Open Access Article
Open Access ArticleBeneficial roles of the AhR ligand FICZ on the regenerative potentials of BMSCs and primed cartilage templates†
Jing Huang a,
Yining Wang
a,
Yining Wang ab and
Yi Zhou
ab and
Yi Zhou *ab
*ab
aThe State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST), Key Laboratory of Oral Biomedicine Ministry of Education, School and Hospital of Stomatology, Wuhan University, 237 Luoyu Road, Wuhan 430079, China. E-mail: dryizhou@whu.edu.cn; Fax: +86 27 87873260; Tel: +86 27 87686318
bDepartment of Prosthodontics, Hospital of Stomatology, Wuhan University, Wuhan 430079, China
First published on 13th April 2022
Abstract
Bone marrow-derived mesenchymal stem cells (BMSCs) are commonly used seed cells, and BMSC-derived primed cartilage templates have been shown to achieve bone regeneration in bone tissue engineering. Aryl hydrocarbon receptor (AhR) is a ligand-activated transcription factor involved in various cellular processes such as osteogenesis and immune regulation. This study investigated the effects of the AhR endogenous ligand 6-formyl (3,2-b) carbazole (FICZ) on the behavior of BMSCs and cartilage templates as well as the possible underlying molecular mechanisms. AhR expressions in rat bone marrow and isolated BMSCs were detected via immunohistochemistry (IHC) and immunofluorescent staining. Alkaline phosphatase staining and alizarin red staining showed that FICZ treatment enhanced the osteogenic potential of BMSCs without influencing their proliferation. FICZ was shown to alleviate the LPS-induced inflammatory cytokines IL-1β, 6 and TNF-α via the quantitative polymerase chain reaction (qPCR). In the chondrogenic process from BMSCs to primed cartilage templates, the expressions of AhR and its target gene cytochrome P450 subfamily B member 1 (CYP1B1) were inhibited. However, IHC staining demonstrated that AhR was still involved in the subcutaneous ossification of cartilage templates. Then, the effects of FICZ on cartilage templates were investigated. The osteogenic markers were upregulated by FICZ administration. The RAW 264.7 treated by condition medium of FICZ-treated cartilage templates exhibited an anti-inflammatory phenotype. Finally, high-throughput sequencing was applied to analyze the differentially expressed genes (DEGs) in the FICZ-treated cartilage templates. The upregulation of cytochrome P450 subfamily A member 1 (CYP1A1) and sphingomyelin phosphodiesterase 3 (Smpd3) were verified by qPCR, which might be the downstream targets of AhR in the cartilage templates promoting osteogenesis and macrophage polarization. These data implied a beneficial role of FICZ in the regenerative potentials of both BMSCs and primed cartilage templates. The FICZ/AhR axis might be a practical target to achieve optimal bone regeneration.
Introduction
Various bone defects are diseases with a high incidence, seriously affecting the health and quality of life of patients and causing a heavy economic burden on society.1 Cell-based bone tissue engineering has become an important method to treat bone defects and the commonly used seed cells are mainly various tissue-derived stem cells, especially bone marrow-derived mesenchymal stem cells (BMSCs).2 Their abilities of proliferation, migration and osteogenic differentiation are the premise of their application in bone regeneration.3 However, as foreign implants, seed cells and materials will contact immune cells such as macrophages before playing their roles in bone regeneration. The prognosis of immune responses such as M1 (pro-inflammatory macrophages)/M2 (anti-inflammatory macrophages) polarization has a significant influence on their osteogenic potency.4 Therefore, the immunoregulatory potentials of BMSCs are an important parameter for bone regeneration.5Despite the great progress made in recent years, BMSC-based bone tissue engineering still faces many challenges due to its uncertain efficiency. The primary reason is that the implanted BMSCs are often exposed to a detrimental pathological microenvironment. Hormonal changes, inflammatory cytokines and metabolic disorders impair the functions of implanted cells, which results in decreased proliferation, damaged differentiation potential and inhibited immunomodulatory properties.6 Accordingly, pre-treatment of seed cells before implantation to make them function better is a hot research frontier, such as the pre-induction of BMSCs into specific cell types to enhance lineage differentiation and the administration of chemical reagents to regulate the expression of target genes or pathways.
Our group has been focused on the pre-chondrogenic induction of BMSCs to repair bone defects, which simulated endochondral bone development.7 BMSCs are induced into hypertrophic chondrocytes into “primed cartilage templates” before being implanted in vivo. Plenty of studies demonstrated that this “developmental engineering” can effectively achieve bone regeneration including mandible defects.8–11 Compared to direction implantation of BMSCs, the implanted primed cartilage templates obtained more new bone areas and higher alveolar bone height in the repair of periodontal defects.8 Moreover, primed cartilage templates were shown to enhance the M2/M1 ratio after the implantation subcutaneously.9
Recently, various kinds of natural and synthetic small molecules targeting specific cellular responses via signaling cascades were analyzed in stems cell biology and bone tissue engineering.12,13 Aryl hydrocarbon receptor (AhR) is a basic helix–loop–helix (bHLH)–PAS (Per–Arnt–Sim domain) protein, which belongs to a member of the bHLH transcription factor superfamily. AhR is a ligand-activated transcription factor that mediates numerous cellular responses.14 Once activated, AhR is translocated to the nucleus and dimerizes with AhR nuclear translocator (ARNT). Then, the heterodimer can recognize a consensus XRE binding site (xenobiotic responsive element, 5′-GCGTC-3′)15 and regulate downstream genes, such as xenobiotic-metabolizing enzymes, including cytochrome P450 subfamily A member 1 (CYP1A1), CYP1B1 and so on, which were considered as the target or responsive genes.16 The activation of AhR-dependent transcription could be inhibited by aryl hydrocarbon receptor repressor (AhRR), which competed with AhR for its interaction with ARNT, limiting the availability of ARNT for AhR-ligand binding.17,18
The latest studies have revealed that AhR is playing an essential role in the biological processes of osteogenesis and immune regulation. In the tibial fracture model, the mineralized callus tissue in the fracture space was visible in wide type (WT) mice after 2 weeks but not in the AhR knockout mice. In the 3rd week, micro-CT analysis also showed that the bone formation of AhR knockout mice was less than that in the WT mice.19 In another study, BMSCs harvested from WT and AhR knockout mice were conducted for osteogenic differentiation. One week later, the mRNA expressions of alkaline phosphatase (ALP) and osteocalcin (OCN) were lower in the AhR knockout group.20 In terms of immune regulation, AhR plays an important role in the phenotypic differentiation of immune cells. Under the stimulation of lipopolysaccharide (LPS), AhR – depleted macrophages produced more pro-inflammatory cytokines. Under the administration of interleukin (IL) – 4, AhR – depleted macrophages showed higher arginase activity, implying AhR would impact the balance of M1/M2 polarization.21 In another study, T cells were co-cultured with AhR ligand kynurenine – treated MSCs or non-treated MSCs. The results showed that the secretion of IL-6 and 17 from T cells co-cultured with kynurenine – treated MSCs were lower, inhibiting the activation of the pro-inflammatory helper T cell (Th) 1 and 17.22 In the latest study, MSCs promoted the polarization of macrophages towards M2 in the co-culture system, during which AhR expression was increased. The treatment of MSCs via AhR antagonist suppressed their capacity to modulate the macrophage phenotypes, resulting in a significant decrease of M2 markers.23
As to small molecules acting as AhR ligands, the firstly identified ones are 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD, dioxin) and dioxin-like compounds (DLCs), which mediate toxic effects after binding to AhR. Subsequently, various kinds of non-DL endogenous ligands have been identified, such as 6-formyl (3,2-b) carbazole (FICZ), 2-(1′-H-indole-3-carbonyl) thiazole-4-carboxylic acid methyl ester (ITE), tryptophan metabolites such as kynurenine, which demonstrated functional diversity.24 In a previous study from our group, the AhR ligand FICZ was observed to enhance the mineralization of periodontal ligament cells (PDLCs) by activating the Wnt/β-catenin signal pathway and inhibiting LPS-induced inflammatory cytokines by regulating the phosphorylation of the signal transducer and the activator of transcription (STAT) 3, while AhR antagonist StemRegenin 1 (SR1) showed an opposite result.25
However, the expression alteration of AhR from undifferentiated BMSCs to primed cartilage templates and the effects of FICZ on regenerative potentials of cartilage templates are lacking investigations. Concerning the essential role of AhR and its chemical ligands such as FICZ in osteogenesis and immune response, it would be of great interest to (i) detect the expression of AhR in BMSCs and the effects of FICZ on BMSCs, (ii) the alteration of AhR from BMSCs to cartilage templates, and (iii) further observe the effects of FICZ in cartilage templates and molecular mechanisms involved.
Experimental
Rat
Four-week-old male Sprague–Dawley (SD) rats were obtained from the Hubei Research Centre of Laboratory Animals (Wuhan, China) and kept under specific pathogen-free (SPF) conditions. All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animal of Wuhan University and approved by the Animal Ethics Committee of the Hospital of Stomatology, Wuhan University.Isolation and primed chondrogenic induction of BMSCs
Rat BMSCs were isolated from 4-week-old male SD rats through the whole femur bone marrow adherent culturing. The primed cartilage templates were prepared using a protocol described before 9. The cells were resuspended in a maintenance medium (DMEM containing 10% fetal bovine serum, 100 U ml−1 penicillin G and 100 μg ml−1 streptomycin, HyClone). In brief, BMSCs were seeded into gelatin sponges (MS0002, Ethicon) and then differentiated into cartilage templates in chondrogenic medium, which contained high-glucose DMEM, 1% FBS, 100 U ml−1 penicillin, 100 μg ml−1 streptomycin (HyClone), 50 μg ml−1 ascorbic acid, 100 nM dexamethasone, 100 mM sodium pyruvate (Sigma), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 insulin-transferrin-selenium (ITS) (BD bioscience), 10 ng ml−1 transforming growth factor beta-2 (TGF-β2) and 100 ng ml−1 bone morphogenetic protein 2 (BMP-2) (R & D systems).
100 insulin-transferrin-selenium (ITS) (BD bioscience), 10 ng ml−1 transforming growth factor beta-2 (TGF-β2) and 100 ng ml−1 bone morphogenetic protein 2 (BMP-2) (R & D systems).
Subcutaneous implantation
A small incision was made in the back of the rats and a subcutaneous pocket was created lateral to each incision. One cartilage template was placed in each pocket. Then, the skin was sutured with 4–0 absorbable ligature. The implants were harvested after 4 weeks.Histological detection
The harvested rat femurs or implants were fixed with 4% paraformaldehyde and then decalcified in 10% ethylene diamine tetraacetic acid (EDTA) for six weeks. The tissues were subsequently processed for paraffin embedding and serial sections (4 μm) were prepared. Then, the sections were dewaxed in xylene and rehydrated through a graded ethanol series to water.For histological and morphometric analysis, the specimens were stained with hematoxylin and eosin (HE). For the immunohistochemistry (IHC) staining, antigen retrieval was conducted in the stomach enzyme antigen repair solution for 30 min at 37 °C. Immunostaining was performed by incubating the sections with anti-AhR (AF6278, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, Affinity) at 4 °C overnight. The slides were then washed with phosphate-buffered saline (PBS) and incubated with secondary antibody (anti-rabbit system, Maxim Biotechnology) for 30 min at 37 °C. Staining was visualized with 3,3-diaminobenzidine and counterstained with hematoxylin.
200, Affinity) at 4 °C overnight. The slides were then washed with phosphate-buffered saline (PBS) and incubated with secondary antibody (anti-rabbit system, Maxim Biotechnology) for 30 min at 37 °C. Staining was visualized with 3,3-diaminobenzidine and counterstained with hematoxylin.
Immunofluorescent staining
BMSCs at the third passage were seeded in a cell dish (801002, NEST). After the cells reached 80% confluence, BMSCs were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.1% Triton X-100 for 20 min. Then, the cells were blocked with bovine serum albumin for 1 h. Subsequently, the cells were incubated with a primary antibody against AhR (AF6278, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, Affinity) at 4 °C overnight. After washing, cells were incubated with daylight 594-conjugated secondary antibody (1
100, Affinity) at 4 °C overnight. After washing, cells were incubated with daylight 594-conjugated secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, A23420, Abbkine) for 1 h at room temperature. Then, the cells were stained with DAPI staining solution (C1005, Beyotime) for 5 min. Finally, the stained cells were observed and photographed under a confocal microscope (Leica-LCS-SP8-STED).
200, A23420, Abbkine) for 1 h at room temperature. Then, the cells were stained with DAPI staining solution (C1005, Beyotime) for 5 min. Finally, the stained cells were observed and photographed under a confocal microscope (Leica-LCS-SP8-STED).
Cell assay
The cell proliferation was analysed using a cell counting kit 8 (CCK8) (Dojindo, Japan) according to the manufacturer's instructions. BMSCs were seeded in 96-well plate (1 × 103 cells per well) and incubated in a culture medium containing 500 nM FICZ (CAS No. 172922-91-7) (HY-12451, MCE), 1 μM SR1 (CAS No. 1227633-49-9) (HY-15001, MCE) or dimethyl sulphoxide (DMSO) as a control. The chemical structures of FICZ and SR1 are shown in Fig. S1.† At consecutive times (day 0, 1, 3, 5), 100 μl of the culture medium containing 10 μl CCK8 was added to each well and the absorbance at 450 nm wavelength was measured using a microplate reader after 1 h incubation.To detect the inflammatory responses of BMSCs, the cells were rendered quiescent by serum starvation and then stimulated with 1 μg ml−1 lipopolysaccharide (LPS) (L4391, Sigma-Aldrich) supplemented with 500 nM FICZ, 1 μM SR1 or dimethyl sulphoxide (DMSO) for 6 h.
Osteogenic induction, alkaline phosphatase (ALP) staining and alizarin red staining (ARS)
For osteogenic induction, BMSCs at passage 3 or primed cartilage templates were cultured in a medium supplemented with 50 μg ml−1 ascorbic acid, 10 mM β-glycerophosphate and 10–8 mol L−1 dexamethasone (Sigma) in addition to 500 nM FICZ, 1 μM SR1 or dimethyl sulphoxide (DMSO) as the control. For BMSCs, cells were fixed in 4% paraformaldehyde for 10 min and stained using an alkaline phosphatase color development kit (C3206, Beyotime) following the manufacturer's instructions. For ARS, cells were fixed and stained with alizarin red solution (Cyagen) for 10 min. Different groups of cartilage templates after osteogenic induction were harvested for subsequent RNA extraction.Treatment of primed cartilage templates and preparation of conditioned medium
The primed cartilage templates were cultured in a normal medium or incubated with 500 nM FICZ for 24 h. The templates were then washed with PBS three times to remove FICZ and cultured in a normal medium for an additional 24 h. The conditioned medium was harvested and centrifuged for 10 min at 1000 rpm and then frozen at −20 °C until used. The macrophage linage RAW 264.7 (ScienceCell) was treated with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 normal medium and conditioned medium for 12 h.
1 normal medium and conditioned medium for 12 h.
Library construction and sequencing
Primed cartilage templates were treated with 500 nM FICZ for 48 h, after which total RNA was extracted by TRIzol reagent (Takara Bio). After total RNA was extracted, messenger RNA (mRNA) was enriched by Oligo(dT) beads (Epicentre, Madison). Then, the enriched mRNA was fragmented into short fragments using the fragmentation buffer and reverse transcription in cDNA with random primers. Then, the cDNA fragments were purified using a QiaQuick PCR extraction kit (Qiagen, Venlo); end-repaired, poly(A) was added and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR amplified and sequenced using Illumina HiSeq2500 by Gene Denovo Biotechnology Co. (Guangzhou, China).Analysis of differentially expressed genes (DEGs)
The transcripts with the parameter of false discovery rate (FDR) below 0.05 and absolute fold change (FC) ≥ 1.5 were considered differentially expressed.Quantitative polymerase chain reaction (qPCR)
Total RNA from different cell samples were isolated using the TRIzol reagent (Takara Bio). The concentration of RNA was determined using the NanoDrop 2000 Spectrophotometer. One microgram of total RNA was reverse transcribed into cDNA using HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (R223, Vazyme). The SYBR Green Reagent (Q311-02, Vazyme) was used to perform qPCR in a BIO-RAD CFX Real-Time PCR system. The primer sequences used in the study are shown in Table 1. The relative gene expression levels were calculated using the 2−ΔΔCt method and normalized with respect to GAPDH. Experiments were duplicated three times.| Gene | Primer sequence (5′–3′) |
|---|---|
| a R, rat; m, mouse. | |
| R-IL-1β | F: GCATCCAGCTTCAAATCTCA |
| R: ACGGGCAAGACATAGGTAGC | |
| R-IL-6 | F: GTGGCTAAGGACCAAGACCA |
| R: AGCACACTAGGTTTGCCGAG | |
| R-TNF-α | F: GAGATGTGGAACTGGCAGAG |
| R: CACGAGCAGGAATGAGAAGA | |
| R-AhR | F: ATGTCCATGTACCAGTGCCAG |
| R: AGCCCTTACCTTGCTTAGGA | |
| R-AhRR | F: CCTCCTCGGCTCTCCTTGTTTTG |
| R: CTTTTGCCCTTGAGTCCATCGTGA | |
| R-CYP1B1 | F: CCATACGTCATGGCTTTTCTTT |
| R: TGCTGGCTAGTGCCTTGTTA | |
| R-ALP | F: GTCCCACAAGAGCCCACAAT |
| R: CAACGGCAGAGCCAGGAAT | |
| R-Col1α1 | F: CTGCCCAGAAGAATATGTATCACC |
| R: GAAGCAAAGTTTCCTCCAAGACC | |
| R-OPN | F: CCAAGCGTGGAAACACACAGCC |
| R: GGCTTTGGAACTCGCCTGACTG | |
| R-Runx2 | F: CCGAGACCAACCGAGTCATT |
| R: CACTGCACTGAAGAGGCTGT | |
| R-OCN | F: CAGTAAGGTGGTGAATAGACTCCG |
| R: GGTGCCATAGATGCGCTTG | |
| R-CYP1A1 | F: CTGGTTCTGGATACCCAGCTG |
| R: CCTAGGGTTGGTTACCAGG | |
| R-Gpc3 | F: TGTGCTGGAACGGACAAGAG |
| R: TGGGCACAGACATGGTTCTC | |
| R-Smpd3 | F: GGTCCTCATGGAATGAAAGG |
| R: ACAGTGCCATTGGTTTAGGG | |
| R-GAPDH | F: GGCAAGTTCAACGGCACAGT |
| R: GCCAGTAGACTCCACGACAT | |
| m-CD86 | F: TCTGCCGTGCCCATTTACAA |
| R: TGTGCCCAAATAGTGCTCGT | |
| m-TNF-α | F: TCTTCTCATTCCTGCTTGTGG |
| R: GGTCTGGGGCATAGAACTGA | |
| m-IL-1β | F: AACCTGCTGGTGTGTGACGTTC |
| R: CAGCACGAGGCTTTTTTGTTGT | |
| m-CD36 | F: AAGCTATTGCGACATGATT |
| R: GATCCGAACACAGCGTAGAT | |
| m-Arg-1 | F: ACATTGGCTTGCGAGACGTA |
| R: ATCACCTTGCCAATCCCCAG | |
| m-IL-10 | F: ATGCTGCCTGCTCTTACTGACTG |
| R: CCCAAGTAACCCTTAAAGTCCTGC | |
| m-GAPDH | F: TGGAAAGCTGTGGCGTGAT |
| R: GTCATCATACTTGGCAGGTTTCT | |
Statistical analysis
All data were expressed as the mean ± SD. For comparison between the two groups, statistical differences were evaluated using a two-tailed Student's t-test. For multiple comparisons, a one-way analysis of variance (ANOVA) followed by Tukey's test was conducted.Results
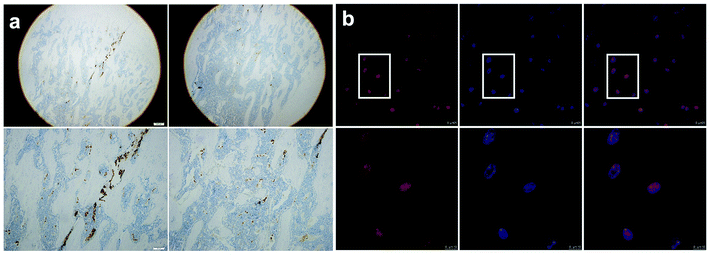
1 The expression of AhR in rat femur and isolated BMSCs
The IHC staining demonstrated that AhR was expressed in parts of rat femur bone marrow (Fig. 1a). In isolated BMSCs, AhR was observed positive via confocal microscopy (Fig. 1b), which was consistent with the observation of IHC detection. The immunofluorescence staining showed that AhR was mostly expressed in the cell nucleus.2 The effects of FICZ and SR1 on the proliferation, osteogenic differentiation, and inflammatory responses of BMSCs
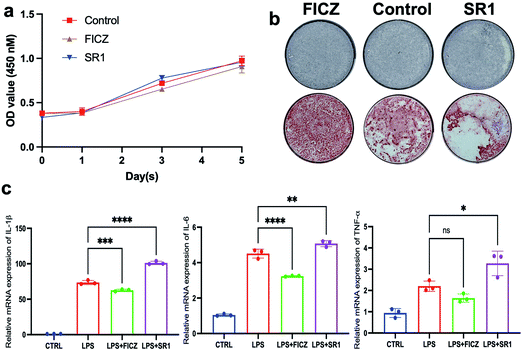
To explore the effect of FICZ and SR1 on the proliferation of BMSCs, the CCK8 assay was used to detect the viability of BMSCs after treatment of 500 nM FICZ, 1 μM SR1 or DMSO for consecutive days. As illustrated in Fig. 2a, the number of viable BMSCs increased during the culture. But there was no significant difference in cell proliferation among the groups at any time point, which demonstrated neither FICZ nor SR1 had an impact on the proliferation of BMSCs. ALP is a membrane-bound enzyme and plays a key role in the early osteogenic differentiation. After osteogenic induction for 7 days, ALP staining demonstrated that FICZ-treated BMSCs showed more ALP-positive nodules than control, while SR1-treated BMSCs showed much less (Fig. 2b upper). Consistently, on day 15 of osteogenic induction, calcium deposits and mineralized nodules visualized by ARS staining also claimed FICZ administration promoted the osteogenic differentiation of BMSCs and SR1 inhibited it (Fig. 2b, lower). To test the influence of FICZ or SR1 on the LPS-induced inflammatory responses in BMSCs, they were stimulated with 1 μg ml−1 LPS supplemented with 500 nM FICZ, 1 μM SR1 or DMSO for 6 h. RT-PCR was used to detect the mRNA expressions of IL-1β, 6 and TNF-α. The results demonstrated that LPS significantly up-regulated the expressions of IL-1β, 6 and TNF-α and FICZ or SR1 prevented or exacerbated the inflammatory responses (Fig. 2c).In conclusion, without influencing the proliferation of BMSCs, the AhR endogenous ligand FICZ promoted the osteogenic differentiation of BMSCs and alleviated LPS-induced inflammatory responses in BMSCs. In contrast, the AhR antagonist SR1 exhibited adverse effects on BMSCs. The results implied that the AhR activation via FICZ was favorable for the regenerative potentials of BMSCs.
3 The expression pattern of AhR from BMSCs to primed cartilage templates to ossification
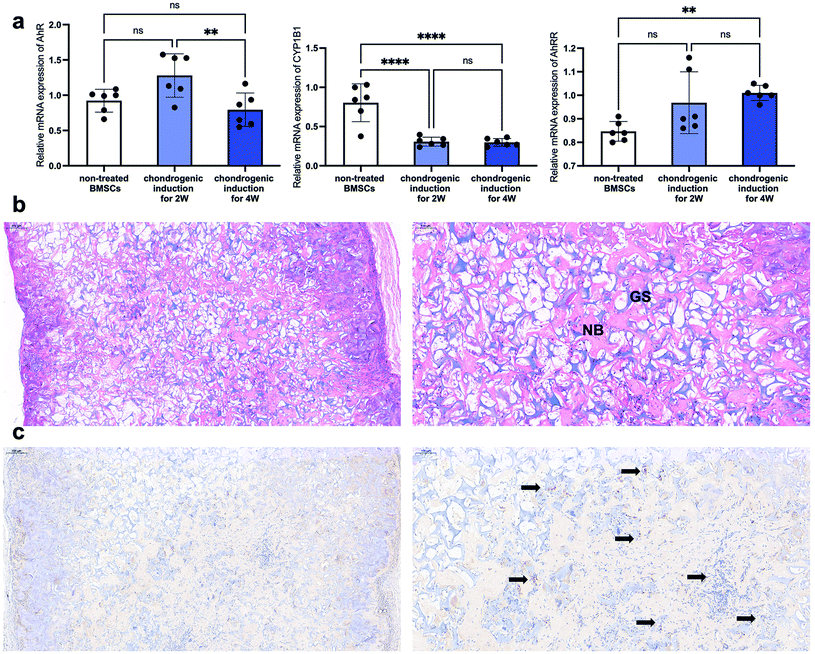
As described in the introduction, a lot of published literature has reported the success of bone regeneration via primed cartilage templates derived from BMSCs. However, the research into the relationship between AhR and cartilage templates is limited. In the present study, the AhR and AhRR mRNA expression pattern of undifferentiated BMSCs and primed cartilage templates were detected via RT-PCR. The results showed that the expression was slightly elevated after chondrogenic induction for 2 weeks, but the elevation was not statistically significant. After chondrogenic induction for 2 more weeks, the AhR mRNA expression was downregulated compared to 2nd week but almost the same when compared to the undifferentiated BMSCs (Fig. 3a left). As to the AhR target gene CYP1B1, a significant decrease from BMSCs to primed cartilage templates was observed in the 2nd or 4th week (Fig. 3a middle). AhRR was detected to be slightly increased in the 2nd week and significantly increased in the 4th week (Fig. 3a right). On a whole, AhR signalling was downregulated from BMSCs to primed cartilage templates, which might be mediated by AhRR.To detect the involvement of AhR in the subsequent ossification of the cartilage templates, they were subcutaneously implanted and harvested for histological detection after four weeks. The images of HE staining showed an extensive distribution of newly formed woven bone-like structure (stained red), which was scattered amongst the remnants of gelatin sponges (stained purple) (Fig. 3b). In the view of IHC staining, plenty of embedded or infiltrated cells were stained positively for AhR in the harvested implants (black arrows in Fig. 3c). The results demonstrated that although AhR was suppressed in cartilage templates, it was still involved in the ossification process via cartilage templates. Therefore, it would be interesting to explore the effects of AhR ligand FICZ on the regenerative potentials of primed cartilage templates, mainly osteogenic differentiation, and immunomodulatory capacity.
4 The effects of FICZ on primed cartilage templates
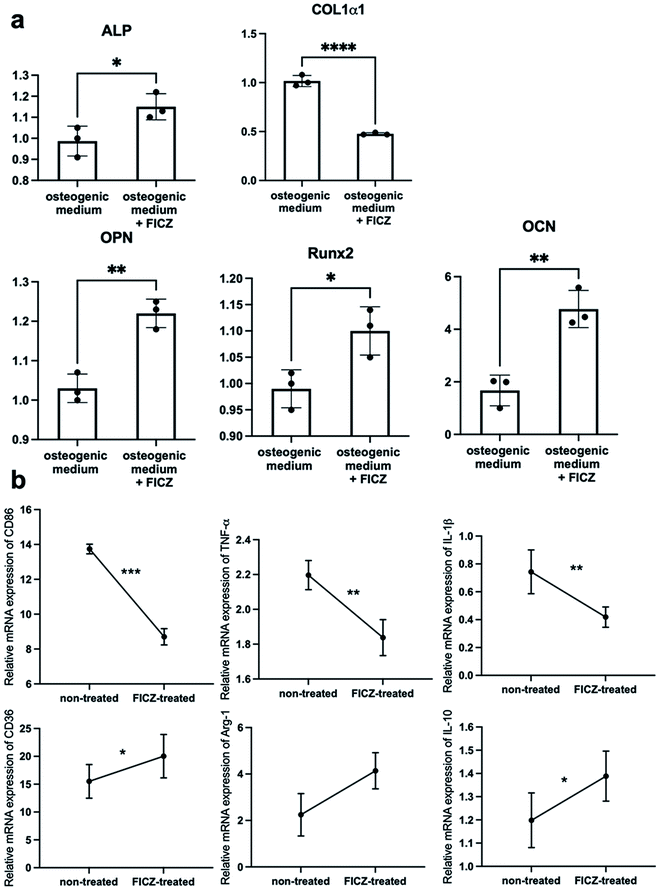
As mentioned above, AhR was suppressed in the process of the cartilage template formation, but it was still involved in the ossification process via cartilage templates. It made us curious about the effects of AhR agonist FICZ on the cartilage templates' osteogenic potential and macrophage polarization capacity.The primed cartilage templates were cultured in an osteogenic medium supplemented with 500 nM FICZ or not. On the 7th day, the cartilage templates were mixed by homogenizer in the TRIzol reagent. Then, the osteogenic markers were analysed via RT-PCR. The data showed that mRNA expressions of ALP, osteopontin (OPN), osteocalcin (OCN), runt-related transcription factor 2 (Runx2) were upregulated in the FICZ supplemented group, apart from collagen type I alpha 1 (Col1α1) (Fig. 4a). On a whole, it could be concluded that FICZ administration promoted the osteogenic potential of the primed cartilage templates.
To investigate the macrophage modulating capacity of FICZ-treated cartilage templates. RAW 264.7 cells were treated in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 normal medium and conditioned medium from FICZ-treated or non-treated templates for 12 h. RT-PCR was applied to test the characteristic markers of M1 and M2. The data showed an upregulation of M2 markers (CD36, Arg-1, IL-10) and downregulation of M1 markers (CD86, TNF-α, IL-1β) in RAW cultured with conditioned medium from FICZ-treated cartilage templates (Fig. 4b). The results demonstrated primed cartilage templates with the FICZ treatment were more prone to polarize macrophages into an M2-like phenotype. In conclusion, similar to BMSCs, FICZ administration was beneficial to the regenerative potential of primed cartilage templates.
1 normal medium and conditioned medium from FICZ-treated or non-treated templates for 12 h. RT-PCR was applied to test the characteristic markers of M1 and M2. The data showed an upregulation of M2 markers (CD36, Arg-1, IL-10) and downregulation of M1 markers (CD86, TNF-α, IL-1β) in RAW cultured with conditioned medium from FICZ-treated cartilage templates (Fig. 4b). The results demonstrated primed cartilage templates with the FICZ treatment were more prone to polarize macrophages into an M2-like phenotype. In conclusion, similar to BMSCs, FICZ administration was beneficial to the regenerative potential of primed cartilage templates.
5 RNA-seq analysis of FICZ on cartilage templates
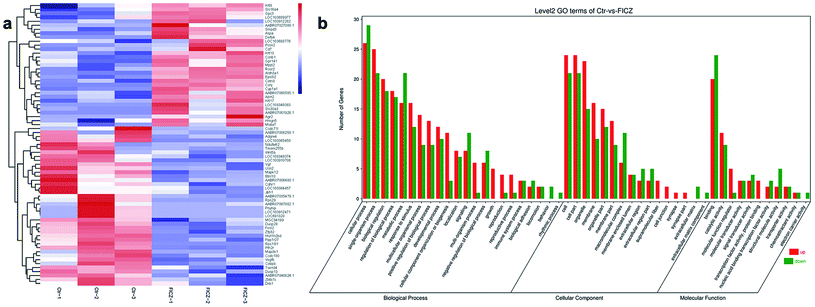
To explore the potential molecular mechanisms of FICZ on cartilage templates, the templates were treated with 500 nM FICZ for 48 h, then, RNA-seq was conducted between FICZ and control groups. Firstly, DEGs were analyzed at the standard of FDR < 0.05 and FC ≥ 1.5. The DEGs were visualized in a heatmap with gene symbols (Fig. 5a). The data showed that 31 genes were upregulated and 40 were downregulated in the FICZ-treated cartilage templates. Gene ontology (GO) is a standardized gene functional classification system that offers a dynamic-updated controlled vocabulary and a strictly defined concept to comprehensively describe the properties of genes and their products.26 The GO enrichment analysis was performed to identify the functions significantly associated with the DEGs in FICZ-treated cartilage templates. The most enriched GO terms in the biological process, cellular component and molecular function domains are shown in Fig. 5b.6 Validation of CYP1A1 and Smpd3 mRNA expressions via qPCR
To further explore the data obtained from the RNA-seq, we analysed the potential function of DEGs by searching published literature in PubMed, trying to find possible genes associated with osteogenesis and immune responses. Among them, we paid particular attention to the genes whose promoter region had binding sequences with AhR/ARNT motif predicted by JASPAR (Fig. S2 and Table S1†).Among the DEGs, sphingomyelin phosphodiesterase 3 (Smpd3) and glypican 3 (Gpc3) could promote osteogenic differentiation and extracellular matrix mineralization of precursor cells.27,28 As to immune responses, Gpc3 was able to recruit more M2 cells29 and CYP1A1 could regulate the resolution of inflammation via lipid molecules.30
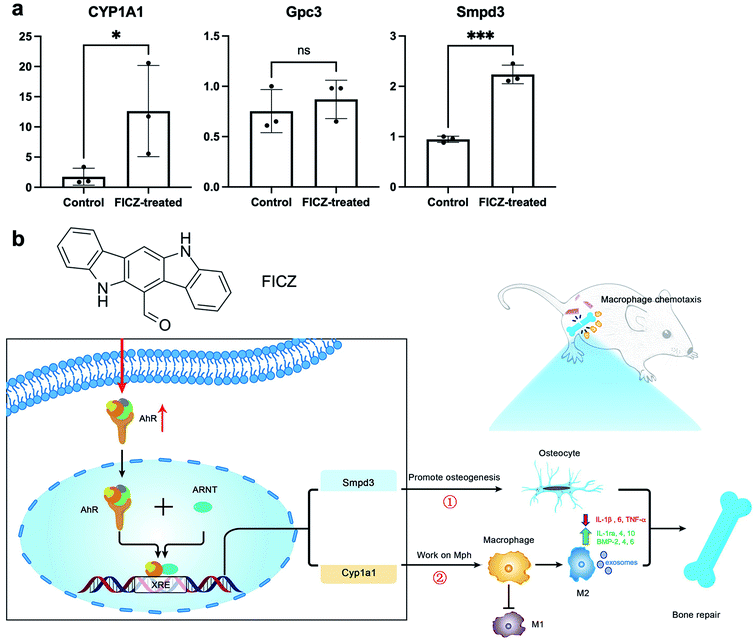
Therefore, qPCR was applied to verify the expressions of CYP1A1, Gpc3 and Smpd3 in the FICZ-treated cartilage templates. The results showed that CYP1A1 and Smpd3 were significantly increased, which was consistent with RNA-seq data, but the upregulation of Gpc3 was not significant (Fig. 6a). Accordingly, it was reasonable to make a scientific hypothesis that AhR agonist FICZ promoted osteogenic potential through Smpd3 and enhanced M2 polarization via CYP1A1 in the primed cartilage templates. M2 could improve the inflammatory microenvironment by downregulating inflammatory cytokines such as IL-1β, 6 and TNF-α.31 Meanwhile, regenerative cytokines such as IL-1ra, 4, 10 and osteogenic factors such as bone morphogenetic protein (BMP)-2, 4, 6 secreted from M2 would enhance the osteogenic differentiation.32,33 Additionally, recent research revealed exosome containing microRNA was positive for the osteogenic process34 (Fig. 6b). However, it would need further experiments such as luciferase reporter assay or chromatin immunoprecipitation (ChIP) to verify the connection between AhR and downstream genes.
Conclusion and discussion
Based on our findings, and within the limitations of the study, it was implied that (i) AhR agonist FICZ administration was beneficial to the regenerative features of BMSCs, (ii) although the AhR signal was suppressed in the process from BMSCs to primed cartilage templates, AhR was still involved in bone generation via templates, and (iii) FICZ played positive roles in the regenerative potentials of cartilage templates and the molecular mechanisms might be downstream CYP1A1 and Smpd3. The present study offered new insights into the roles of AhR and its ligand FICZ in the primed cartilage templates, which might be a target and small molecule for achieving optimal bone regeneration.“Developmental bone tissue engineering” was an endochondral ossification approach, in which a cartilage intermediate was created in vitro before being transformed in vivo for bone regeneration.35 The approach had various advantages such as vascularized bone formation, hypoxia tolerance and so on.36 Primed cartilage templates kept the features of osteogenic differentiation and immunomodulating like BMSCs. In terms of osteogenic differentiation, genetic markers were used to track hypertrophic chondrocytes. The chondrocytes were found to directly differentiate into Col1α1+ osteoblasts and sclerostin (SOST)+ osteocytes.37 In another study, primed cartilage templates derived from human BMSCs were implanted into immunodeficient mice. In the newly formed bone tissue, human Alu repeats could be observed even after weeks, which proved that new bone was partly differentiated from implants.36 The above studies have confirmed the important role of the direct osteogenic differentiation of the primed cartilage templates in the process of bone regeneration. In terms of immune regulation, it has been confirmed that cartilage templates could inhibit the proliferation and activation of T cells and dendritic cells, suppressing the secretion of inflammatory factors.38 However, according to our experience, the process of preparing primed cartilage templates was time-consuming, which might be the restriction of its clinical applications. In the present study, the templates were prepared via chondrogenic induction for 4 weeks. It had been shown that 2 weeks might be enough to form a cartilage template, but a longer priming time of up to 4 weeks would lead to a more homogeneous bone formation.39
AhR was observed in both osteoblasts and osteoclasts in bone and other hard tissues, and the osteogenic process was mediated by the AhR signal through cytokines.40 In this research, AhR was also positively detected in bone marrow tissue via IHC staining and isolated BMSCs via immunofluorescent staining, which laid a foundation for further experiments.
AhR was first recognized as an environmental chemical sensor and played a role in biological rhythms and oxidative stress.14 Nowadays, mounting evidence has proved the presence of AhR in various biological processes. But the functional roles of AhR might be diverse depending on dioxin-like compounds (DLCs) such as TCDD, and non-DL endogenous ligands such as FICZ.24
As to proliferation, AhR might exhibit adverse effects due to different AhR ligands and different cell types.41 Even FICZ in particular, its effect on cell proliferation was the biphasic effect depending on the concentration. In HepG2 cells, 0.01 nM FICZ treatment for 24 h significantly enhanced the cell growth while 1000 nM FICZ obviously inhibited it via the tetrazolium salt (MTT) assay.42 Similarly, in MG-63 cells, the proliferation was promoted by low-concentration (1, 10, 100 nM) FICZ while inhibited by high-concentration (1000 nM) FICZ application for 24 h.43 In the present study, an intermedia concentration (500 nM) was chosen, and the treatment period was consecutive 1, 3, 5 days. The CCK8 assay demonstrated that FICZ or SR1 had no influence on the proliferation of BMSCs. But the results verified that the effects of FICZ on osteogenic differentiation or inflammatory responses of BMSCs were not induced by the changes in the cell number.
The dioxin-like compound ligands of AhR were basically harmful to the osteogenic process, but the effect of endogenous ligands such as FICZ was controversial. In a 10 nM dioxin-containing osteogenic medium, the ALP activity and calcium depositions were inhibited compared to DMSO vehicle control. The qPCR and western blot demonstrated that dioxin downregulated a wide array of osteogenic marker genes, such as ALP, Col1α1 and so on.44 Similarly, exposure to 10 nM TCDD inhibited mineralization of hBMSCs and the expressions of osteogenic markers.45 In a most recent study, two other AhR activators benzo[α]pyrene (B[α]P) and FICZ were analyzed in experimental temporomandibular joint osteoarthritis (TMJ-OA).46 The results showed that B[α]P-induced mandibular subchondral bone resorption was in an AhR-dependent way. However, FICZ played a therapeutic function and rescued the bone loss, no matter at low (100 μg kg−1) or high (100 mg kg−1) concentrations. Meanwhile, in vitro, the treatment with FICZ (200 ng ml−1) enhanced the osteoblastogenesis on MC3T3 E1 cells, manifesting in more obvious ALP and ARS staining, and increased ALP, OCN and Col1α1 mRNA expression. Our results were mostly consistent with those of previous research. In BMSCs, FICZ treatment promoted the ALP and ARS staining after osteogenic induction. In primed cartilage templates, the FICZ supplement enhanced the mRNA expressions of several osteogenic markers including ALP, OPN, Runx2 and OCN, but with the exception of Col1α1. FICZ was found to downregulate the collagen synthesis including Col1α1 in UV-induced photo-aging of skin,47,48 Crohn's disease fibroblasts,49 systemic sclerosis.50 Therefore, AhR signalling was considered a potential anti-fibrotic target. The cellular mechanism might be that FICZ-led AhR activation inactivated p38, ERK1/2, p65 and p-Smad2/3 signalling.49 However, the promotive effect on osteogenic differentiation of AhR activation might be attributed to the impairment of self-renewal.51,52 To sum up, we believe that the role of AhR and its ligands in osteogenesis and Col1α1 expression should be specifically studied in different diseases or models.
The role of AhR in chondrogenic phenotype has been relatively less investigated. In smooth muscle cells of (SMC)-specific AhR knockout (KO) mice, an increased chondrogenic phenotype was detected in SMC, which implied AhR inhibited the chondrogenic fate of SMC.53 In the present study in chondrogenic induction of rBMSCs in vitro, a slight increase in AhR mRNA expression was observed in the 2nd week and an insignificant decrease in the 4th week. But CYP1B1 mRNA was significantly downregulated, which demonstrated inhibition of the AhR signal. However, IHC showed the AhR-positive staining in harvest after implantation of primed cartilage templates. It made us reasonably wonder about the effects of the AhR ligand FICZ on cartilage templates.
Compared with controversy over the osteogenic effects of FICZ and AhR activation, its protective roles in inflammatory responses and immune regulations were basically consensus. FICZ administration alleviated the acute kidney injury,54 periodontitis,25 and colitis55 in vivo and inhibited inflammatory cytokines such as IL-1β, IL-6, IL-7 and TNF-α in LPS-treated cells in vitro. Consistently, FICZ treatment suppressed the IL-1β, 6 and TNF-α expressions in LPS-stimulated BMSCs and SR1 intensified it, demonstrating that the effect was AhR-dependent.
FICZ intervention attenuated calcium oxalate nephrocalcinosis in a mice model. In vitro, a transwell system was used to co-culture bone marrow-derived macrophages (BMDMs) and calcium oxalate monohydrate (COM) (100 μg ml−1)-treated renal tubular epithelial cells (TECs). FICZ with increasing concentrations (0, 100, 200, 300 nM) were added to the system. The results showed that the FICZ promoted the M2 markers (Arg-1, Chi3l3, Fizz 1) and diminished M1 markers (iNOS, IL-6, CllTA) in BMDMs, and the effect was stronger as the FICZ concentration was increased. The molecular mechanism was that AhR directly targeted downstream microRNA-142a-3p, which suppressed IRF-1 and HIF-1 by binding to their 3′UTR.56 In another study, mouse BMSCs were co-cultured with macrophages, and BMSC promoted macrophages to M2 polarization, during which the AhR expression was increased in BMSCs. The treatment of BMSCs with TCDD enhanced their ability to modulate the phenotype of macrophages, resulting in significantly increased M2 markers and decreased M1 markers in macrophages. AhR antagonist CH223191 had an opposite effect.23 Primed cartilage templates were proven to induce the macrophage to M2 polarization.9 Consistent with the above experiments, the FICZ treatment had a trend of polarizing macrophages into an M2-like phenotype in the present study.
High-throughput sequencing technique was next-generation sequencing, which allowed deep analysis in a short time. In the present study, to clarify the alteration of gene expressions in FICZ-treated cartilage templates, RNA-seq was applied. In DEGs, combining the molecular functions and binding prediction with AhR/ARNT, CYP1A1, Gpc3 and Smpd3 were selected as the potential target genes. After validation of qPCR, CYP1A1 and Smpd3 might be the downstream genes contributing to the immunomodulating capacity and osteogenic differentiation of primed cartilage templates. However, luciferase reporter assay or chromatin immunoprecipitation (ChIP) and loss-and-gain function assays were needed to test the molecular hypothesis.
As to the therapeutic potential of FICZ in humans, immune-modulatory effects were observed. The AhR activation by FICZ downregulated the expression of co-stimulatory molecules including HLA-DR, CD80 and CD86 in dendritic cells (DCs) from Behcet's disease patients and normal controls. Also, FICZ significantly inhibited the production of pro-inflammatory cytokines like IL-1β, 6 and TNF-α while increasing the IL-10 in DCs. Meanwhile, the FICZ-treated DCs could suppress the T helper (Th) 1 and 17.57 Consistently, another study showed that FICZ treatment decreased CD83 expression during the maturation of DCs and inhibited the production of IL-6 and TNF-α and upregulated the enzyme IDO. What is more, FICZ-DCs could induce naïve T cells into Treg-like cells.58 In Langerhans cells, FICZ-induced AhR activation reduced the high-affinity receptor for IgE, FcεRI and increased indoleamine (2,3-dioxygenase) IDO expression, mediating anti-inflammatory feedback.59 As a tryptophan-derived photoproduct, FICZ was widely analyzed as a solar ultraviolet (UV) photosensitizer in human skin. In human HaCaT and primary epidermal keratinocytes, photodynamic cell death was induced upon UVA and FICZ cotreatment, accompanied by cellular heat shock, endoplasmic reticulum stress, and oxidative stress response gene expression.60 Similarly, reactive oxygen species (ROS) generation and extensive protein damage were observed in HaCaT human keratinocytes, and nucleotide excision repair was impaired.61 On the other hand, the feature of photosensitizer could be therapeutically used for photodynamic elimination of human skin cancer cells in vivo and in vitro.62 From our point of view, despite the reported immune regulation and osteogenesis promotion of FICZ, its biphasic effects in different cells and models should also be put into consideration before future applications of FICZ in clinical therapy practice.
Author contributions
Conceptualization: Jing Huang, Yi Zhou; methodology: Yining Wang, Yi Zhou; project administration: Jing Huang; formal analysis: Yining Wang, Yi Zhou; writing – original draft: Jing Huang, Yining Wang; writing – review & editing: Yining Wang, Yi Zhou; funding acquisition: Jing Huang, Yi Zhou.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 82001014, J. H., and 82071090, Y. Z.); and the Fundamental Research Funds for the Central Universities (No. 2042020kf0178, J. H.). We appreciate the technical assistance from Gene Denovo.Notes and references
- X. Zhou, X. Xu, J. Li, D. Hu, T. Hu, W. Yin, Y. Fan and X. Zhang, Int. J. Oral Sci., 2018, 10, 1 CrossRef PubMed.
- F. Moreno Sancho, Y. Leira, M. Orlandi, J. Buti, W. V. Giannobile and F. D'Aiuto, Stem Cells Transl. Med., 2019, 8, 1286–1295 CrossRef PubMed.
- A. M. Yousefi, P. F. James, R. Akbarzadeh, A. Subramanian, C. Flavin and H. Oudadesse, Stem Cells Int., 2016, 2016, 6180487 CrossRef PubMed.
- Z. Chen, T. Klein, R. Z. Murray, R. Crawford and X. Yin, Mater. Today, 2016, 19, 304–321 CrossRef CAS.
- N. Kim and S. G. Cho, Internet J. Hematol., 2016, 103, 129–137 CrossRef CAS PubMed.
- C. Zheng, J. Chen, S. Liu and Y. Jin, Int. J. Oral Sci., 2019, 11, 23 CrossRef PubMed.
- J. E. Zins and L. A. Whitaker, Plast. Reconstr. Surg., 1983, 72, 778–785 CrossRef CAS PubMed.
- X. Cai, F. Yang, X. Yan, W. Yang, N. Yu, D. A. Oortgiesen, Y. Wang, J. A. Jansen and X. F. Walboomers, J. Clin. Periodontol., 2015, 42, 380–389 CrossRef CAS PubMed.
- J. Huang, Y. Zhou, Y. Wang, X. Cai and Y. Wang, RSC Adv., 2018, 8, 23679–23687 RSC.
- Y. Wang, X. Cai, J. Huang, Y. Zhou, T. Jiang and Y. Wang, RSC Adv., 2018, 8, 31745–31754 RSC.
- W. Yang, F. Yang, Y. Wang, S. K. Both and J. A. Jansen, Acta Biomater., 2013, 9, 4505–4512 CrossRef CAS PubMed.
- A. Aravamudhan, D. M. Ramos, J. Nip, A. Subramanian, R. James, M. D. Harmon, X. Yu and S. G. Kumbar, Curr. Pharm. Des., 2013, 19, 3420–3428 CrossRef CAS PubMed.
- A. Kochegarov, Expert Opin. Ther. Pat., 2009, 19, 275–281 CrossRef CAS PubMed.
- V. Rothhammer and F. J. Quintana, Nat. Rev. Immunol., 2019, 19, 184–197 CrossRef CAS PubMed.
- D. C. Bersten, A. E. Sullivan, D. J. Peet and M. L. Whitelaw, Nat. Rev. Cancer, 2013, 13, 827–841 CrossRef CAS PubMed.
- Y. Y. Wo, J. Stewart and W. F. Greenlee, J. Biol. Chem., 1997, 272, 26702–26707 CrossRef CAS PubMed.
- T. Haarmann-Stemmann and J. Abel, 2006.
- S. Sakurai, T. Shimizu and U. Ohto, J. Biol. Chem., 2017, 292, 17609–17616 CrossRef CAS PubMed.
- T. Izawa, R. Arakaki, H. Mori, T. Tsunematsu, Y. Kudo, E. Tanaka and N. Ishimaru, J. Immunol., 2016, 197, 4639–4650 CrossRef CAS PubMed.
- M. Korkalainen, E. Kallio, A. Olkku, K. Nelo, J. Ilvesaro, J. Tuukkanen, A. Mahonen and M. Viluksela, Bone, 2009, 44, 1134–1142 CrossRef CAS PubMed.
- S. Climaco-Arvizu, O. Domínguez-Acosta, M. A. Cabañas-Cortés, M. Rodríguez-Sosa, F. J. Gonzalez, L. Vega and G. Elizondo, Life Sci., 2016, 155, 76–84 CrossRef CAS PubMed.
- L. Hinden, R. Shainer, O. Almogi-Hazan and R. Or, Stem Cells, 2015, 33, 2256–2267 CrossRef CAS PubMed.
- Z. Cui, Y. Feng, D. Li, T. Li, P. Gao and T. Xu, J. Immunotoxicol., 2020, 17, 21–30 CrossRef PubMed.
- S. Safe, U. H. Jin, H. Park, R. S. Chapkin and A. Jayaraman, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- J. Huang, X. Cai, Y. Ou, L. Fan, Y. Zhou and Y. Wang, J. Clin. Periodontol., 2019, 46, 882–893 CrossRef CAS PubMed.
- M. Ashburner, C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin and G. Sherlock, Nat. Genet., 2000, 25, 25–29 CrossRef CAS PubMed.
- L. M. Haupt, S. Murali, F. K. Mun, N. Teplyuk, L. F. Mei, G. S. Stein, A. J. van Wijnen, V. Nurcombe and S. M. Cool, J. Cell. Physiol., 2009, 220, 780–791 CrossRef CAS PubMed.
- G. Manickam, P. Moffatt and M. Murshed, Mol. Cell. Biol., 2019, 39, e00370-18 CrossRef PubMed.
- H. Takai, M. Ashihara, T. Ishiguro, H. Terashima, T. Watanabe, A. Kato and M. Suzuki, Cancer Biol. Ther., 2009, 8, 2329–2338 CrossRef CAS PubMed.
- S. Divanovic, J. Dalli, L. F. Jorge-Nebert, L. M. Flick, M. Gálvez-Peralta, N. D. Boespflug, T. E. Stankiewicz, J. M. Fitzgerald, M. Somarathna, C. L. Karp, C. N. Serhan and D. W. Nebert, J. Immunol., 2013, 191, 3347–3357 CrossRef CAS PubMed.
- X. Sun, Z. Ma, X. Zhao, W. Jin, C. Zhang, J. Ma, L. Qiang, W. Wang, Q. Deng, H. Yang, J. Zhao, Q. Liang, X. Zhou, T. Li and J. Wang, Bioact. Mater., 2021, 6, 757–769 CrossRef CAS PubMed.
- O. R. Mahon, D. C. Browe, T. Gonzalez-Fernandez, P. Pitacco, I. T. Whelan, S. Von Euw, C. Hobbs, V. Nicolosi, K. T. Cunningham and K. H. Mills, Biomaterials, 2020, 239, 119833 CrossRef CAS PubMed.
- Q. Zhang, B. Wu, Y. Yuan, X. Zhang, Y. Guo, P. Gong and L. Xiang, Arch. Biochem. Biophys., 2021, 697, 108697 CrossRef CAS PubMed.
- Z. Li, Y. Wang, S. Li and Y. Li, Front. Endocrinol., 2021, 12, 680328 CrossRef PubMed.
- D. Gawlitta, E. Farrell, J. Malda, L. B. Creemers, J. Alblas and W. J. Dhert, Tissue Eng., Part B, 2010, 16, 385–395 CrossRef PubMed.
- C. Scotti, B. Tonnarelli, A. Papadimitropoulos, A. Scherberich, S. Schaeren, A. Schauerte, J. Lopez-Rios, R. Zeller, A. Barbero and I. Martin, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7251–7256 CrossRef CAS PubMed.
- L. Yang, K. Y. Tsang, H. C. Tang, D. Chan and K. S. Cheah, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 12097–12102 CrossRef CAS PubMed.
- A. Longoni, L. Knežević, K. Schepers, H. Weinans, A. Rosenberg and D. Gawlitta, npj Regener. Med., 2018, 3, 22 CrossRef CAS PubMed.
- W. Yang, S. K. Both, G. J. van Osch, Y. Wang, J. A. Jansen and F. Yang, Acta Biomater., 2015, 13, 254–265 CrossRef CAS PubMed.
- Y. Miki, S. Hata, R. Saito, K. Ono, H. Sasano and H. Kumamoto, in Interface Oral Health Science 2011, Springer, 2012, pp. 134–136 Search PubMed.
- J. Yin, B. Sheng, Y. Qiu, K. Yang, W. Xiao and H. Yang, Cell Proliferation, 2016, 49, 554–560 CrossRef CAS PubMed.
- A. Mohammadi-Bardbori, F. Bastan and A. R. Akbarizadeh, Arch. Toxicol., 2017, 91, 3365–3372 CrossRef CAS PubMed.
- N. Montazeri-Najafabady, M. H. Dabbaghmanesh, N. Chatrabnous and M. R. Arabnezhad, Nutr. Cancer, 2020, 72, 1400–1410 CrossRef CAS PubMed.
- C. Yun, J. A. Weiner, D. S. Chun, J. Yun, R. W. Cook, M. S. Schallmo, A. S. Kannan, S. M. Mitchell, R. D. Freshman, C. Park, W. K. Hsu and E. L. Hsu, Bone Rep., 2017, 6, 51–59 CrossRef PubMed.
- A. T. D. Watson, R. C. Nordberg, E. G. Loboa and S. W. Kullman, Toxicol. Sci., 2019, 167, 145–156 CrossRef CAS PubMed.
- Y. Yoshikawa, T. Izawa, Y. Hamada, H. Takenaga, Z. Wang, N. Ishimaru and H. Kamioka, Sci. Rep., 2021, 11, 14927 CrossRef CAS PubMed.
- M. Murai, G. Tsuji, A. Hashimoto-Hachiya, Y. Kawakami, M. Furue and C. Mitoma, J. Dermatol. Sci., 2018, 89, 19–26 CrossRef CAS PubMed.
- M. Murai-Mizote, G. Tsuji, C. Mitoma, A. Hashimoto-Hachiya, M. Nakahara, T. Nakahara, H. Uchi and M. Furue, J. Dermatol. Sci., 2017, 86, e77 CrossRef.
- I. Monteleone, F. Zorzi, I. Marafini, D. Di Fusco, V. Dinallo, R. Caruso, R. Izzo, E. Franze, A. Colantoni, F. Pallone and G. Monteleone, Eur. J. Immunol., 2016, 46, 1047–1057 CrossRef CAS PubMed.
- Y. Shi, B. Tang, J. Yu, Y. Luo, Y. Xiao, Z. Pi, R. Tang, Y. Wang, T. Kanekura, Z. Zeng and R. Xiao, Int. Immunopharmacol., 2020, 88, 106886 CrossRef CAS PubMed.
- D. Kondrikov, A. Elmansi, R. T. Bragg, T. Mobley, T. Barrett, N. Eisa, G. Kondrikova, P. Schoeinlein, A. Aguilar-Perez, X.-M. Shi, S. Fulzele, M. M. Lawrence, M. Hamrick, C. Isales and W. Hill, Exp. Gerontol., 2020, 130, 110805 CrossRef CAS PubMed.
- S. Mulero-Navarro and P. M. Fernandez-Salguero, Front. Cell Dev. Biol., 2016, 4, 45 Search PubMed.
- J. B. Kim, Q. Zhao, T. Nguyen, M. Pjanic, P. Cheng, R. Wirka, S. Travisano, M. Nagao, R. Kundu and T. Quertermous, Circulation, 2020, 142, 575–590 CrossRef CAS PubMed.
- S. Tao, F. Guo, Q. Ren, J. Liu, T. Wei, L. Li, L. Ma and P. Fu, J. Cell. Mol. Med., 2021, 25, 1035–1047 CrossRef CAS PubMed.
- Q. Wang, K. Yang, B. Han, B. Sheng, J. Yin, A. Pu, L. Li, L. Sun, M. Yu, Y. Qiu, W. Xiao and H. Yang, Int. J. Mol. Med., 2018, 41, 868–876 CAS.
- X. Yang, H. Liu, T. Ye, C. Duan, P. Lv, X. Wu, J. Liu, K. Jiang, H. Lu, H. Yang, D. Xia, E. Peng, Z. Chen, K. Tang and Z. Ye, Theranostics, 2020, 10, 12011–12025 CrossRef CAS PubMed.
- C. Wang, Z. Ye, A. Kijlstra, Y. Zhou and P. Yang, Clin. Exp. Immunol., 2014, 177, 521–530 CrossRef CAS PubMed.
- B. B. Jurado-Manzano, D. Zavala-Reyes, E. A. Turrubiartes-Martinez, D. P. Portales-Perez, R. Gonzalez-Amaro and E. Layseca-Espinosa, Immunol. Lett., 2017, 190, 84–92 CrossRef CAS PubMed.
- S. Koch, T. Stroisch, J. Vorac, N. Herrmann, N. Leib, S. Schnautz, H. Kirins, I. Förster, H. Weighardt and T. Bieber, Allergy, 2017, 72, 1686–1693 CrossRef CAS PubMed.
- S. L. Park, R. Justiniano, J. D. Williams, C. M. Cabello, S. Qiao and G. T. Wondrak, J. Invest. Dermatol., 2015, 135, 1649–1658 CrossRef CAS PubMed.
- R. Brem, P. Macpherson, M. Guven and P. Karran, Sci. Rep., 2017, 7, 4310 CrossRef PubMed.
- R. Justiniano, L. de Faria Lopes, J. Perer, A. Hua, S. L. Park, J. Jandova, M. S. Baptista and G. T. Wondrak, Photochem. Photobiol., 2021, 97, 180–191 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra00622g |
| This journal is © The Royal Society of Chemistry 2022 |