Open Access Article
Open Access ArticleElectrospray ionization of native membrane proteins proceeds via a charge equilibration step†
Hsin-Yung Yen*ab,
Mia L. Abramsson c,
Mark T. Agasida,
Dilraj Lamac,
Joseph Gaulta,
Idlir Likoa,
Margit Kaldmäec,
Mihkel Saluric,
Abdul Aziz Qureshiad,
Albert Suadesd,
David Drew
c,
Mark T. Agasida,
Dilraj Lamac,
Joseph Gaulta,
Idlir Likoa,
Margit Kaldmäec,
Mihkel Saluric,
Abdul Aziz Qureshiad,
Albert Suadesd,
David Drew d,
Matteo T. Degiacomi
d,
Matteo T. Degiacomi e,
Erik G. Marklund
e,
Erik G. Marklund f,
Timothy M. Allison
f,
Timothy M. Allison g,
Carol V. Robinson
g,
Carol V. Robinson a and
Michael Landreh
a and
Michael Landreh *c
*c
aDepartment of Chemistry, University of Oxford, South Parks Road, Oxford, OX1 3QZ, UK. E-mail: hsinyungyen@gate.sinica.edu.tw
bInstitute of Biological Chemistry, Academia Sinica, 128, Academia Road Sec. 2, Nankang, Taipei 115, Taiwan
cDepartment of Microbiology, Tumor and Cell Biology, Karolinska Institutet, Tomtebodavägen 23A, 17165 Stockholm, Sweden. E-mail: michael.landreh@ki.se
dDepartment of Biochemistry and Biophysics, Stockholm University, 10691 Stockholm, Sweden
eDepartment of Physics, Durham University, South Road, Durham, DH1 3LE, UK
fDepartment of Chemistry – BMC, Uppsala University, Box 576, 75123 Uppsala, Sweden
gBiomolecular Interaction Centre, School of Physical and Chemical Sciences, University of Canterbury, Christchurch 8140, New Zealand
First published on 1st April 2022
Abstract
Electrospray ionization mass spectrometry is increasingly applied to study the structures and interactions of membrane protein complexes. However, the charging mechanism is complicated by the presence of detergent micelles during ionization. Here, we show that the final charge of membrane proteins can be predicted by their molecular weight when released from the non-charge reducing saccharide detergents. Our data indicate that PEG detergents lower the charge depending on the number of detergent molecules in the surrounding micelle, whereas fos-choline detergents may additionally participate in ion–ion reactions after desolvation. The supercharging reagent sulfolane, on the other hand, has no discernible effect on the charge of detergent-free membrane proteins. Taking our observations into the context of protein-detergent interactions in the gas phase, we propose a charge equilibration model for the generation of native-like membrane protein ions. During ionization of the protein-detergent complex, the ESI charges are distributed between detergent and protein according to proton affinity of the detergent, number of detergent molecules, and surface area of the protein. Charge equilibration influenced by detergents determines the final charge state of membrane proteins. This process likely contributes to maintaining a native-like fold after detergent release and can be harnessed to stabilize particularly labile membrane protein complexes in the gas phase.
Introduction
Native mass spectrometry (MS) is rapidly gaining importance as a tool for structural biology.1,2 By preserving protein complexes during native MS analysis, we can obtain information about their composition, oligomeric states, relative stabilities, and binding interactions.1,2 In recent years, applications of this technique have been extended to native membrane protein complexes, providing definite insights and a notable ability to probe the role of lipid interactions.3–7 Native MS requires the ionization and transfer of intact protein complexes to the gas phase, which is commonly achieved via electrospray ionization (ESI). Here, small, charged droplets containing the folded protein are created from bulk solution by an applied voltage. These droplets gradually evaporate, and undergo fission events when their charge becomes too high for their volume, until the last solvent evaporates leaving the protein and the remaining charge – predominantly excess protons in positive ionization mode – on the protein surface, as described by the charged residue model.8–10 The resulting protein ion charge is largely controlled by the Rayleigh limit, which determines the maximum number of charges that can be accommodated on the surface of a droplet.10–12 As the final droplet should approach the size of the protein itself, the number of charges acquired during ESI can be approximated using the protein's solvent accessible surface area (SASA) or its molecular weight (MW).11–14 Bush et al. have reported an average density of 0.63 g cm−3 based on drift-tube ion mobility MS measurements of near-spherical globular proteins.15,16 The relationship between size, SASA, and therefore ESI charge, is well-established and predictable for soluble proteins.The conditions for analyzing membrane proteins by native MS are distinctly different from those used for soluble proteins. This difference is because membrane proteins are commonly ionized while embedded in detergent micelles,17 which stabilize these highly amphipathic proteins in solution by mimicking the membrane environment. For the mass measurement of these proteins, the micelle is removed through collisional activation inside the mass spectrometer leaving behind the intact protein complex.17,18 This process implies that the natively membrane-embedded regions of the membrane protein's surface area (SA) are covered by detergent molecules during ionization. Ergo, the charge that membrane proteins could potentially acquire should depend on, and be best predicted by, the size of the full complex including the additional size and mass of the detergent micelle.
Interestingly, previous studies have indicated that membrane proteins consistently carry lower average charge than soluble proteins of similar SASA, lower than the prediction for both just the protein and the protein plus detergent micelle. This observation led to the hypothesis that only the non-transmembrane (TM) surface may determine the charge.19 However, the hypothesis was developed based on the analysis of membrane proteins with large soluble domains, therefore it remains unclear whether the hydrophobic transmembrane domains can contribute to the mechanism of lower-than-expected average charge. Moreover, Reading and co-workers demonstrated that membrane proteins ejected from non-ionic detergents such as n-dodecyl-β-D-maltoside (DDM), have maximum charge states close to the Rayleigh limit, and thus follow the same trend as for soluble proteins.20 These observations suggest that the entire protein surface, both trans- and intramembrane regions contribute to the ion's final charge despite being partially covered in detergent molecules during desolvation and ionization.20 Overall, the similarity to soluble proteins in this respect is inconsistent with membrane proteins ionizing while encapsulated in detergent micelles, which raises the question of how exactly membrane proteins acquire their final ESI charge states.
Results
Integral membrane proteins charge according to molecular weight in ESI-MS
To investigate out how the charge and SA of membrane proteins may be related, we considered monomeric membrane proteins without soluble domains as representing the simplest scenario. Mass spectra of these proteins should be able to show whether the surface area of the transmembrane regions is still important in determining the final ion charge. We selected three different membrane proteins that share this structural criterion, the sugar transporter Glut5 from Rattus norvegicus, a thermostabilized neurotensin receptor 1 (NTR1) from Rattus norvegicus, and the hexose transporter 1 from the malaria parasite Plasmodium falciparum (PfHT1). All three proteins are functional monomers and have the majority of their surface area buried in the bilayer21–23 (Fig. 1A–C). We performed native MS analysis of these proteins solubilized in the commonly used saccharide detergent DDM, which the presence or concentration of does not affect the charge of soluble proteins20 (Fig. 1A–C and S1†). Detergent molecules were removed from the ionized proteins by collisional activation in the ion trap of the mass spectrometer, revealing narrow charge state distributions that indicate compact and native-like solution conformations. We then calculated the expected average charge state [Zavg(SA)] using the crystal structure of each protein and the correlation described for the SA of soluble proteins (Table 1).24 Notably, the experimental Zavg of NTR1 was slightly lower than predicted, whereas those of Glut5 and PfHT1 matched the expected values (Fig. 1 and Table 1). We also predicted Zavg(MW) and the maximum charge Zmax(MW) based on the proteins' molecular weights instead of SA. This approach, which assumes a spherical protein shape, matched the average and maximum charges of PfHT1 very well and yielded only marginally higher or lower values for Glut5 and NTR1, respectively (Table 1). Our observations are thus generally consistent with those reported earlier.19,20 We then calculated the solvent- and membrane-accessible surface areas (SASA and MASA, respectively) for the proteins and determined the ratio (Table S1†). For all proteins, the membrane-embedded region accounts for 35 to 50% of the total surface. Since the MASA will be covered by detergent, the observation of ESI charge states only moderately lower than expected strongly suggests the transmembrane regions must contribute towards the charge on the protein once it is released from the micelle.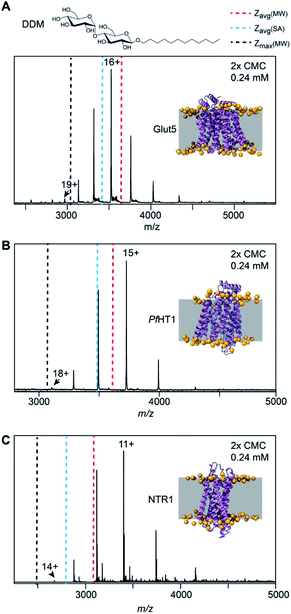 | ||
| Fig. 1 Mass spectra of (A) Glut5 (PDB ID 4YB9), (B) PfHT1 (PDB ID 6RW3), and (C) NTR1 (PDB ID 4BWB) after release from DDM micelles show agreement with the average (Zavg(MW)) and maximum charges (Zmax(MW)) predicted based on their MW, which are indicated by blue and black dashed lines. The average charge predicted by the SA (Zavg(SA)) of the crystal structures (blue dashed line) is moderately higher. The structure of DDM is shown at the top. The crystal structures are shown as cartoon renderings, with the TM region indicated by a grey box. Phosphates are shown as orange spheres. Membrane-embedded models were obtained from MemProtMD database.49 | ||
| Protein | MW (Da) | Zavg (SA) | Zavg (MW) | Zmax (MW) | Zavg (Exp) | Zmax (Exp) |
|---|---|---|---|---|---|---|
| Glut5 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 875 875 |
16.6 | 15.3 | 18.6 | 16.1 | 19 |
| NTR1 | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399 399 |
13.4 | 12.3 | 15.2 | 11.2 | 14 |
| PfHT1 | 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460 460 |
16.0 | 15.4 | 18.7 | 15.4 | 18 |
| GlpGMono | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 529 529 |
— | 11.5 | 14.2 | 11.4 | 15 |
| GlpGDimer | 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 058 058 |
— | 16.6 | 20.1 | 12.3 | 14 |
Detergents limit membrane protein charge
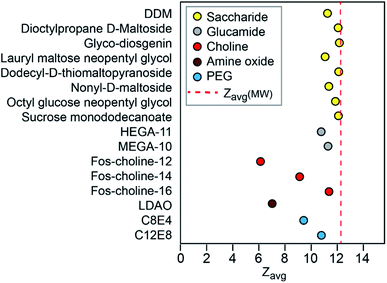
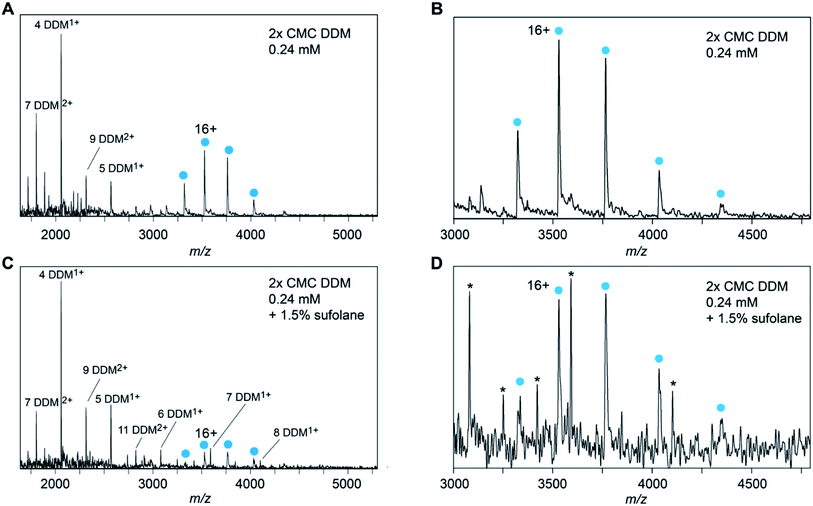
To assess if other detergents yield a similar correlation between charge and protein MW, we recorded spectra of NTR1 in 16 different MS-compatible detergents and determined the experimental Zavg (Fig. 2, S2 and Table S2†). The detergents used here can be grouped into saccharide, glucamide, polyethylene glycol (PEG), fos-choline, and amine oxide detergents, based on their head group structures, and feature many detergents for which membrane protein mass spectra have not before been published. Strikingly, all spectra collected with saccharide and glucamide detergents yielded Zavg close to each other. The experimental Zavg in each case was close to that predicted based on protein MW. We then asked whether the size of the detergent micelle could influence charging. Unfortunately, aggregation numbers are not available for most of the detergents in this study. We therefore used the molecular weight of the detergents as a proxy, assuming larger detergent molecules yield larger micelles. We did not observe a correlation between detergent MW and protein charge (Fig. S3†), which suggests that it is unlikely that the size of the micelle has any notable effect on charging. The similar average charge of NTR1 in the different detergents depends on the presence of mono- or di-saccharide head groups and is independent of alkyl tail length. These observations indicate that in general, and expanding upon publication of membrane proteins recorded with other detergents,3,20,25 the alkyl chain of the detergent has little role in membrane protein ionization.Notably, even though each membrane protein must ionize with the additional mass and SA of the detergent micelle, for all detergents the observed charge states are lower and only match the Zavg or Zmax predictions based on protein-only MW or SA. To help probe how the anticipated ‘extra’ charge is eliminated, we wondered what effect a common supercharging reagent, sulfolane, would have on the observed charge states. Sulfolane accumulates on the surface of ESI droplets, “trapping” excess charges inside which are ultimately deposited on the protein surface to yield highly charged ions.26,27 If these additional charges are likewise eliminated, then it may be possible to build a clearer picture of how the final charge of membrane protein ions arises. Mass spectra of DDM-solubilized Glut5 recorded in the presence of 1.5% sulfolane, show no significant shift in charge state compared to in DDM alone (Fig. 3A–D). However, sulfolane addition resulted in a notable increase in the observation of DDM clusters in spectra recorded under similar conditions (Fig. 3C). In fact, at sulfolane concentrations of 2% and above, the detergent clusters suppressed the Glut5 signal almost completely. From these observations, we conclude that sulfolane does not lead to notable supercharging effects but appears to increase the generation of charged detergent species. Thus, if more charges are present that can be accommodated by the protein, they can be expelled before or during release of the protein from the micelle, presumably with detergent molecules or clusters.
Detergents charge-reduce in the gas phase
In the mass spectra of NTR1 recorded in PEG, fos-choline, and amine oxide detergents, we observed lower charge states than for the glucamide and saccharide detergents. Reading et al. suggested that proton transfer can occur from the protein to PEG and amine oxide due to the high proton affinity of these detergent molecules (927 kJ mol−1 for tetraethylene glycol28) which will reduce the charge of the free protein.20 We therefore investigated further how charge transfer may occur in these systems. First, we compared mass spectra of NTR1 recorded in the PEG detergents C8E4 and C12E8 (Fig. 4A–C). Interestingly, despite similar head-group chemistry, the shorter-chain C8E4 exhibited more potent charge reduction properties than C12E8, reducing the average charge to 9.5, compared to 10.8 for C12E8. However, due to its higher critical micelle concentration (CMC), C8E4 was present at a roughly 100-fold higher concentration than C12E8. We thus wondered whether the higher concentration of C8E4 gives rise to the lower charge. Mass spectra of NTR1 in 100× CMC (9 mM) C12E8 show no change in charge state distributions, suggesting that the excess of detergent does not influence the final ion charge. This observation suggests that only the detergent molecules that constitute the proteomicelle affect the charge of the protein, whereas empty micelles or DDM molecules that might be present in the same droplet do not have a notable effect on charging. We then examined the effects of fos-choline detergents, which, like PEG ones, result in charge reduction with a clear dependence on alkyl chain length (Fig. 4D–F). With an average charge of 6.2, fos-choline-12 has more potent charge reducing capabilities than C8E4, whilst analysis in fos-choline-16 gave spectra with similar average charge as those recorded in C12E8. Unlike C12E8, however, increasing the fos-choline-16 concentration further reduced NTR1 average charge to 9.4. We conclude that PEG and fos-choline detergents, although both charge-reducing, affect the final charge of membrane proteins via different mechanisms.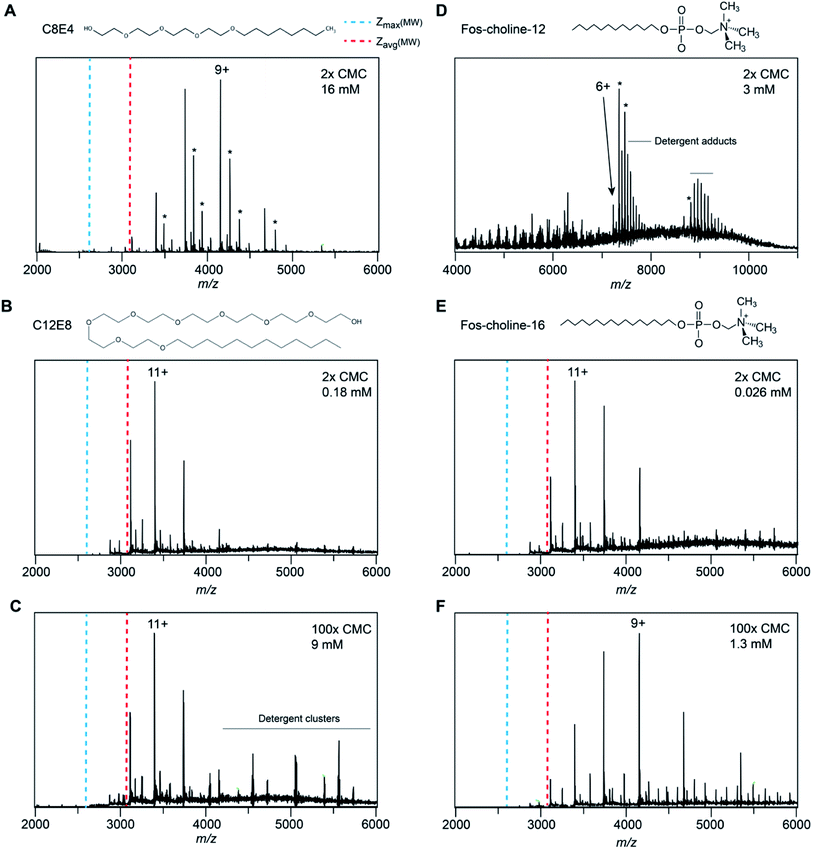 | ||
| Fig. 4 Spectra of NTR1 in different PEG and fos-choline detergents have different charge-reducing properties. (A) The native mass spectrum of NTR1 in 2× CMC (16 mM) C8E4 shows clear charge reduction compared to DDM (see Fig. 1C). (B) The presence of C12E8 resulted in only minor charge reduction compared to DDM. (C) Increasing the C12E8 concentration to 100× CMC (9 mM) does not affect the charge state distribution of NTR1. (D) Release from fos-choline-12 micelles produces highly charge-reduced ions with lipid- and detergent adducts. (E) At 2× CMC (0.026 mM), the longer-chain fos-choline-16 exhibits similar charge-reducing properties as C12E8. (F) Increasing the fos-choline-16 concentration to 100× CMC (1.3 mM) produces NTR1 ions with a similar charge as in C8E4. Asterisks denote lipid adducts. | ||
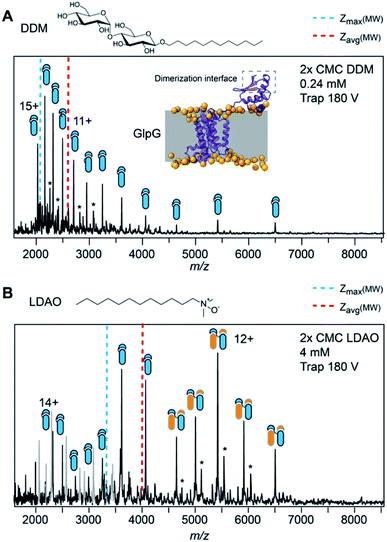
Charge-reducing detergents preserve labile GlpGdimers
Reducing the charge of membrane protein complexes by chemical additives has been shown to reduce dissociation in the gas phase.29,30 We thus speculated that by choosing a detergent with high proton affinity, we might be able to preserve particularly labile interactions for MS analysis. To test this idea, we selected the E. coli protease GlpG, which has been extensively studied by MS. DDM-solubilized GlpG exists predominantly as dimers, and a small soluble domain that is attached to the protein via a flexible linker has been found to participate in partial domain-swapping.31,32 However, the extent of dimerization in the membrane, and thus its biological relevance, are debated.33 To date, only monomeric GlpG has been observed by native MS, even under the most native-like conditions.34 When released from DDM, we detect GlpG monomers with a broad charge state distribution and comparatively high charges, which indicate some degree of unfolding either during ionization or during detergent release. In line with our previous observations, the Zavg and Zmax remain close the predicted values (Table 1). We then exchanged DDM for lauryldimethylamine oxide (LDAO), one of the most charge-reducing detergents in our dataset. Mass spectra recorded under identical MS conditions, including the same trap voltage of 180 V, show a large dimer population with an average charge of 12.3, significantly below the expected value of 15.4 (Fig. 5B). Interestingly, we also detect a single adduct with a mass of 1460 Da, consistent with a bound cardiolipin. The adduct signal is stronger for the dimer, suggesting that the interaction is preserved under charge-reducing conditions. Taken together, our results underscore the lability of the full-length GlpG dimer, providing a potential rationale for the conflicting observations regarding its oligomeric state in the membrane and in detergent micelles.33,35 We therefore conclude that detergents with a high proton affinity can be adept at preserving particularly labile interactions in the gas phase.Discussion
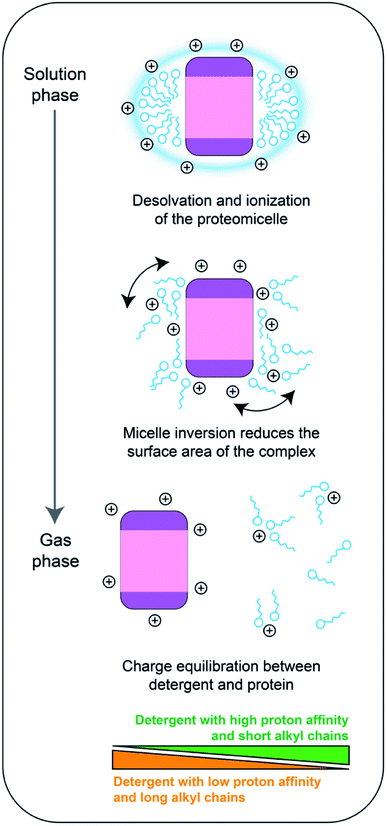
Here, in agreement with previous observations,20 we find that the ESI of membrane proteins generates ions with a similar number of charges as soluble proteins, and that the charge states are lower than expected for proteins encapsulated in detergent micelles at the time of ionization. Based on these observations, we propose that membrane proteins in detergent micelles initially ionize with charge states consistent with charged residue model predictions, before some, but not all the charges acquired by the protein-detergent complex are transferred to the protein upon detergent removal.The distribution of charges on a protein-detergent complex ion, and how this distribution relates to the final protein charge, has several contributing factors related to gas-phase basicities of amino acid residues and detergent molecules, and coulombic repulsion. To place our results in context, and to understand the charging mechanism in more detail, we have considered previous investigations into what happens to detergent molecules in a protein-detergent complex after ionization. Extensive computational and MS studies have revealed that once in the vacuum environment inside the mass spectrometer, detergent micelles tend to invert, that is, the head-groups rather than the alkyl chains, form the core of the assembly.36,37 Similar changes can be observed in MD simulations of desolvated protein-detergent complexes: contacts between detergent head-groups and the protein increase, while the hydrophobic alkyl chains extend away from the surface into the vacuum.38,39 Additionally, the inversion process is promoted by thermal activation, which is typically used to remove detergent from the protein ions during mass spectrometry analysis.37
In the results we present here, the detergent head-groups have the most pronounced effect on the final charge of a membrane protein, which indicates that the head groups are the most likely sites for protonation on a detergent molecule. Inversion of the micelle around the protein would bring the head-groups, and therefore the ESI charges that reside on the micelle, both closer together and closer to the protein. There would then be two clear potential routes to mitigate the resulting increase in coulombic repulsion, the protein could unfold to shift the charges further away from each other, or some of the charges could be removed via dissociation of detergent molecules. The first scenario would result in ions of higher average charge, but we observed no increase in ion charge for NTR1 above what can be expected for compact, soluble proteins (Fig. 2), arguing in this case for the latter scenario. Strengthening this proposition, since the detergent belt encapsulating the protein is less stable than the protein itself,19 the protein-detergent complex could at this stage shed detergent molecules, or clusters, and the maximum number of charges that stay on the complex would be limited by the protein surface. Hence, the average charge of the protein will approach that of a soluble protein when the detergent is removed. This model is further supported by the fact that we did not observe native supercharging by addition of sulfolane. Sulfolane acts by trapping charges on the surface of the droplet, which are subsequently transferred to the protein surface.27 In our model, the detergent micelle inversion and partial dissociation occurs after ionization of the protein-detergent complex, enabling removal of the excess charge as the protein is released (Fig. 6).
For mass spectra recorded in PEG detergents, we observed lower average charge states when the detergent is removed. Since the PEG detergents have a higher propensity to leave as charged species than saccharides,20 the dissociation we speculate to be induced by micelle inversion would reduce ion charges below the limit set by the SA. The charge-reducing effect is more pronounced for C8E4, which has just over half the chain length of C12E8, and independent on the amount of free detergent in solution. We therefore suggest that charge reduction is instead controlled by the number of detergent molecules that are included in the protein-detergent complex, which will depend on the detergent chain length. Shorter chain lengths may increase the propensity of the detergent to dissociate as clusters, which may be better charge carriers than individual molecules. Similarly, a higher number of molecules in the micelle means an increase in the number of head groups that can act as charge carriers.
The picture is more complex for fos-choline: the structurally related lipid phosphatidylcholine has the highest gas-phase basicity of all common lipids and is prone to causing charge reduction upon dissociation from a protein complex, in line with our findings for fos-choline.40,41 However, the charge-reducing effect of fos-choline detergents is concentration-dependent, even above the CMC and therefore this effect must not solely rely on interactions between protein and micelle. Interestingly, the amine oxide detergent LDAO as well as its head-group trimethylamine-N-oxide (TMAO) have been found to reduce the charge of protein ions through gas-phase collisions.20,42,43 We speculate that fos-choline additionally charge-reduces through ion–ion reactions in the gas phase, a mechanism consistent with observations that the charge-reducing effects of C8E4 and TMAO are additive.29 We conclude that fos-choline may affect membrane protein charging through a combination of charge equilibration upon dissociation, and ion–ion reactions in the gas phase.
Overall, we propose that the charge states of membrane proteins are determined by a combination of the surface area of the free protein, the proton affinity of the detergent, and the number of detergent molecules in contact with the protein. Together, these factors control a charge equilibration step that occurs between initial ionization and detergent removal, which distributes the ESI charges between the protein surface and the dissociating detergent molecules. An important implication of this model is that the membrane protein is unable to be charged above the limits dictated by its surface area in the native state. Charge equilibration may thus help to minimize coulombic unfolding and give rise predominantly to native-like, compact ions. Based on this model, it will be possible to design detergents with charge-reducing properties, as well as exploit the use of charge-reducing additives to preserve labile protein interactions in native MS.29,30,44,45
Experimental
Glut5, PfHT1, GlpG, and NTR1 were expressed and purified as described.21,22,46,47 All proteins were purified in 2× CMC (0.24 mM) DDM (Anatrace, CA) except for NTR1, which was purified in LMNG, and stored at −80 °C until analysis. Before MS analysis, all proteins were exchanged into 100 mM AmAc, pH 6.9 and the indicated detergent using Biospin 6 columns. nESI capillaries were purchased from Thermo. Mass spectra of Glut5 and PfHT1 were acquired on a Micromass LCT ToF modified for analysis of intact protein complexes (MS Vision, The Netherlands) equipped with an offline nanospray source. For the LCT, the capillary voltage was 1.5 kV and the RF lens 1.5 kV. The cone voltage was 300 V for collisional activation. The pressure in the ion source was maintained at 9.0 mbar. All data were analyzed using MassLynx V 4.1 (Waters, UK). MS of NTR1 was performed on a modified Q Exactive (Thermo Scientific, UK) mass spectrometer as previously reported.48 Briefly, a gentle voltage gradient was applied to avoid the collisional activation of the ions before transferring into the higher-energy collisional dissociation (HCD) cell. The detergent was removed by ramping the HCD voltage from 80 to 180 V until well-resolved spectra were obtained. Backing pressure was maintained at 1.05 × 10−9 mbar for optimal transmission of protein ions. Data were analyzed using the Xcalibur 2.2 software package.Surface area calculations
Structure models for the three membrane proteins embedded in lipid membranes were downloaded from MemProtMD.49 The surface calculations were carried out using the Gromacs tool gmx sasa with a default 1.4 Å probe radius.50 The full SA of the protein was calculated without any additional options. In order to get the separate SASA and MASA, the full protein was used for the calculations, but the area output was restricted to the atoms in the vicinity of the solvent and the membrane, respectively. Since hydrogen bonds are up to 3.5 Å between donor and acceptor, a 2.5 Å distance would capture the H-acceptor distance and therefore most solvent interactions. A 3.0 Å distance was used for PfHT1 and 4.0 Å for Glut5 and NTR1, since they were surrounded by different lipids (DPPC and DPP, respectively), based on radial distribution functions calculated from the protein surface (Fig. S4†). This approach inevitably leads to some atoms contributing to both SASA and MASA but given the low cut-off distance for the SASA we expect it to be a relatively small population.Charge state predictions
Zavg(SASA) was calculated based on the empirical data by Kaltashov and Mohimen12 by fitting the reported SASA and average charge values. The resulting correlation y = 0.6189x + 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 337 (R2 = 0.9775) was used to predict average charge states based on SASA. The correlation for Zavg(MW) = 0.0467x0.53 was reported by Stengel et al.51 based on the data of De La Mora.11 Zmax(MW) was calculated according to the correlation of Zmax(MW) = 0.778x0.5 reported by De La Mora.11
337 (R2 = 0.9775) was used to predict average charge states based on SASA. The correlation for Zavg(MW) = 0.0467x0.53 was reported by Stengel et al.51 based on the data of De La Mora.11 Zmax(MW) was calculated according to the correlation of Zmax(MW) = 0.778x0.5 reported by De La Mora.11
Abbreviations
| ESI | Electrospray ionization |
| (SA)SA | (Solvent-accessible) surface area |
| TM | Transmembrane |
| CMC | Critical micelle concentration |
| TEA | Triethylamine |
| TMA | Trimethylamine |
| TMAO | Trimethylamine-N-oxide |
| (PEG) | Polyethylene glycol |
Author contributions
HYY, EGM, TMA, CVR and ML designed the study. HYY, MLA, MK and MS performed MS experiments. DL, MTD and EGM performed computational simulations. MTA, JG, and IL provided additional data and expertise. AAQ, AS, and DD purified proteins. GM, TMA, and ML wrote the paper with input from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
ML is supported by a KI faculty-funded Career Position, an Ingvar Carlsson Award from the Foundation for Strategic Research (SSF), a Cancerfonden Project grant (190480), a VR Starting Grant (2019-01961), and a VR Research Environment Grant 2019-02433_VR (with DD). DL is supported by a Swedish Research Council grant for Internationally Recruited Scientists (2013-08807) to Prof. Sir David P. Lane, Karolinska Institutet. EGM acknowledges funding from MS SPIDOC by the European Union's Horizon 2020 Research and Innovation Program (801406).References
- P. Lössl, M. van de Waterbeemd and A. J. Heck, The diverse and expanding role of mass spectrometry in structural and molecular biology, EMBO J., 2016, 35, 2634–2657 CrossRef PubMed.
- J. L. P. Benesch and B. T. Ruotolo, Mass spectrometry: come of age for structural and dynamical biology, Curr. Opin. Struct. Biol., 2011, 21, 641–649 CrossRef CAS PubMed.
- A. Laganowsky, et al., Membrane proteins bind lipids selectively to modulate their structure and function, Nature, 2014, 510, 172–175 CrossRef CAS PubMed.
- H. Y. Yen, et al., PtdIns(4,5)P2 stabilizes active states of GPCRs and enhances selectivity of G-protein coupling, Nature, 2018, 559, 423–427 CrossRef CAS PubMed.
- C. Bechara and C. V. Robinson, Different modes of lipid binding to membrane proteins probed by mass spectrometry, J. Am. Chem. Soc., 2015, 137, 5240–5247 CrossRef CAS PubMed.
- E. Pyle, et al., Structural Lipids Enable the Formation of Functional Oligomers of the Eukaryotic Purine Symporter UapA, Cell Chem. Biol., 2018, 25, 840–848.e4 CrossRef CAS PubMed.
- R. A. Corey, et al., Specific cardiolipin–SecY interactions are required for proton-motive force stimulation of protein secretion, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 7967–7972 CrossRef CAS PubMed.
- P. Kebarle and U. H. Verkcerk, Electrospray: from Ions in solution to Ions in the gas phase, what we know now, Mass Spectrom. Rev., 2009, 28, 898–917 CrossRef CAS PubMed.
- R. G. McAllister, H. Metwally, Y. Sun and L. Konermann, Release of Native-like Gaseous Proteins from Electrospray Droplets via the Charged Residue Mechanism: Insights from Molecular Dynamics Simulations, J. Am. Chem. Soc., 2015, 137, 12667–12676 CrossRef CAS PubMed.
- L. Konermann, E. Ahadi, A. D. Rodriguez and S. Vahidi, Unraveling the mechanism of electrospray ionization, Anal. Chem., 2013, 85, 2–9 CrossRef CAS PubMed.
- J. F. De La Mora, Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism, Anal. Chim. Acta, 2000, 406, 93–104 CrossRef.
- I. Kaltashov and A. Mohimen, Estimates of protein surface areas in solution by electrospray ionization mass spectrometry, Anal. Chem., 2005, 77, 5370–5379 CrossRef CAS PubMed.
- Z. Hall, A. Politis, M. F. Bush, L. J. Smith and C. V. Robinson, Charge-state dependent compaction and dissociation of protein complexes: insights from ion mobility and molecular dynamics, J. Am. Chem. Soc., 2012, 134, 3429–3438 CrossRef CAS PubMed.
- A. D. Rolland and J. S. Prell, Computational insights into compaction of gas-phase protein and protein complex ions in native ion mobility-mass spectrometry, TrAC, Trends Anal. Chem., 2019, 116, 282–291 CrossRef CAS PubMed.
- M. F. Bush, et al., Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology, Anal. Chem., 2010, 82, 9557–9565 CrossRef CAS PubMed.
- M. Landreh, et al., Predicting the Shapes of Protein Complexes Through Collision Cross Section Measurements and Database Searches, Anal. Chem., 2020, 92, 12297–12303 CrossRef CAS PubMed.
- N. P. Barrera, N. Di Bartolo, P. J. Booth and C. V. Robinson, Micelles protect membrane complexes from solution to vacuum, Science, 2008, 321, 243–246 CrossRef CAS PubMed.
- M. Landreh, et al., Controlling release, unfolding and dissociation of membrane protein complexes in the gas phase through collisional cooling, Chem. Commun., 2015, 51, 15582–15584 RSC.
- N. Morgner, F. Montenegro, N. P. Barrera and C. V. Robinson, Mass spectrometry – from peripheral proteins to membrane motors, J. Mol. Biol., 2012, 423, 1–13 CrossRef CAS PubMed.
- E. Reading, et al., The Role of the Detergent Micelle in Preserving the Structure of Membrane Proteins in the Gas Phase, Angew. Chem., Int. Ed., 2015, 54, 4577–4581 CrossRef CAS PubMed.
- N. Nomura, et al., Structure and mechanism of the mammalian fructose transporter GLUT5, Nature, 2015, 526, 397–401 CrossRef CAS PubMed.
- A. A. Qureshi, et al., The molecular basis for sugar import in malaria parasites, Nature, 2020, 578, 321–325 CrossRef CAS PubMed.
- P. Egloff, et al., Structure of signaling-competent neurotensin receptor 1 obtained by directed evolution in Escherichia coli, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, E655–E662 CrossRef CAS PubMed.
- I. A. Kaltashov and A. Mohimen, Estimates of protein surface areas in solution by electrospray ionization mass spectrometry, Anal. Chem., 2005, 77, 5370–5379 CrossRef CAS PubMed.
- A. Laganowsky, E. Reading, J. T. S. Hopper and C. V. Robinson, Mass spectrometry of intact membrane protein complexes, Nat. Protoc., 2013, 8, 639–651 CrossRef CAS PubMed.
- R. R. Ogorzalek Loo, R. Lakshmanan and J. A. Loo, What protein charging (and supercharging) reveal about the mechanism of electrospray ionization, J. Am. Soc. Mass Spectrom., 2014, 25, 1675–1693 CrossRef CAS PubMed.
- H. Metwally, R. G. McAllister, V. Popa and L. Konermann, Mechanism of Protein Supercharging by Sulfolane and m-Nitrobenzyl Alcohol: Molecular Dynamics Simulations of the Electrospray Process, Anal. Chem., 2016, 88, 5345–5354 CrossRef CAS PubMed.
- A. Onigbinde, G. Nicol and B. Munson, Gas chromatography/mass spectrometry of polyethylene glycol oligomers, Eur. J. Mass Spectrom., 2001, 7, 279–291 CrossRef CAS.
- J. W. Patrick and A. Laganowsky, Generation of Charge-Reduced Ions of Membrane Protein Complexes for Native Ion Mobility Mass Spectrometry Studies, J. Am. Soc. Mass Spectrom., 2019, 30, 886–892 CrossRef CAS PubMed.
- S. Mehmood, et al., Charge reduction stabilizes intact membrane protein complexes for mass spectrometry, J. Am. Chem. Soc., 2014, 136, 17010–17012 CrossRef CAS PubMed.
- P. Sampathkumar, et al., Oligomeric state study of prokaryotic rhomboid proteases, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 3090–3097 CrossRef CAS PubMed.
- C. Lazareno-Saez, E. Arutyunova, N. Coquelle and M. J. Lemieux, Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease, J. Mol. Biol., 2013, 425, 1127–1142 CrossRef CAS PubMed.
- A. J. B. Kreutzberger and S. Urban, Single-Molecule Analyses Reveal Rhomboid Proteins Are Strict and Functional Monomers in the Membrane, Biophys. J., 2018, 115, 1755–1761 CrossRef CAS PubMed.
- N. Hellwig, et al., Native mass spectrometry goes more native: investigation of membrane protein complexes directly from SMALPs, Chem. Commun., 2018, 54, 13702–13705 RSC.
- E. Arutyunova, et al., Allosteric regulation of rhomboid intramembrane proteolysis, EMBO J., 2014, 33, 1869–1881 CrossRef CAS PubMed.
- L. Ceraulo, et al., Mass spectrometry of surfactant aggregates, Eur. J. Mass Spectrom., 2011, 17, 525–541 CrossRef CAS PubMed.
- A. J. Borysik, Structure and Dynamics of a Protein-Surfactant Assembly Studied by Ion-Mobility Mass Spectrometry and Molecular Dynamics Simulations, Anal. Chem., 2015, 87, 8970–8976 CrossRef CAS PubMed.
- S. L. Rouse, J. Marcoux, C. V. Robinson and M. S. P. Sansom, Dodecyl maltoside protects membrane proteins in vacuo, Biophys. J., 2013, 105, 648–656 CrossRef CAS PubMed.
- R. Friemann, D. S. D. Larsson, Y. Wang and D. Van Der Spoel, Molecular dynamics simulations of a membrane protein-micelle complex in vacuo, J. Am. Chem. Soc., 2009, 131, 16606–16607 CrossRef CAS PubMed.
- Z. M. Miller, J. D. Zhang, W. A. Donald and J. S. Prell, Gas-Phase Protonation Thermodynamics of Biological Lipids: Experiment, Theory, and Implications, Anal. Chem., 2020, 92, 10365–10374 CrossRef CAS PubMed.
- M. T. Donor, J. W. Wilson, S. O. Shepherd and J. S. Prell, Lipid head group adduction to soluble proteins follows gas-phase basicity predictions: dissociation barriers and charge abstraction, Int. J. Mass Spectrom., 2021, 469, 116670 CrossRef CAS PubMed.
- M. Kaldmäe, Gas-phase collisions with trimethylamine-N-oxide enable activation-controlled protein ion charge reduction, J. Am. Soc. Mass Spectrom., 2019, 30, 1385–1388 CrossRef PubMed.
- J. Gault, et al., Mass Spectrometry Reveals the Direct Action of a Chemical Chaperone, J. Phys. Chem. Lett., 2018, 9, 4082–4086 CrossRef CAS PubMed.
- J. Lyu, et al., Discovery of Potent Charge-Reducing Molecules for Native Ion Mobility Mass Spectrometry Studies, Anal. Chem., 2020, 92, 11242–11249 CrossRef CAS PubMed.
- L. H. Urner, et al., Modular detergents tailor the purification and structural analysis of membrane proteins including G-protein coupled receptors, Nat. Commun., 2020, 11, 564 CrossRef CAS PubMed.
- H. Y. Yen, et al., Ligand binding to a G protein-coupled receptor captured in a mass spectrometer, Sci. Adv., 2017, 3, e1701016 CrossRef PubMed.
- S. Zoll, et al., Substrate binding and specificity of rhomboid intramembrane protease revealed by substrate-peptide complex structures, EMBO J., 2014, 33, 2408–2421 CrossRef CAS PubMed.
- J. Gault, et al., High-resolution mass spectrometry of small molecules bound to membrane proteins, Nat. Methods, 2016, 13, 333–336 CrossRef PubMed.
- T. D. Newport, M. S. P. Sansom and P. J. Stansfeld, The MemProtMD database: a resource for membrane-embedded protein structures and their lipid interactions, Nucleic Acids Res., 2019, 47, D390–D397 CrossRef CAS PubMed.
- M. J. Abraham, et al., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- F. Stengel, et al., Quaternary dynamics and plasticity underlie small heat shock protein chaperone function, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 2007–2012 CrossRef CAS PubMed.
- E. F. Pettersen, et al., UCSF Chimera—A Visualization System for Exploratory Research and Analysis, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra01282k |
| This journal is © The Royal Society of Chemistry 2022 |