prepared using a previously described method.21 The extracts were used directly as a source of insects' AChE. For electrode modification, the electrodes were electrochemically cleaned using 0.1 M H2SO4 and after washing with dH2O and drying at RT, the functionalized Ni/GCN composite was adsorbed on the electrode surface by dipping in 2.5 μL of its aqueous suspension for 15 minutes. After drying and washing, the modified electrodes were immersed in 0.05 mL of AChE/EDC/NHS/MES extracts of T. castaneum, A. mellifera and Z. nevadensis in separate Eppendorf tubes for 15 minutes to allow for enzyme immobilization. The modified electrodes were then removed, dried at room temperature and washed carefully with deionized water to remove any unbound enzyme and impurities. The fabricated bioelectrodes were again dried and used for electrochemical measurements. The bonding mechanism depends on the activation of the carboxyl group functionalized material by EDC/NHS coupled enzyme molecules to make amide bonds between the amino group of the enzyme and carboxyl group of the material. Fig. 1 provides a schematic illustration of steps involved in fabrication of the biosensor.3.6. Electrochemical measurement procedure
The electrochemical behaviour of the material on the surface of the electrode was determined through cyclic voltammetry using ferro-ferricyanide electrolyte solution at a potential ranging from −0.8 to +0.8 V. Similarly, the electro-catalytic ability of as-prepared biosensors was analysed through cyclic voltammetry using 0.1 M pH 7.4 PBS buffer electrolyte solution comprising acetylthiocholine chloride (ATCl), at a potential ranging from 0.2 to 0.9 V and 50 mV s−1 scan rate. The response of bare electrodes and modified controls was recorded and compared by observing the oxidation peak potential of Tch.For enzyme inhibition by the pesticide, the modified electrodes were pre-incubated with malathion. The optimum inhibition time was determined by incubating the electrodes with malathion solution for 5 to 60 minutes, and 30 min was selected as the optimum incubation time for malathion. CV was recorded afterwards, and percentage inhibition was calculated from the following formula:
| [% Inhibition = I1 − I0/I1 × 100] |
3.7. Detection of malathion in wheat flour
The prepared biosensors were employed to detect malathion in whole wheat flour as a real sample using the procedure described previously.21 The pesticide was spiked in 2 grams of wheat flour to obtain three concentrations covering the whole range, and the pesticide was extracted in acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS (6
PBS (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 v/v) extraction solvent. After 10 minutes of incubation and centrifugation, the resulting supernatant was used as a pesticide mother solution to determine the percentage recovery of the pesticide using the same measurement procedure described above. The obtained results have been compared with a standard HPLC method using procedure as described in ref. 21.
4 v/v) extraction solvent. After 10 minutes of incubation and centrifugation, the resulting supernatant was used as a pesticide mother solution to determine the percentage recovery of the pesticide using the same measurement procedure described above. The obtained results have been compared with a standard HPLC method using procedure as described in ref. 21.
3.8. Ligand and protein structure preparation
The structure for ligand, malathion in SDF format (CID: 4004) was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov). The homology models for Am-AChE (A. mellifera), Tc-AChE (T. castaneum), and Zn-AChE (Z. nevadensis) were built using YASARA (Yet Another Scientific Artificial Reality Application) Structure version 20.1.18 (ref. 21 and 71) and further refined by loop region optimization and steepest descent, simulated annealing minimization. The homology models for Tc-AChE, Am-AChE1, and Zn-AChE were obtained. Models were validated by YASARA z-scores, Ramachandran plot, PROCHECK server and VERIFY 3D showing acquisition of good quality homology modelled structures.3.9. Molecular docking simulations
For Malathion molecular docking against three different AChE, YASARA Structure ver. 20.1.18 (ref. 71) with a modified AutoDock LGA algorithm and an AMBER03 force field was used.21 For improved accuracy and better prediction of ligand–protein interactions, 100 global docking runs were obtained, rescored, and analyzed for dissociation constants, binding energy calculations and active site mapping as described in ref. 21. PyMol, Maestro, and PLIP were used to map the ligand–protein interactions.4. Conclusions
In conclusion, the proposed work utilized the electrochemical biosensing approach to compare the malathion sensitivity based on three different insect and pest AChE enzymes that were immobilized on a potential NiCr2O4/g-C3N4 composite material. The surface and electrochemical properties of NiCr2O4/g-C3N4 favoured the immobilization and electrocatalytic activity of insects' enzyme, thus amplifying the sensitivity and amperometric response of the developed biosensors. The results suggested that the composite provides a biocompatible environment and potential binding sites for enzyme immobilization, a large surface area and high electrical conductivity, which resulted in good electrochemical performance of electrodes. The proposed platform was expected to be utilized to determine the sensitivity of OP insecticides and evaluate their toxicity against insects. Among three insect species, A. mellifera AChE was found to be immobilized with the highest efficiency and kinetic analysis showed its high affinity for the substrate. The electrocatalytic behavior of Am-AChE was also better than that of Tc-AChE and Zn-AChE; however, the malathion inhibition was observed at high concentrations compared to Tc-AChE and Zn-AChE, which showed that Tc-AChE and Zn-AChE are more sensitive to malathion than Am-AChE. The results were further confirmed through computational docking and the binding interactions of malathion with AChE of these insects were explored. These initial studies have opened ways for the rational designs of electrochemical biosensors and their use in the study of the insecticides' toxicity to insects. This study provides new development in the biosensor design, which offers considerable promise in several agriculture and environmental applications.Author contributions
Sehrish Bilal: data curation, writing original draft, software, reviewing and editing; Muhammad Nasir: methodology, resources, and investigation; M. Mudassir Hassan: data curation and software; Muhammad Fayyaz ur Rehman: computational studies; Amtul Jamil Sami: supervision, methodology, investigation, and validation; Akhtar Hayat: supervision, methodology, resources, investigation, and validation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge Pakistan Science Foundation for providing research grant to carry out this study under project PSF-MSRTII/AGR/P-COMSATS-LHR (09)/2019.References
- N. Gallai, J.-M. Salles, J. Settele and B. E. Vaissière, Ecol. Econ., 2009, 68, 810–821 CrossRef.
- N. W. Calderone, PLoS One, 2012, 7, e37235 CrossRef CAS PubMed.
- F. Sánchez-Bayo, D. Goulson, F. Pennacchio, F. Nazzi, K. Goka and N. Desneux, Enviorn. Int., 2016, 89, 7–11 CrossRef.
- A. J. Sami, S. Bilal, M. Khalid, M. T. Nazir and A. Shakoori, Pak. J. Zool., 2018, 50, 725–733 CrossRef CAS.
- S. B. Walsh, T. A. Dolden, G. D. Moores, M. Kristensen, T. Lewis, A. L. Devonshire and M. S. Williamson, Biochem. J., 2001, 359, 175–181 CrossRef CAS.
- K. L. Newhart, Environmentalfate of Malathion, California Environmental Protection Agency, 2006, pp. 1–20 Search PubMed.
- N. E. Gary and E. C. Mussen, Environ. Entomol., 1984, 13, 711–717 CrossRef CAS.
- E. A. Parkin, J. Sci. Food Agric., 1958, 9, 370–375 CrossRef CAS.
- R. K. Upadhyay, G. Jaiswal and S. Ahmad, Acta Univ. Sapientiae, Agric. Environ., 2010, 2, 58–79 Search PubMed.
- F. Bughio and R. J. Wilkins, J. Stored Prod. Res., 2004, 40, 65–75 CrossRef CAS.
- N. White and R. J. Bell, J. Econ. Entomol., 1988, 81, 381–386 CrossRef.
- E. Haubruge, M. Amichot, A. Cuany, J.-B. Berge and L. Arnaud, Insect Biochem. Mol. Biol., 2002, 32, 1181–1190 CrossRef CAS PubMed.
- D. Yong, C. Liu, D. Yu and S. Dong, Talanta, 2011, 84, 7–12 CrossRef CAS.
- S. Rodriguez-Mozaz, M. J. de Alda and D. Barceló, Anal. Bioanal. Chem., 2006, 386, 1025–1041 CrossRef CAS.
- A. J. Baeumner, Anal. Bioanal. Chem., 2003, 377, 434–445 CrossRef CAS.
- P. Walter, A. Pepłowski, D. Janczak and M. Jakubowska, Microelectron. J., 2019, 120–126 CrossRef.
- A. El-Moghazy, E. Soliman, H. Ibrahim, T. Noguer, J.-L. Marty and G. Istamboulie, Food Chem., 2016, 203, 73–78 CrossRef CAS PubMed.
- N. Chauhan, J. Narang and C. Pundir, Biosens. Bioelectron., 2011, 29, 82–88 CrossRef CAS.
- X. Liu, R. Sakthivel, W.-C. Liu, C.-W. Huang, J. Li, C. Xu, Y. Wu, L. Song, W. He and R.-J. Chung, Food Chem., 2020, 126889 CrossRef CAS.
- S. Liu, Z. Zheng and X. Li, Bioanal. Chem., 2013, 405, 63–90 CrossRef CAS.
- S. Bilal, M. M. Hassan, M. F. ur Rehman, M. Nasir, A. J. Sami and A. Hayat, Food Chem., 2021, 346, 128894 CrossRef CAS PubMed.
- J. Wang and E. Katz, Anal. Bioanal. Chem., 2010, 398, 1591–1603 CrossRef CAS.
- X. Zhu, L. Fan, S. Wang, C. Lei, Y. Huang, Z. Nie and S. Yao, Anal. Chem., 2018, 90, 6742–6748 CrossRef CAS PubMed.
- J. Xie, D. Cheng, P. Li, Z. Xu, X. Zhu, Y. Zhang, H. Li, X. Liu, M. Liu and S. Yao, ACS Appl. Nano Mater., 2021, 4, 4853–4862 CrossRef CAS.
- S. Zhang, R. Li, X. Liu, L. Yang, Q. Lu, M. Liu, H. Li, Y. Zhang and S. Yao, Biosens. Bioelectron., 2017, 92, 457–464 CrossRef CAS PubMed.
- G. T. Anand, L. J. Kennedy and J. J. Vijaya, J. Alloys Compd., 2013, 581, 558–566 CrossRef CAS.
- L. Shen, Q. Che, H. Li and X. Zhang, Adv. Funct. Mater., 2014, 24, 2630–2637 CrossRef CAS.
- N.-H. Li, Y.-H. Chen, C.-Y. Hu, C.-H. Hsieh and S.-L. Lo, J. Hazard. Mater., 2011, 198, 356–361 CrossRef CAS PubMed.
- S. A. Bakar, N. Soltani, W. M. M. Yunus, E. Saion and A. Bahrami, Solid State Commun., 2014, 192, 15–19 CrossRef CAS.
- A. K. Geim and K. S. Novoselov, in Nanoscience and technology: a collection of reviews from nature journals, World Scientific, 2010, pp. 11–19 Search PubMed.
- Z. Zhao, Y. Sun and F. Dong, Nanoscale, 2015, 7, 15–37 RSC.
- A. I. Cooper and M. J. J. Bojdys, Mater. Today, 2014, 17, 468–469 CrossRef.
- Y. Liu, M. Huang, J. Zhao, M. Lu, X. Zhou, Q. Lin, P. Wang and J. Zhu, J. Electrochem. Soc., 2020, 167, 066520 CrossRef CAS.
- S. Zhuiykov, T. Nakano, A. Kunimoto, N. Yamazoe and N. Miura, Electrochem. Commun., 2001, 3, 97–101 CrossRef CAS.
- H. Gao, J. Guo, Y. Li, C. Xie, X. Li, L. Liu, Y. Chen, P. Sun, F. Liu and X. Yan, Sens. Actuators, B, 2019, 284, 305–315 CrossRef CAS.
- J. Tang, S. Ni, Q. Chen, X. Yang and L. Zhang, J. Alloys Compd., 2017, 698, 121–127 CrossRef CAS.
- L. P. Martin, A.-Q. Pham and R. S. Glass, MRS Online Proc. Libr., 2002, 756, 113 CrossRef.
- M. Sabri, A. Habibi-Yangjeh and A. Khataee, Chemosphere, 2022, 134594 CrossRef CAS PubMed.
- R. Shafique, A. Mahmood, K. Batool, A. Ahmad, T. Yaqoob, M. Jabeen, A. U. Shah, U. Asjad and M. Rani, ECS J. Solid State Sci. Technol., 2021, 10, 101005 CrossRef CAS.
- R. Shafique, M. Rani, A. Mahmood, S. Khan, N. Janjua, M. Sattar, K. Batool and T. Yaqoob, Int. J. Enviorn. Sci. Technol., 2021, 1–10 Search PubMed.
- R. S. Prakasham, G. S. Devi, C. S. Rao, V. Sivakumar, T. Sathish and P. Sarma, Appl. Biochem. Biotechnol., 2010, 160, 1888–1895 CrossRef CAS PubMed.
- H. Wang, W. He, X. Dong, H. Wang and F. Dong, Sci. Bull., 2018, 63, 117–125 CrossRef CAS.
- S. A. Shobeiri, M. Mousavi-Kamazani and F. Beshkar, J. Mater. Sci.: Mater. Electron., 2017, 28, 8108–8115 CrossRef CAS.
- L. Huang, H. Xu, Y. Li, H. Li, X. Cheng, J. Xia, Y. Xu and G. Cai, Dalton Trans., 2013, 42, 8606–8616 RSC.
- X. Han, D. Xu, L. An, C. Hou, Y. Li, Q. Zhang and H. Wang, Int. J. Hydrogen Energy, 2018, 43, 4845–4855 CrossRef CAS.
- S. Yang, Y. Gong, J. Zhang, L. Zhan, L. Ma, Z. Fang, R. Vajtai, X. Wang and P. M. Ajayan, Adv. Mater., 2013, 25, 2452–2456 CrossRef CAS PubMed.
- Y. Che, B. Lu, Q. Qi, H. Chang, J. Zhai, K. Wang and Z. Liu, Sci. Rep., 2018, 8, 1–12 CAS.
- J.-y. Tang, R.-t. Guo, W.-g. Zhou, C.-y. Huang and W.-g. Pan, Appl. Catal. B, 2018, 237, 802–810 CrossRef CAS.
- I. Papailias, T. Giannakopoulou, N. Todorova, D. Demotikali, T. Vaimakis and C. Trapalis, Appl. Surf. Sci., 2015, 358, 278–286 CrossRef CAS.
- S. Sun, E. Fan, H. Xu, W. Cao, G. Shao, B. Fan, H. Wang and R. Zhang, Nanotechnology, 2019, 30, 315601 CrossRef CAS PubMed.
- B. Palanivel, C. Ayappan, V. Jayaraman, S. Chidambaram, R. Maheswaran and A. Mani, Mater. Sci. Semicond. Process., 2019, 100, 87–97 CrossRef CAS.
- T. Beenish, T. Muhammad and M. G. M. Nawawi, Chem. Eng. Trans., 2021, 83, 247–252 Search PubMed.
- B. Wang, Y. Li, H. Hu, W. Shu, L. Yang and J. Zhang, PLoS One, 2020, 15, e0231981 CrossRef CAS PubMed.
- Y. Yang, Y. Zhao, F. Sun, T. You, Y. Gao and P. Yin, Microchem. J., 2020, 159, 105427 CrossRef CAS.
- X. Lu, L. Tao, Y. Li, H. Huang and F. Gao, Sens. Actuators, B, 2019, 284, 103–109 CrossRef CAS.
- M. Kesik, F. E. Kanik, J. Turan, M. Kolb, S. Timur, M. Bahadir and L. Toppare, Sens. Actuators, B, 2014, 205, 39–49 CrossRef CAS.
- Y. Al Naggar, G. Codling, A. Vogt, E. Naiem, M. Mona, A. Seif and J. P. Giesy, Ecotoxicol. Enviorn. Saf., 2015, 114, 1–8 CrossRef CAS.
- V. Bommuraj, Y. Chen, M. Birenboim, S. Barel and J. A. Shimshoni, Chemosphere, 2021, 266, 128974 CrossRef CAS PubMed.
- A. D. Noi, S. Casini, T. Campani, G. Cai and I. Caliani, Int. J. Environ. Res. Public Health, 2021, 18, 1863 CrossRef PubMed.
- A. J. Sami, S. Bilal, M. Khalid, M. T. Nazir and A. R. Shakoori, Pak. J. Zool., 2018, 50, 725–733 CAS.
- B. M. Woldegebriel, ECS Meeting Abstracts, 2020 Search PubMed.
- C. S. Kushwaha and S. K. Shukla, J. Mater. Sci., 2019, 54, 10846–10855 CrossRef CAS.
- M. Guler, V. Turkoglu and A. Kivrak, Environ. Sci. Pollut. Res., 2016, 23, 12343–12351 CrossRef CAS PubMed.
- S. Ebrahim, R. El-Raey, A. Hefnawy, H. Ibrahim, M. Soliman and T. M. Abdel-Fattah, Synth. Met., 2014, 190, 13–19 CrossRef CAS.
- O. Yamauchi, A. Odani and M. Takani, J. Chem. Soc., Dalton Trans., 2002, 3411–3421 RSC.
- G. Faccio, Sensors, 2018, 18, 1204 CrossRef PubMed.
- K. Khaldi, S. Sam, A. Gouget-Laemmel, C. Henry de Villeneuve, A. Moraillon, F. Ozanam, J. Yang, A. Kermad, N. Ghellai and N. Gabouze, Langmuir, 2015, 31, 8421–8428 CrossRef CAS PubMed.
- Y. H. Kim, D. J. Cha, J. W. Jung, H. W. Kwon and S. H. Lee, PLoS One, 2012, 7, e48838 CrossRef CAS PubMed.
- A. J. Sami, S. Bilal, M. Khalid, F. R. Shakoori, F. Rehman and A. R. Shakoori, Pak. J. Zool., 2016, 48, 881–886 CAS.
- M. A. Saleem and A. R. Shakoori, Arch. Insect Biochem. Physiol., 1987, 5, 45–55 CrossRef CAS.
- E. Krieger and G. Vriend, Bioinformatics, 2014, 30, 2981–2982 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |