Open Access Article
Open Access ArticleInsights into substrate behavior in a solvent-free protein liquid to rationalize its reduced catalytic rate†
Sudarshan Behera and
Sundaram Balasubramanian
and
Sundaram Balasubramanian *
*
Chemistry and Physics of Materials Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore 560 064, India. E-mail: bala@jncasr.ac.in
First published on 19th April 2022
Abstract
When proteins are engineered with a polymer surfactant coating on their surface, they can form a liquid phase by themselves, without the need for a solvent, such as, say, water. However, such solvent-free protein liquids (SFPL), despite their capability to function at temperatures above those in aqueous solutions, exhibit much reduced catalytic rates. A comprehensive understanding of the nature of substrates in such liquids is crucial to reason out the reduced catalytic activity of enzymes as SFPL media, and thus identify the means to improve the same. Employing atomistic molecular dynamics simulations of lipase A from Bacillus subtilis in its SFPL form, we demonstrate that at low concentrations, the substrate molecules are located mostly in the hydrophilic layer of the surfactant shell that ensheaths the enzyme; substrates in this SFPL are present in various conformations with similar propensities as in the aqueous solution. Slower translational diffusion and reorientational dynamics, as well as the reduced tendency of a substrate molecule to closely interact with the enzymes in the SFPL medium have been identified herein as the contributing factors for the reduced activity of enzymes in this hybrid liquid. At high concentrations of substrates corresponding to those used in in vitro experiments, the formation of an enzyme-substrate complex is observed. Microscopic insights reported here can aid in the choice of surfactants to improve the catalytic rate of enzymes in SFPL.
Introduction
The use of a solvent medium (aqueous or non-aqueous) for enzyme catalysis is limited by the low stability of enzymes, unwanted side reactions, low solubilities, etc.1–5 Inspired by the work on nanoparticle fluids,6,7 hybrid protein-surfactant nanoconstructs have been synthesized, in which a solvent medium is no longer needed for the realization of a stable liquid phase.8–16 The preparation of the solvent-free protein liquid (SFPL) from an aqueous protein solution involves many steps: (i) cationization of the surface-exposed negatively charged amino acid residues (aspartate and glutamate), (ii) non-covalent linking of negatively charged surfactant molecules to the positively charged sites on the surface of the proteins, (iii) freeze-drying of the sample to remove water, and (iv) thermal annealing of the freeze-dried sample to obtain a hybrid, viscous liquid. Freeze-drying does not assure the complete removal of water, and hence a minuscule amount of water may still be present in the SFPL. However, it is far less than what is required for the formation of a hydration shell (∼40 water molecules are present per enzyme in the case of the solvent-free liquid of Bacillus subtilis lipase A (LipA) enzyme).15 Similar protocols have also been followed to synthesize protein-polymer surfactant liquid crystals and films.10,17–20SFPLs have shown promise not only in enhancing the thermal stability of proteins12,13 (even up to 150 °C), but also to act as a platform for biochemical reactions.21 Although the thermal stability of proteins in the SFPL form has been enhanced to a great extent over aqueous protein solutions, the high viscosity of the SFPL reduces the mass transfer of substrates to such a degree that the catalytic rate in the SFPL at a high temperature is still much less than that in the aqueous systems at room temperature.13
Besides the chemical transformation step (reactant to product transformation), a catalytic process also depends on other biophysical and biochemical steps such as substrate (and protein) diffusion, substrate binding, conformational flexibility (of both the enzyme and the substrate), and product release.22,23 If any of these steps is hindered, the overall catalytic rate will reduce. Hence a clear understanding of the microscopic behaviour of substrates in the SFPL is needed for the progress of this exotic field. Although many experiments and a few molecular dynamics simulations24–26 have been reported on various SFPLs, the microscopic behavior of substrates in such liquids is yet to be investigated. The questions which need investigations are: (i) how much conformational flexibility do substrates have in the SFPL? (ii) How is the mobility (translational and rotational) of the substrate affected in the absence of solvent? (iii) How does the substrate interact with the proteins? (iv) Where are the substrates located in the surfactant medium of the SFPL? Herein, we address these questions using extensive molecular dynamics simulations of substrates in the solvent-free liquid of a mutant of LipA enzyme. For the sake of comparison, the enzyme with the substrate is also modeled in an aqueous solution.27
LipA is a small lidless lipase enzyme (Kw = 19.8 kDa) of dimensions 35 × 36 × 42 Å.28,29 It comprises 181 amino acid residues with an α/β hydrolase fold and a solvent-exposed catalytic triad, Ser77-His156-Asp133. Recently, solvent-free liquids using wild type (WT) and various mutants of LipA enzyme have been synthesized and studied experimentally.15 In a recent report,24 we had studied extensively the structure of the WT SFPL and the interesting dynamics exhibited by the trace amounts of water present therein. The current work is devoted to understanding substrate behavior in SFPL using atomistic MD simulations. Zhou et al.15 showed that the catalytic activity depends on the number and distribution of positively charged sites on the LipA surface. The SFPL of a four-site mutant, 4M2 (F17E, A20E, G111D and M134E), showed the highest activity among the SFPLs of all variants for the hydrolysis of p-nitrophenyl butyrate (PNB, the substrate). Following this work, here we have investigated the behavior of PNB in the solvent-free liquid of the 4M2 mutant of LipA. We show that PNB in SFPL is displays different conformations with a similar propensity as in the aqueous solution. However, both the translational and rotational dynamics of the substrate molecules are much restricted in the SFPL. The translational mobility of PNB is observed to be sub-diffusive in nature. Further, a few substrate molecules have been observed to transit between enzymes during the course of the simulation, an elementary step in the recognition of the active site of enzymes. We also demonstrate that PNB molecules interact with the enzyme via residues present close to the active site, both in aqueous and SFPL media. In the SFPL, substrate molecules are found to be predominantly located in the polar, polyethylene glycol (PEG) part of the surfactant layer around the enzyme. The tendency of a substrate molecule to move towards an enzyme is reduced in the SFPL relative to that in an aqueous medium. At a high concentration of the substrate corresponding to typical in vitro experimental conditions, the formation of Michaelis complexes stabilized by the pre-formed oxyanion hole in the SFPL are also observed. On the whole, we have delineated possible reasons for the reduced catalytic activity of enzymes in the SFPL from the perspective of the behaviour of the substrate. These microscopic insights provide better understanding of the behaviour of the substrates in the hybrid liquid which can help engineer newer SFPLs with improved characteristics.
Computational methods
The crystal structure of the 4M2 mutant (ESI Fig. S1†) of Bacillus subtilis lipase A (LipA) is not available, and hence it was modeled from that of WT LipA. The crystal structure of WT LipA was taken from Protein Data Bank (PDB ID – 1i6w). The structure (obtained employing X-ray diffraction with 1.4 Å resolution) has 2 residues missing from the N-terminus.28 These were added and subsequently the 4M2 mutant was modeled, both using Pymol.30 The four site mutations were done sequentially, choosing the best rotamer for each mutated residue as the one having the least number of hard contacts with the enzyme (refer to ESI Section S2.1† for more detail). The protonation states of the titrable residues of the mutant, at pH 7.4, were generated using the ProteinPrepare web application.31Simulation of aqueous 4M2 mutant
Hydrogen atom coordinates were added to the structure of the mutant based on the protonation states of different amino acid residues at pH 7.4. The modeled 4M2 mutant, along with the water molecules which were present in the crystal structure (PDB ID – 1i6w), was solvated in a cubic water box of linear dimensions large enough as to avoid contact between periodic images of the enzyme. Appropriate number of counter ions were added to neutralize the system. The following steps were employed prior to the final production run: (i) energy minimization, (ii) simulated annealing in the NVT ensemble, with the temperature rising from 0 to 300 K during the course of the initial 500 ps simulation, followed by 500 ps of equilibration at 300 K, (iii) equilibration under NPT conditions at T = 300 K and P = 1 bar for 5 ns. The final configuration was subjected to NVT equilibration for 1 ns with the converged cubic box length and reinitialized velocities at 300 K to generate two more independent configurations. Each of these three independent configurations was simulated for 100 ns of production run in the NVT ensemble at T = 300 K.Simulation of aqueous cationized 4M2 mutant
The protocol followed for the cationization of 4M2 mutant is the same as that outlined for the WT enzyme in our earlier report.24 Since the 4M2 mutant has four additional negatively charged residues on the surface than the WT enzyme, cationization leads to a total of +31 charge on the surface of the 4M2 mutant, thus creating 31 binding sites for surfactants.15 The simulation protocols employed for this system is the same as described for the aqueous 4M2 mutant.Simulation of the aqueous cationized 4M2 mutant along with 31 surfactants (the surfactant used in the experimental report by Zhou et al.15 and hence used in this study is carboxylated Brij-L23, ESI Fig. S11†) has also been carried out with the same protocol. Modeling the aqueous cationized mutant system attached with 31 surfactants is similar to the one narrated in our earlier report.24
Simulation of aqueous 4M2 mutant with the substrate
Five different initial configurations were generated, each having a substrate (p-nitrophenyl butyrate; PNB) manually placed at different positions relative to the enzyme in a cubic box. Every configuration was subjected to the same simulation protocol as followed in the simulation of the aqueous 4M2 mutant and hence five trajectories each of 100 ns were generated. Three configurations of the system were extracted from each trajectory at the time frames of 0 ns, 50 ns and 100 ns. Hence a total of 15 new configurations were extracted from these five trajectories. All these fifteen configurations were subjected to NVT equilibration at T = 300 K with reinitialized velocities. Later, production trajectory of 100 ns in the NVT ensemble was generated from each of these fifteen independent initial configurations, making the total number of independent simulations to twenty.Simulation of solvent-free protein liquid
The modeling of SFPL is similar to the one detailed in our earlier report.24 In the present work, 31 surfactant molecules per enzyme were considered as there are 31 positively charged sites on the cationized 4M2 mutant. The construction of the initial configuration of surfactant is as explained earlier.24 31 surfactant molecules were placed manually around a 4M2 mutant LipA in such a manner that the positively charged site of the enzyme and the negatively charged carboxylate group of the surfactant are in close contact in the initial structure. The forty slowest moving water molecules (around forty water molecules per enzyme are found in the SFPL of LipA, observed experimentally15) as determined in our earlier report24 were retained in the present study as well. A configuration of 64 copies of Protein–Surfactant–Water complex (PSW complex; the cationized 4M2 mutant with 31 surfactant and 40 water molecules) was created using Packmol.32 The initial configuration was generated in a large cubic box in such a way that the PSW complexes are well separated, and hence free to rotate and translate during the initial phase of equilibration. The generated configuration was then subjected to (i) energy minimization, (ii) NVT at 300 K and (iii) NPT at T = 300 K, P = 100 bar and (iv) NPT at T = 300 K, P = 1 bar. It was then equilibrated at the NPT ensemble with T = 333 K (to match the experimental conditions15) and P = 1 bar until the box volume converged. The equilibrated system was then used for the final production run at NVT ensemble with T = 333 K. Three independent MD trajectories of length 200 ns were generated, each following the same strategy. Refer to ESI Fig. S12† for the flowchart of the protocol.Simulation of substrate in SFPL
The configuration after the first NVT equilibration of the 64 copies of PSW-complex (i.e., post step (ii) of simulation of SFPL) was taken. Either 32 and 128 substrate molecules were inserted into this structure using packmol32 to generate two systems with substrates at different concentrations. The substrate molecules were then inspected, translated and rotated manually, using VMD, in such a manner that they are present between PWS-complexes in the intervening, vacant spaces.33 Two snapshots showing the initial configurations of the SFPL with 32 and 128 substrate molecules are presented in ESI Fig. S13.† The same simulation strategy as used in the simulation of SFPL was employed to generate three independent MD trajectories for the SFPL with 32 PNB molecules and one trajectory for the SFPL with 128 PNB molecules. The length of each of these trajectories was of 1 μs duration. In the activity measurements reported by Zhou et al.,15 0.2 μl of desiccated PNB was added to a sample of SFPL containing 0.05 mg of protein. Converting these amounts to number of molecules leads us to the following stoichiometry: Protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNB = 1
PNB = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472 (Mw of the cationized 4M2 mutant = 20
472 (Mw of the cationized 4M2 mutant = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 751.7 g mol−1, Mw of PNB = 209.2 g mol−1, density of the liquid PNB at 20 °C = 1.19 g mL−1). In the current study, Protein
751.7 g mol−1, Mw of PNB = 209.2 g mol−1, density of the liquid PNB at 20 °C = 1.19 g mL−1). In the current study, Protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNB stoichiometries of 1
PNB stoichiometries of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (hereinafter referred to as SFPL-PNB-0.5), and 1
0.5 (hereinafter referred to as SFPL-PNB-0.5), and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (hereinafter referred to as SFPL-PNB-2) were studied to obtain an understanding of the system at dilute substrate concentrations without much inter-PNB interactions. Substrate concentrations in in vitro experiments are typically 2–3 orders of magnitude higher than that of the enzyme so as to be able to measure the rate of enzymatic catalysis in the concentration-independent regime of rate (Michaelis–Menten kinetics), using the stopped-flow technique. Thus, MD simulations were also carried out for PNB in SFPL at the same concentration as in the in vitro experiments15 (Protein
2 (hereinafter referred to as SFPL-PNB-2) were studied to obtain an understanding of the system at dilute substrate concentrations without much inter-PNB interactions. Substrate concentrations in in vitro experiments are typically 2–3 orders of magnitude higher than that of the enzyme so as to be able to measure the rate of enzymatic catalysis in the concentration-independent regime of rate (Michaelis–Menten kinetics), using the stopped-flow technique. Thus, MD simulations were also carried out for PNB in SFPL at the same concentration as in the in vitro experiments15 (Protein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNB = 1
PNB = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
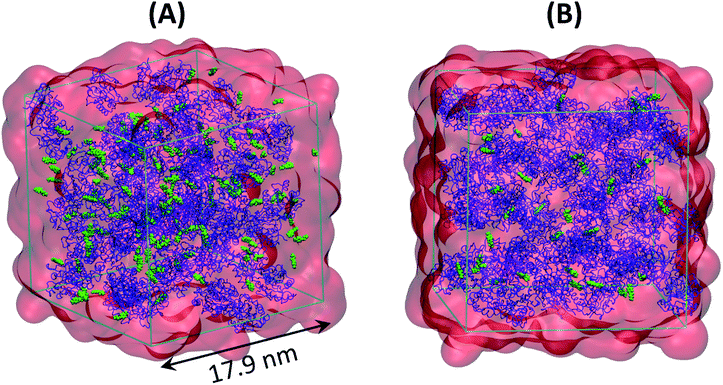
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472, hereinafter referred to as SFPL-PNB-472), using the same protocol. Two snapshots of the equilibrated systems of SFPL-PNB-0.5 and SFPL-PNB-2 are shown in Fig. 1 and ESI Fig. S14.† An ESI movie (refer to ESI Section S2.8.1 and SM1.mp4†) displays the trajectory (1 μs) of an SFPL-PNB-0.5 simulation focusing on the mobilities of substrates.
472, hereinafter referred to as SFPL-PNB-472), using the same protocol. Two snapshots of the equilibrated systems of SFPL-PNB-0.5 and SFPL-PNB-2 are shown in Fig. 1 and ESI Fig. S14.† An ESI movie (refer to ESI Section S2.8.1 and SM1.mp4†) displays the trajectory (1 μs) of an SFPL-PNB-0.5 simulation focusing on the mobilities of substrates.
Refer to ESI Table S1† for a summary of all the simulated systems (number of molecules of each kind, trajectory lengths, number of independent trajectories etc.) and the additional computational details section for more information on computational methods.
Additional computational details
AMBER99SB-ILDN34 force field parameters were used for standard amino acid residues. The cationized aspartate and glutamate, and the surfactant molecule were modeled with the same parameter set as used earlier.24 To obtain the force field parameters for p-Nitrophenyl Butyrate (PNB), it was first subjected to geometry optimization in gas phase using Gaussian0935 at HF/6-31G* level of theory, followed by GAFF36 parameters and RESP37 partial atomic charges generation using antechamber38 and acpype.39 TIP3P model40 was employed for water. GROMACS-2018.341 was used for energy minimization with conjugate gradient algorithm,42 and molecular dynamics simulations were performed using the Leap-frog integrator43 and a 2 fs time step with three-dimensional periodic boundary condition taken into account. Both the coordinates and velocities were written every 10 ps. All bonds were constrained using the LINCS algorithm,44 and particle mesh Ewald (PME)45 was used for the calculation of electrostatic interactions. A 10 Å cut off was used for both van der Waals and electrostatic interactions,34 and long-range corrections were added to both energy and pressure. Bussi–Donadio–Parrinello velocity rescaling thermostat46 and Berendsen barostat47 were employed for temperature and pressure coupling, respectively. VMD33 was used for visualization. Python scripting with MDAnalysis package48 and in-built modules of GROMACS41 were used for various analyses. Matplotlib49 was employed for plotting and movies were made using Molywood.50Unless otherwise stated, the results reported in this work are presented from the SFPL-PNB-0.5 and SFPL-PNB-2 systems, and averaged over all the independent simulations. Results from the SFPL-PNB-472 simulations are discussed briefly in Section 3.6 and elaborated in ESI Section S2.7.† The results of SFPL-PNB-472 are presented for the last 100 ns of simulation, after the convergence of Protein-PNB g(r) (refer to ESI Section S2.7†).
Results and discussion
The root-mean-square deviation (RMSD) of backbone atomic positions of the enzymes in the SFPL and aqueous media with respect to the crystal structure converges to a value less than 2 Å (ESI Fig. S15A†). This result indicates that the force field parameters used in this study stabilizes the native state of the enzyme fairly well, which is also supported by the convergence of the radius of gyration values (ESI Fig. S15B†).Location of the substrates
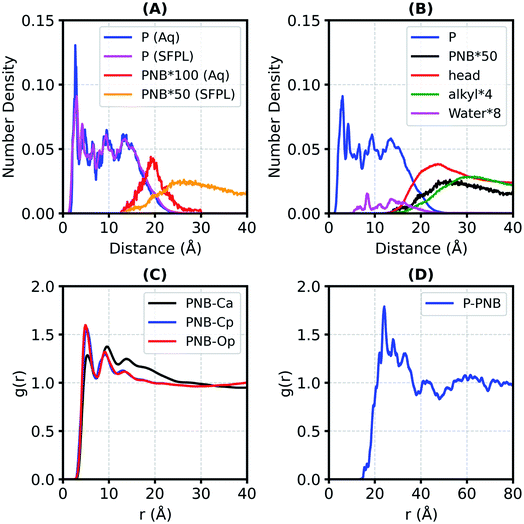
In order to identify the location of the substrate molecules in the SFPL, the number density profiles (nDP, the number of non-hydrogen atoms per Å3) with respect to the radial distance from the center of mass of enzyme for various constituents of the aqueous protein system and of the SFPL were obtained (Fig. 2A and B). In both, the nDP for the protein behaves similarly; it starts decaying at ∼15 Å (Fig. 2A), the same value as the radius gyration (Rg) of LipA. The nDP for PNB (Fig. 2A) illustrates that the substrate molecules are present in the vicinity of the protein surface in both the aqueous and SFPL systems. In the case of the aqueous solution, the substrate is present closer to the enzyme (the nDP starts at ∼13 Å and peaks at ∼19.5 Å) than in the SFPL (the nDP starts at ∼13 Å and peaks at ∼26 Å). The reduced propensity of the substrate to be present near the enzyme in the SFPL than in the aqueous case may also be a contributory factor to the reduced catalytic rate of enzymes in the SFPL – an observation that was hitherto unrecognized. Further discussion on this aspect is presented in the “Interaction of the substrate molecules with the enzymes” section.The nDP for the hydrophilic head group of surfactant (PEG and carboxylate group) in the SFPL starts at ∼13 Å and peaks at 23.6 Å with a number density value of 0.036 Å−3 suggesting that the hydrophilic head groups interact directly with the enzyme and form almost a complete layer around it (for comparison, the non-hydrogen atomic density for liquid ethylene glycol at ambient conditions is 0.045 Å−3). The nDP for the hydrophobic alkyl tail peaks at 29 Å with a number density value of 0.007 Å−3, which is much less than the same for liquid tetradecane at ambient conditions (0.032 Å−3). It is deduced from this observation that the hydrophobic alkyl tail of the surfactant cannot form a complete layer around the enzyme.24 Water molecules are present close to the enzyme (either buried in it or on its surface). The behavior of nDP of the substrate in the SFPL is similar to that of the hydrophilic head group of the surfactant. Their peak positions are close to each other, near ∼25 Å, which suggests that the PNB molecules are mostly present in the hydrophilic layer of the surfactant.
Support for the substrate molecules being located in the hydrophilic head layer of the surfactant comes from the radial distribution functions (RDFs) between the center of mass (COM) of PNB and different types of non-hydrogen atoms from the surfactant in the SFPL (Fig. 2C). The first peaks of PNB-Cp and PNB-Op RDFs are taller than that of PNB-Ca (Cp: C of the PEG part, Op: O of the PEG part, Ca: C of the alkyl part), suggesting an increased tendency for the PEG group to be present closer to the PNB molecules than the alkyl part of the surfactant. The coordination number of Cp, Op and Ca around the COM of PNB (ESI Fig. S16A and S16B†) are 32, 16 and 8, respectively (at the first minimum in the RDFs, 7.4 Å). This demonstrates that for every carbon atom of the alkyl part of the surfactant present in the first coordination shell of PNB, two PEG groups are present. In the SFPL, the RDF between the COMs of the protein and PNB (Fig. 2D) peaks at 24 Å, which too confirms the presence of the substrate molecules mostly in the hydrophilic layer of the surfactant (as the nDP of hydrophilic part of the surfactant peaks at 24 Å).
Conformations of the substrates
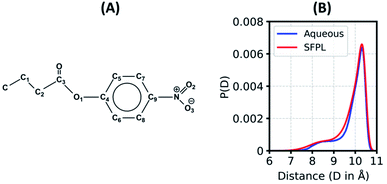
In an enzymatic catalysis process that proceeds via covalent linkages and dissociation, the substrate molecules need to adopt various conformations before binding to the active site.51 The ensemble of conformations helps a substrate molecule to recognize the best binding pose. In a highly viscous liquid such as the solvent-free protein liquid, the substrate (PNB; Fig. 3A) may find it rather difficult to explore all different conformations as found in the aqueous medium. To investigate this aspect, we calculated the equilibrium distribution of the end-to-end distance (Fig. 3B) as well as the distributions of various flexible dihedral angles (ESI Fig. S17†) of the substrate. The distributions for the substrate in the SFPL medium match well with those in the aqueous medium. Substrate molecules in the SFPL system exhibit various conformations with similar propensities as in the aqueous system. These observations demonstrate that the conformational plasticity of the substrate molecules is retained in the SFPL. The hump around 8.5 Å in the end-to-end distance distribution (Fig. 3B) appears due to the peak around 70° in the distribution of C3–C2–C1–C dihedral angle (ESI Fig. S17C†).Translational dynamics of the substrate
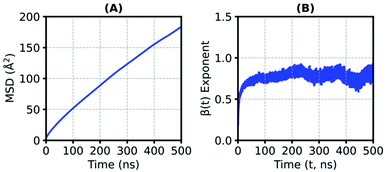
The translational mobility of a substrate is substantially reduced in the SFPL medium, relative to that in the aqueous solution. In the aqueous system, the mean-squared-displacement (MSD) values are two orders of magnitude higher than that in the SFPL (the MSD values at 50 ns are ∼30 Å2 and ∼12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Å2 for the SFPL and aqueous media, respectively (Fig. 4 and ESI Fig. S18†)). On an average, in the SFPL, a substrate molecule exhibits a linear displacement of just around 13.5 Å over a duration of 500 ns (MSD value is around 180 Å2) (Fig. 4A). The translational motion of a substrate molecule in the SFPL medium is sub-diffusive in nature, as can be deduced from the β(t)-exponent plot (Fig. 4B). These results are to be expected, since even a small molecule like water has been shown to be sub-diffusive in the solvent-free hybrid liquid24 over these time scales. This hindered motion of the substrates (and proteins24) is yet another cause for the reduced catalytic activity of enzymes in the SFPL relative to that in aqueous solution. Within the overall sub-diffusive character, substrate molecules exhibit diverse translational mobilities (ESI Fig. S19†) in SFPL, some being almost immobile, while some exhibit relatively large displacements. ESI Fig. S20† shows the COM motion of four highly mobile PNB molecules, which move more than 30 Å in 1 μs, while ESI Fig. S21† shows the same for four other PNB molecules whose displacements are minimal over the same time period.
000 Å2 for the SFPL and aqueous media, respectively (Fig. 4 and ESI Fig. S18†)). On an average, in the SFPL, a substrate molecule exhibits a linear displacement of just around 13.5 Å over a duration of 500 ns (MSD value is around 180 Å2) (Fig. 4A). The translational motion of a substrate molecule in the SFPL medium is sub-diffusive in nature, as can be deduced from the β(t)-exponent plot (Fig. 4B). These results are to be expected, since even a small molecule like water has been shown to be sub-diffusive in the solvent-free hybrid liquid24 over these time scales. This hindered motion of the substrates (and proteins24) is yet another cause for the reduced catalytic activity of enzymes in the SFPL relative to that in aqueous solution. Within the overall sub-diffusive character, substrate molecules exhibit diverse translational mobilities (ESI Fig. S19†) in SFPL, some being almost immobile, while some exhibit relatively large displacements. ESI Fig. S20† shows the COM motion of four highly mobile PNB molecules, which move more than 30 Å in 1 μs, while ESI Fig. S21† shows the same for four other PNB molecules whose displacements are minimal over the same time period.
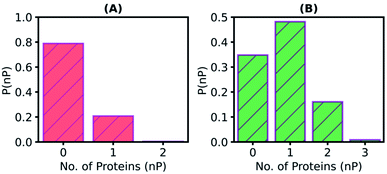
Since some substrate molecules show relatively high mobility, do they transit between the enzymes? To answer this question, we define a PNB molecule to be interacting with a particular enzyme molecule if any of the non-hydrogen atoms of the enzyme falls within 5 Å of any non-hydrogen atom of the substrate. The number of enzymes a substrate molecule interacts with simultaneously (i.e., in the same time frame) was calculated, and the histogram is plotted in Fig. 5A. Most substrate molecules (∼80%) are found to be present in the surfactant medium without interacting with any enzyme. Also to note is the fact that no PNB molecule ‘bridges' two enzymes simultaneously (value of the histogram for abscissa value of two is zero). When a similar histogram (Fig. 5B) for the number of enzymes a substrate molecule visits during the entire course of the simulation was constructed, the transit of the substrate molecules between enzymes was clearly demonstrated. About 20% of the substrate molecules transit between enzymes (i.e., visit more than one enzyme). Transits of substrate molecules between enzymes may help a substrate to locate and bind to the catalytic site of an available nearby enzyme, readily.
Rotational dynamics of the substrate
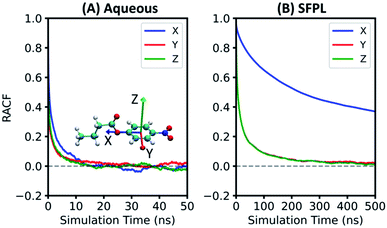
The catalytic rate is dependent not only on the translational mobility of the substrate, but also on its reorientational dynamics.23 Herein, the rotational dynamics of PNB has been characterized through the reorientational time autocorrelation functions (RACFs) of three of its internal vectors (Fig. 6 and refer to ESI Section S2.4 for more details†). In both the aqueous and SFPL cases, the RACF for Vector X decays slower as it requires rotation along the axes with large moment of inertia. The RACFs for Vectors Y and Z decay faster and with comparable rates, for similar reasons. It should however be noted that the rotational dynamics of the substrates as captured by the RACFs of Vectors Y and Z in the SFPL medium is about an order of magnitude slower than that in the aqueous medium. The X vector of the substrate in the SFPL medium does not decay to zero even over 500 ns. The overall slower rotational dynamics of the substrate in the SFPL could also be one of the contributing factors for the lower catalytic activity of enzymes in this liquid than in the aqueous systems.Interaction of the substrate molecules with the enzymes
During the function of an enzyme, the substrate has to recognize the binding site (pocket) effectively which is followed by covalent linking with the enzyme via its catalytic site.51,52 In the case of LipA, which is a lidless enzyme, the catalytic triad is always exposed to the solvent. To characterize the tendency of a substrate molecule to interact with the catalytic region or with any amino acid residue (standard or cationized), herein a quantity P(Rid) is used. P(Rid) of a residue (Rid) is defined as the number of MD simulation frames in which the residue interacts with PNB molecule(s) divided by both the P![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PNB stoichiometry (0.5, 2, and 472 for SFPL-PNB-0.5, SFPL-PNB-2, and SFPL-PNB-472 systems, respectively) and the total number of frames. As before, a residue is defined to be interacting with a PNB molecule if any of the non-hydrogen atoms of the residue falls within 5 Å of any non-hydrogen atom of the PNB molecule. Cutoff values of 0.1 and 0.033 were chosen for the aqueous and SFPL media, respectively (the top two panels of ESI Fig. S9†), to identify few residues which interact with the enzyme(s), the most. It is observed that the residues located around the catalytic site (see Fig. 7 for the corresponding residue numbers) and few residues farthest from the catalytic site interact the most with substrates, both in the aqueous and SFPL media (Fig. 7 and ESI Fig. S25†). These results demonstrate that the ability of the substrate molecules to recognize the active site of the enzyme even in the highly viscous SFPL medium is similar to that in the aqueous system. Most of these residues, whose P(Rid) values lie above the cutoff, are hydrophobic in nature for the aqueous case (ten out of fifteen are hydrophobic, four are polar, and one charged). In the case of the SFPL, these residues are a mixture of charged (two cationized glutamate, Gluc), polar (six in number) and non-polar (eight in number). An SI movie (refer to ESI Section S2.8.2† and SM2.mp4) shows four independent trajectories of the aqueous enzyme with PNB, where the substrate molecule successfully binds to the active site (and later leaves the site) within simulation time scales. However, such a binding event was not observed in the simulations of the SFPL (in SFPL-PNB-0.5 and SFPL-PNB-2).
PNB stoichiometry (0.5, 2, and 472 for SFPL-PNB-0.5, SFPL-PNB-2, and SFPL-PNB-472 systems, respectively) and the total number of frames. As before, a residue is defined to be interacting with a PNB molecule if any of the non-hydrogen atoms of the residue falls within 5 Å of any non-hydrogen atom of the PNB molecule. Cutoff values of 0.1 and 0.033 were chosen for the aqueous and SFPL media, respectively (the top two panels of ESI Fig. S9†), to identify few residues which interact with the enzyme(s), the most. It is observed that the residues located around the catalytic site (see Fig. 7 for the corresponding residue numbers) and few residues farthest from the catalytic site interact the most with substrates, both in the aqueous and SFPL media (Fig. 7 and ESI Fig. S25†). These results demonstrate that the ability of the substrate molecules to recognize the active site of the enzyme even in the highly viscous SFPL medium is similar to that in the aqueous system. Most of these residues, whose P(Rid) values lie above the cutoff, are hydrophobic in nature for the aqueous case (ten out of fifteen are hydrophobic, four are polar, and one charged). In the case of the SFPL, these residues are a mixture of charged (two cationized glutamate, Gluc), polar (six in number) and non-polar (eight in number). An SI movie (refer to ESI Section S2.8.2† and SM2.mp4) shows four independent trajectories of the aqueous enzyme with PNB, where the substrate molecule successfully binds to the active site (and later leaves the site) within simulation time scales. However, such a binding event was not observed in the simulations of the SFPL (in SFPL-PNB-0.5 and SFPL-PNB-2).
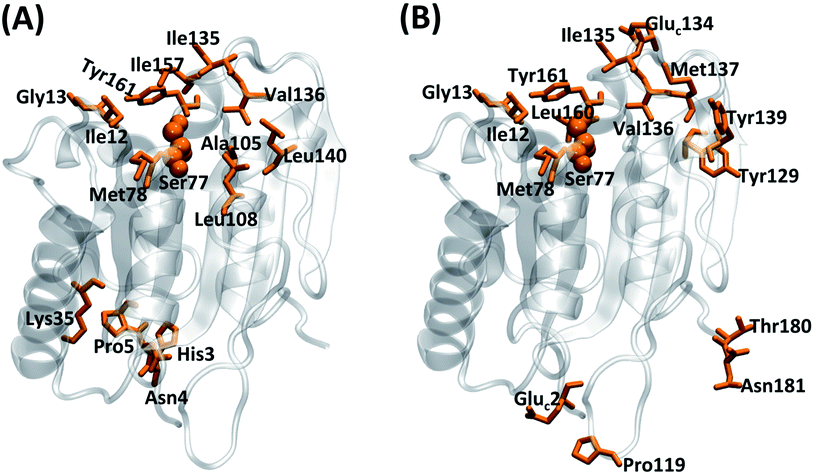 | ||
| Fig. 7 Residues whose P(Rid) values are more than 0.1 and 0.033 (refer to Fig. S9†) for (A) the aqueous and (B) the SFPL (the mean of SFPL-PNB-0.5 and SFPL-PNB-2 systems), respectively, highlighted in orange. The residues for the aqueous system are His3, Asn4, Pro5, Ile12, Gly13, Lys35, Ser77, Met78, Ala105, Leu108, Ile135, Val136, Leu140, Ile157 and Tyr161, and for the SFPL are Gluc2, Ile12, Gly13, Ser77, Met78, Pro119, Tyr129, Gluc134, Ile135, Val136, Met137, Tyr139, Leu160, Tyr161, Thr180 and Asn181. The three letter code “Gluc” is used for the cationized glutamate residue. The catalytic Ser77 is shown in ball representation to easily identify the active site. The enzymes are represented as transparent cartoons with silver color. | ||
In the aqueous solution, the substrate is less likely to reside completely in bulk water because of the n-propyl part and thus is likely to be present near the hydrophobic surface residues of the enzymes such as those near the catalytic region. It should however be noted that lipases are catalytically most active at oil-water interfaces, wherein the substrates dissolved in the oil phase approach the active site of the enzyme which is primarily located at the interface.27,53,54 The surfactant medium of the SFPL, unlike water, has both polar (PEG part) and non-polar groups (alkyl tail). As a consequence, the substrate molecule resides in the surfactant medium in the SFPL (particularly within the PEG layer) rather than on the surface of the enzyme as was the case in the aqueous solution. This result is also supported by Fig. 5A, wherein ∼80% of the substrates in the SFPL did not interact with the enzyme at all. Fig. 2A also confirms the presence of the PNB closer to the enzymes in the aqueous medium than in the SFPL. Overall, the less hydrophilic nature of the surfactant medium decreases the tendency of the substrate molecules to approach the enzymes in the SFPL. The reduced propensity of the substrate molecules to interact with the enzymes can also be counted as another reason for the hampered activity of enzymes in the SFPL, relative to that in aqueous solution.
Formation of the Michaelis complex in SFPL-PNB-472
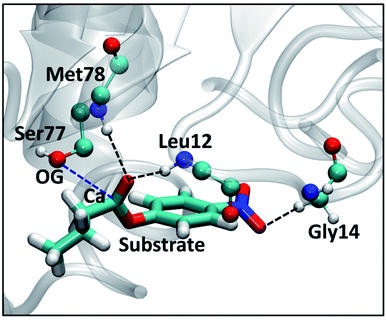
Results from the SFPL-PNB-472 system are discussed elaborately in ESI Section S2.7.† Briefly, ∼80% of the substrate molecules are observed to be in a bulk liquid PNB-like environment, and the rest ∼20% are present in the surfactant layer and/or close to enzymes. Both the translational and rotational dynamics of the substrate are faster in SFPL-PNB-472 compared to that in the SFPL-PNB-0.5 and SFPL-PNB-2 systems, due to the preponderance of substrate molecules away from either the enzyme or the surfactants. However, they are significantly slower compared to that in the aqueous solution, which explains the reduced catalytic activity of SFPL than the aqueous solution. Interestingly, we observed the formation of a Michaelis complex (the noncovalent enzyme-substrate complex prior to catalysis) in the SFPL-PNB-472 system, which is narrated below.The first step (the acylation step) in the hydrolysis reaction catalyzed by a lipase is a nucleophilic attack of the hydroxyl oxygen (referred to as OG) of a serine (Ser77 in the case of LipA) to the carboxyl carbon (referred to as Ca) of the substrate.55 Prior to the catalysis, the OG of serine and the carboxylate carbon of the substrate (Ca) come close to form a Michaelis complex. The RDF between the OG of Ser77 and the Ca of substrate for three different simulation runs of SFPL-PNB-472 are presented in ESI Fig. S10A.† The small peaks around 3.5 Å unveil the presence of a few Michaelis complexes in the system. The coordination number (ESI Fig. S10B†) at 5 Å shows the presence of two (CN at 5 Å (0.068) × total number of enzymes in the box (32) = 2.2), one (0.03 × 32 = 0.96), and two (0.052 × 32 = 1.7) such complexes in the three independent MD trajectories respectively (out of the thirty-two possible complexes in a single configuration, as there are thirty-two enzymes). These five complexes were observed to remain intact (OG-Ca distance to be less than 5 Å) for 100%, 100%, 72%, 69%, and 67% of the simulation time (100 ns). The nature of interactions that stabilizes the Michaelis complex was also examined (refer to Fig. 8). The major contribution is from the pre-formed oxyanion hole in LipA. The backbone N–H groups of residue Leu12 and Met78 (the oxyanion hole) form strong hydrogen bonds with the carbonyl oxygen (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of the substrate. Other contributing factors are the weak C–H⋯O hydrogen bond between a hydrogen of Cα of Gly14 and an oxygen of the NO2 group of the substrate, and a number of van der Waals interactions between the substrate and the enzyme active site.
O) of the substrate. Other contributing factors are the weak C–H⋯O hydrogen bond between a hydrogen of Cα of Gly14 and an oxygen of the NO2 group of the substrate, and a number of van der Waals interactions between the substrate and the enzyme active site.
Conclusions
The behavior of substrate molecules in the medium plays an important role in determining the overall catalytic rate of enzymes.23 The catalytic rate of industrially relevant enzymes such as lipases can, in principle be enhanced if the operating temperature can be increased without denaturation of the protein. This has been achieved experimentally, upon the complete removal of the solvent (water) and suitably ‘protecting’ the enzymes from aggregating with each other by encasing each of them in a polymer surfactant shell. The process yields a solvent-free protein liquid which can function beyond 100 °C, although with catalytic rates much reduced when compared to that in the aqueous solution. A fundamental understanding of the reduction in the rate requires a study of substrate behavior in these exotic liquids. Atomistic molecular dynamics simulations of the substrate, p-nitrophenyl butyrate (PNB) in a solvent-free liquid of lipase A, reported here, have provided the same. For the sake of comparison, the substrate behavior in an aqueous solution of lipase A has also been studied.Our findings include
• PNB molecules are primarily located in the surfactant medium of the SFPL and mostly in its hydrophilic (PEG) layer, which was discerned through number density profiles and radial distribution functions.• Despite being a highly viscous liquid, the SFPL facilitates conformational flexibility of the substrate to exhibit different dihedral angles with similar probabilities as in the aqueous medium.
• The tendency of a substrate to interact with enzymes is reduced in the SFPL compared to that in the aqueous medium. This is a significant difference from its behavior in the aqueous solution, and underlies the hampered catalytic activity of the enzymes as an SFPL medium.
• The translational displacement of a substrate is substantially hindered in the SFPL relative to that in the aqueous solution. The motion of the PNB molecule is sub-diffusive in nature, and the displacement magnitudes in the SFPL are two orders of magnitude smaller than that in the aqueous system, for any given time duration. The overall hampered motion of the substrates is a major reason for the lower catalytic rate of enzymes in the SFPL compared to that in the aqueous system.
• Despite their overall sluggishness in the SFPL, substrate molecules display a variety of translational mobilities, from being almost immobile to ones which move reasonably well. Substrates with the high mobility are shown to interact lesser with the enzymes and more with the surfactant than their low mobile counterparts. A few substrate molecules (15%) have even been seen to transit between enzymes during the course of the simulation – a crucial event in molecular recognition.
• The reorientational dynamics of a substrate molecule, which also impacts the catalytic rate, is slower in the SFPL than in the aqueous form and hence could also contribute to the reduction in the activity of enzymes in this hybrid liquid.
• The active site recognition ability of the substrate molecules is found to be largely retained in the SFPL. On occasions when the PNB molecules interact with the enzyme, they interact mostly with amino acid residues close to the catalytic triad. In simulations with high substrate concentrations comparable to those used in in vitro experiments, the formation of the enzyme-substrate Michaelis complex was observed.
In summary, we have investigated various features displayed by substrates in an engineered solvent free protein liquid using extensive all-atom molecular dynamics simulations and have presented possible reasons for the experimentally observed reduction in the catalytic activity of enzymes in this hybrid liquid, from the perspective of the substrates. These include: (a) the reduced translational diffusion of the substrates, (b) their hindered reorientational dynamics, and (c) the decreased tendency of substrates to interact with enzymes, in the SFPL. These microscopic insights into the behavior of substrate in the SFPL can potentially aid in designing SFPLs with improved properties.
Conflicts of interest
The authors have no conflicts to disclose.Acknowledgements
We acknowledge the Department of Science and Technology, India, for its support. S. Behera thanks the Council of Scientific and Industrial Research for a fellowship. This work was also supported by grants from the Department of Biotechnology, India (Project No. BT/INF/22/SP27679/2018). The support and the resources provided by ‘PARAM Yukti Facility’ under the National Supercomputing Mission, Government of India at the Jawaharlal Nehru Centre For Advanced Scientific Research are gratefully acknowledged.References
- M. Ghaffari-Moghaddam, H. Eslahi, Y. A. Aydin and D. Saloglu, J. Microbiol. Methods, 2015, 2, e25 Search PubMed.
- S. Wang, X. Meng, H. Zhou, Y. Liu, F. Secundo and Y. Liu, Catalysts, 2016, 6, 32 CrossRef.
- S. Ghosh, S. Chattoraj, R. Chowdhury and K. Bhattacharyya, RSC Adv., 2014, 4, 14378–14384 RSC.
- F. J. Deive, D. Ruivo, J. V. Rodrigues, C. M. Gomes, M. A. Sanroman, L. P. N. Rebelo, J. M. S. S. Esperanca and A. Rodriguez, RSC Adv., 2015, 5, 3386–3389 RSC.
- K. P. Ghanta, S. Mondal, S. Mondal and S. Bandyopadhyay, J. Phys. Chem. B, 2021, 125, 9441–9453 CrossRef CAS PubMed.
- S. C. Warren, M. J. Banholzer, L. S. Slaughter, E. P. Giannelis, F. J. DiSalvo and U. B. Wiesner, J. Am. Chem. Soc., 2006, 128, 12074–12075 CrossRef CAS PubMed.
- R. Rodriguez, R. Herrera, L. A. Archer and E. P. Giannelis, Adv. Mater., 2008, 20, 4353–4358 CrossRef CAS.
- A. W. Perriman, A. P. S. Brogan, H. Cölfen, N. Tsoureas, G. R. Owen and S. Mann, Nat. Chem., 2010, 2, 622–626 CrossRef CAS PubMed.
- A. W. Perriman and S. Mann, ACS Nano, 2011, 5, 6085–6091 CrossRef CAS PubMed.
- K. Liu, C. Ma, R. Göstl, L. Zhang and A. Herrmann, Acc. Chem. Res., 2017, 50, 1212–1221 CrossRef CAS PubMed.
- A. W. Perriman, H. Cölfen, R. W. Hughes, C. L. Barrie and S. Mann, Angew. Chem., Int. Ed., 2009, 48, 6242–6246 CrossRef CAS PubMed.
- A. P. S. Brogan, G. Siligardi, R. Hussain, A. W. Perriman and S. Mann, Chem. Sci., 2012, 3, 1839–1846 RSC.
- A. P. Brogan, K. P. Sharma, A. W. Perriman and S. Mann, Nat. Commun., 2014, 5, 1–8 RSC.
- B. Pérez, A. Coletta, J. N. Pedersen, S. V. Petersen, X. Periole, J. S. Pedersen, R. B. Sessions, Z. Guo, A. Perriman and B. Schiøtt, Sci. Rep., 2018, 8, 1–13 Search PubMed.
- Y. Zhou, N. C. Jones, J. Nedergaard Pedersen, B. Pérez, S. Vrønning Hoffmann, S. Vang Petersen, J. Skov Pedersen, A. Perriman, P. Kristensen, R. Gao and Z. Guo, ChemBioChem, 2019, 20, 1266–1272 CrossRef CAS PubMed.
- K. P. Sharma, K. Bradley, A. P. Brogan, S. Mann, A. W. Perriman and D. J. Fermin, J. Am. Chem. Soc., 2013, 135, 18311–18314 CrossRef CAS PubMed.
- P. M. Naveenkumar, S. Mann and K. P. Sharma, Adv. Mater. Interfaces, 2019, 6, 1–9 CAS.
- K. P. Sharma, A. M. Collins, A. W. Perriman and S. Mann, Adv. Mater., 2013, 25, 2005–2010 CrossRef CAS PubMed.
- K. P. Sharma, R. Harniman, T. Farrugia, W. H. Briscoe, A. W. Perriman and S. Mann, Adv. Mater., 2016, 28, 1597–1602 CrossRef CAS PubMed.
- T. Farrugia, A. W. Perriman, K. P. Sharma and S. Mann, Chem. Commun., 2017, 53, 2094–2097 RSC.
- A. Mukhopadhyay, T. Das, A. Datta and K. P. Sharma, Biomacromolecules, 2018, 19, 943–950 CrossRef CAS PubMed.
- Z. E. Sauna and S. V. Ambudkar, Proc. Natl. Acad. Sci., 2000, 97, 2515–2520 CrossRef CAS PubMed.
- J. Dong, Processes, 2021, 9, 271 CrossRef.
- S. Behera, S. Das and S. Balasubramanian, Phys. Chem. Chem. Phys., 2021, 23, 7302–7312 RSC.
- G. Schirò, Y. Fichou, A. P. S. Brogan, R. Sessions, W. Lohstroh, M. Zamponi, G. J. Schneider, F.-x. Gallat, A. Paciaroni, D. J. Tobias, A. Perriman and M. Weik, Phys. Rev. Lett., 2021, 126, 88102 CrossRef PubMed.
- A. P. Brogan, R. B. Sessions, A. W. Perriman and S. Mann, J. Am. Chem. Soc., 2014, 136, 16824–16831 CrossRef CAS PubMed.
- Lipase A is an enzyme whose catalytic rate is the highest when it is present at an oil-water interface. Generic substrates of Lipase A, being lipids, are poorly soluble in water and hence require surfactants as additives to increase their solubility. Alternately, the lipids are dissolved in the oil phase and the catalysis proceeds at the oil-water interface..
- G. Van Pouderoyen, T. Eggert, K. E. Jaeger and B. W. Dijkstra, J. Mol. Biol., 2001, 309, 215–226 CrossRef CAS PubMed.
- K. Kawasaki, H. Kondo, M. Suzuki, S. Ohgiya and S. Tsuda, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 1168–1174 CrossRef PubMed.
- W. L. DeLano, CCP4 Newsl. Protein Crystallogr., 2002, 40, 82–92 Search PubMed.
- G. Martínez-Rosell, T. Giorgino and G. De Fabritiis, J. Chem. Inf. Model., 2017, 57, 1511–1516 CrossRef PubMed.
- L. Martinez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- K. Lindorff-Larsen, S. Piana, K. Palmo, P. Maragakis, J. L. Klepeis, R. O. Dror and D. E. Shaw, Proteins: Struct., Funct., Bioinf., 2010, 78, 1950–1958 CrossRef CAS PubMed.
- M. J. Frischet al., Gaussian 09, Revision B.01, 2009 Search PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- C. I. Bayly, P. Cieplak, W. D. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269–10280 CrossRef CAS.
- J. Wang, W. Wang, P. A. Kollman and D. A. Case, J. Mol. Graph. Model., 2006, 25, 247–260 CrossRef CAS PubMed.
- A. W. Sousa da Silva and W. F. Vranken, BMC Res. Notes, 2012, 5, 1–8 CrossRef PubMed.
- D. J. Price and C. L. Brooks, J. Chem. Phys., 2004, 121, 10096–10103 CrossRef CAS PubMed.
- H. J. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- R. Fletcher, Comput. J., 1964, 7, 149–154 CrossRef.
- W. F. Van Gunsteren and H. J. Berendsen, Mol. Simul., 1988, 1, 173–185 CrossRef.
- B. Hess, H. Bekker, H. J. Berendsen and J. G. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- H. J. Berendsen, J. P. Postma, W. F. Van Gunsteren, A. Dinola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- N. Michaud-Agrawal, E. J. Denning, T. B. Woolf and O. Beckstein, J. Comput. Chem., 2011, 32, 2319–2327 CrossRef CAS PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- M. Wieczór, A. Hospital, G. Bayarri, J. Czub and M. Orozco, Bioinformatics, 2020, 36, 4660–4661 CrossRef PubMed.
- D. L. Mobley and K. A. Dill, Structure, 2009, 17, 489–498 CrossRef CAS PubMed.
- A. R. Fersht and M. F. Perutz, Proc. R. Soc. London, Ser. B, 1974, 187, 397–407 CAS.
- S. Das and S. Balasubramanian, J. Phys. Chem. B, 2018, 122, 4802–4812 CrossRef CAS PubMed.
- S. Das, S. Behera and S. Balasubramanian, J. Phys. Chem. Lett., 2020, 11, 2977–2982 CrossRef CAS PubMed.
- L. Casas-Godoy, S. Duquesne, F. Bordes, G. Sandoval and A. Marty, in Lipases: An Overview, ed. G. Sandoval, Humana Press, Totowa, NJ, 2012, pp. 3–30 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI text and figures in a PDF. A movie (SM1.mp4) displaying a trajectory (1 μs) of the SFPL system focusing on the mobilities of substrates. Another movie (SM2.mp4) showing binding of the substrate to the catalytic region of LipA in the aqueous solution. See https://doi.org/10.1039/d2ra00666a |
| This journal is © The Royal Society of Chemistry 2022 |