Open Access Article
Open Access ArticleIn silico prediction and interaction of resveratrol on methyl-CpG binding proteins by molecular docking and MD simulations study†
Ram Krishna Sahuab,
Ved Vrat Vermac,
Amit Kumarc,
Simran Tandonb,
Bhudev Chandra Dasb and
Suresh T. Hedau *a
*a
aDivision of Molecular Oncology, National Institute of Cancer Prevention and Research, ICMR, I – 7, Sector – 39, Noida – 201 301, Utter Pradesh, India. E-mail: hedaus62@gmail.com
bAmity Institute of Molecular Medicine & Stem Cell Research, Amity University, Noida – 201301, Utter Pradesh, India
cInformation System for Research Management Division, ICMR, Aruna Asif Ali Marg, New Delhi – 110029, India
First published on 13th April 2022
Abstract
Resveratrol enhances the BRCA1 gene expression and the MBD family of proteins bind to the promoter region of the BRCA1 gene. However, the molecular interaction is not yet reported. Here we have analyzed the binding affinity of resveratrol with MBD proteins. Our results suggest that resveratrol binds to the MBD proteins with higher binding affinity toward MeCP2 protein (ΔG = −6.5) by sharing four hydrogen bonds as predicted by molecular docking studies. Further, the molecular dynamics simulations outcomes showed that the backbones of all three protein–ligand complexes are stabilized after the period of 75 ns, constantly fluctuating around the deviations of 0.4 Å, 0.5 Å and 0.7 Å for MBD1, MBD2 and MeCP2, respectively. The inter-molecular hydrogen bonding trajectory analysis for protein–ligand complexes also support the strong binding between MeCP2–resveratrol complex. Further, binding free energy calculations showed binding energy of −94.764 kJ mol−1, −53.826 kJ mol−1 and −36.735 kJ mol−1 for MeCP2–resveratrol, MBD2–resveratrol and MBD1–resveratrol complexes, respectively, which also supported our docking results. Our study also highlighted that the MBD family of proteins forms a binding interaction with other signaling proteins that are involved in various cancer initiation pathways.
Introduction
DNA methylation, histone modification, nucleosome remodeling and RNA-mediated targeting proteins regulate many biological processes that are not only essential for normal development and gene expression but also fundamental to the genesis of cancer.1 Epigenetic modification plays an important role in regulation of transcription, DNA repair and replication.2 At the time of chromatin regulation, expression patterns or genomic alterations can lead to the induction and maintenance of various cancers.2–5 The presence of mCpG dinucleotide in a DNA sequence directly inhibits transcription or it recruits proteins that specifically recognize methylated DNA and initiate the remodeling of euchromatin into a heterochromatin structure in the genome to form a spatial obstacle that is unable to bind transcription factors to promoter sequences. The DNA methylation pattern is believed to be ‘read’ by a conserved MBD family of proteins.6,7 These proteins share a common motif, the methyl CpG binding domain (MBD).8,9 Currently, the NCBI Conserved Domain Database lists 11 human proteins containing the methyl binding domain derived from methyl CpG binding protein 2.10,11 Based on the presence of other domains, these are further divided into 3 groups within the MBD superfamily according to the CDD30: the histone methyl transferases, the MeCP2_MBD proteins, and the histone acetyl transferases. The MBD protein family includes MeCP2, MBD1, MBD2, MBD3, MBD4, and the uncharacterized Kaiso complex, which binds to methylated DNA. MBD1 binds to symmetrically methylated CpG dinucleotides and inhibits gene expression by blocking transcription factors' interaction with the promoter.12,13The MBD1 protein is the largest member of the family of proteins. It has a complex expression profile as there are 13 isoforms of the gene expressed on chromosome 18. The main difference between the isoforms is the presence of 2 or 3 CXXC-type zinc fingers present in the protein.14 The isoforms containing the first 2 CXXC domains preferentially repress methylated promoters, whereas those with the third CXXC domain are capable of DNA binding regardless of methylation status.15,16 MBD2 may bind to methylated DNA and mediates the methylated DNA binding functions for 2 different transcriptional repressor complexes, MeCP1 and Mi2/NuRD.16–19 Both these complexes use MBD2 to direct HDACs and chromatin remodelers to methylated promoters where they effect transcriptional repression (Fig. S1†). Once again, this protein has been shown to silence genes in a variety of cancers: colorectal, lung, prostate, and renal cancer.20–26 The structures of MBD motifs from three different MBD proteins have been solved and their overall similarity indicates that all MBD-containing proteins are likely to adopt a similar folding of protein chain.27–30 The MBD forms a wedge-shaped structure composed of a β-sheet superimposed over an α-helix and loop. Amino acid side chains in two of the β-strands along with residues immediately next to the N-terminal to the α-helix interact with the cytosine methyl groups within the major groove, providing the structural basis for selective recognition of methylated CpG dinucleotide.31,32
Resveratrol is the common term for 3,5,4′-hydroxystilbene (Fig. S2A†) which is produced naturally by several plants in response to injury or when the plant is under attack by pathogens such as bacteria or fungi.33,34 Food sources of resveratrol include the skin of grapes, blueberries, raspberries and mulberries.35 Resveratrol was first reported to exert anti-tumor activities in 1997.33 Since then, the antioxidant, anti-inflammatory, anti-proliferative and anti-angiogenic effects of resveratrol have been widely studied. It has been shown that it exhibits anti-oxidative and anti-inflammatory activity and reverses the effects of aging in rats.36 Resveratrol suppresses proliferation of several types of cancers, such as colon, breast, pancreas, prostate, ovarian and endometrial cancers, as well as lymphoma, and affects diverse molecular targets.33 Resveratrol has been used in many studies not only for its preventive effects but also anti-tumor effects against various cancers and its ability to suppress cell proliferation, apoptosis, metastasis and invasion.37 Resveratrol is found widely in nature and a number of its natural and synthetic analogues and their isomer adducts, derivatives and conjugates are available.38–40 It is an off-white powder (extracted by methanol) with a melting point of 253–255 °C and molecular weight of 228.25. Resveratrol is insoluble in water but dissolves in ethanol and dimethylsulphoxide.41
Earlier, it was reported that resveratrol enhances the BRCA1 gene expression in breast cancer cells and MBD proteins binding to the BRCA1 promoter region.42,43 However the molecular interaction or mechanism has not yet been reported. In this study, we have used MBD proteins as an emerging biomarker. Here, we have analyzed the binding affinity of resveratrol on MBD proteins with a high binding affinity score of resveratrol against MBD1, MBD2, and MeCP2 proteins by docking. Further, the study has been followed by MD simulation and binding free energy calculations for these protein–ligand complexes. Additionally, protein–protein interaction analysis of MBD proteins with their neighboring counterparts has been carried out to identify crucial interacting signaling proteins which are directly or indirectly involved in cancer initiation pathways.
Materials and methods
Structural optimization of target proteins and ligand
The three-dimensional structures of Homo sapiens MBD1 (PDB ID: 6d1t), MBD2 (PDB ID: 6c1a) and MeCP2 (PDB ID: 5bt2) proteins were retrieved from the RCSB protein data bank (https://www.rcsb.org/pdb/home/home.do). The chemical structure of resveratrol (Pubchem CID: 445154) was retrieved from the PubChem compound database. Further, bio-molecule visualizer Chimera 3.1 was used to optimize the molecule and convert it into a PDBQT file format.44 Further, for performing docking studies, the PDB coordinates of the MBD1, MBD2 and MeCP2 protein and resveratrol molecules were optimized by using the protein visualization tool UCSF chimera (added missing residues to minimize energy level and removed unwanted molecules). The optimized three-dimensional coordinates of all proteins were saved at a minimum energy and stable conformation. Chimera allows for building of chemical structure, visualization, molecular analysis and structure optimization (Fig. S3†).45Prediction of binding sites
COACH is a meta-server approach to protein–ligand binding site prediction. Starting from the given structure of target proteins, COACH will generate complementary ligand binding site predictions using two comparative methods, TM-SITE and S-SITE, which recognize ligand-binding templates from the BioLiP protein function database by binding-specific substructure and sequence profile comparisons. These predictions will be combined with results from other methods (including COFACTOR, FINDSITE and ConCavity) to generate final ligand binding site predictions. The COACH algorithm was ranked as the best method in the weekly CAMEO ligand binding site prediction experiments.46,47Molecular docking between MBD proteins and resveratrol molecule
Molecular docking studies were performed to understand the binding affinity behavior of resveratrol with various proteins as MBD1, MBD2 and MeCP2 protein. Here in the present study Auto dock vina was used to perform docking studies. Further, the molecular visualization tool Chimera was used to visualize the detailed protein–ligand binding interactions and final image formations. We have created an active binding grid using position center and size on the X, Y and Z axis by auto dock vina. For MBD1 the grid was centered at: −45.7294, 14.6662, −1.54214, and maintained a size of grid as 24.0191, 33.5461, and 37.6976. For MBD2 the grid was centered at: −47.7262, 13.1876, −0.072281 and maintained a size of grid as 32.647, 35.0381, and 37.7098. For MeCP2 the grid was centered at: 6.51469, −5.95367, −19.5398, and maintained a size of grid as 30.7524, 46.6142, and 34.3735. Further, we have added charge and H-bonds on the receptor protein for stable conformation and energy minimization. Finally, docking was run by auto dock vina to calculate high affinity docking scores and optimize the final binding pose of the protein–ligand complexes. The energy of interaction of resveratrol with the MBD1, MBD2 and MeCP2 protein is assigned as the “grid point”, and at each step of the simulation, the energy of interactions between protein and ligand was evaluated using atomic affinity potentials computed on a grid. The remaining parameters were set as the default.48 Further, PyMOL software was used for visualizing protein–ligand binding interactions and for calculating their hydrogen bond lengths. We have used UCSF chimera structure analysis tools for comparison of protein conformational change and sequence similarity after ligand binding on the MBDs protein's structure.Molecular dynamic simulations and binding free energy calculations between protein–ligand complexes
Molecular dynamic (MD) simulations for all three protein–ligand complexes MBD1–resveratrol, MBD2–resveratrol and MeCP2–resveratrol were performed by the GROMACS 2019 package using force field GROMACS96 43a1. The topology parameters for resveratrol were determined by using the PRODRG server (https://prodrg1.dyndns.org/). The topology parameters of all three proteins and resveratrol were merged to build up the topology of MBD1–resveratrol, MBD2–resveratrol and MeCP2–resveratrol complexes to initialize the next stage of simulations. Individually, MBD1–resveratrol, MBD2–resveratrol and MeCP2–resveratrol complexes were centered in the dodecahedron box by maintaining the distance of 1.2 nm from the wall and the boxes were solvated by explicit water using the TIP3P model.49 The solvated systems were electrically neutralized by adding 0.1 M concentration of sodium ions (NaCl) to all systems.50 Prior to MD simulations, solvated systems were minimized by using 1000 steps of the steepest descent algorithm followed by a conjugate gradient algorithm, allowing the whole MBD1–resveratrol, MBD2–resveratrol and MeCP2–resveratrol complexes environment to relax the system by removing close contacts in the environment.51 The LINCS algorithm was used to constraint the bond length and bond angles and the time step throughout the MD simulations was set to 2 femtoseconds (fs).52 The Particle Mesh Ewald (PME) algorithm was used to calculate long range electrostatic interactions and the cut-off for non-bonded van der Waal interactions was set to 10 Å.53 The heavy atoms were restrained during equilibration at a constant temperature of 300 K with 1 atmospheric pressure for 1 nano-second (ns) together with the method given by Parrinello and Rahman.54 Before the production of MD simulations run at NPT ensembles of 2 ns, all position restrains were cleared from the systems and the data was saved after an interval of every 10 picoseconds (ps). Finally, all minimized and equilibrated complex systems were subjected to a final run of MD simulations for 100 ns. The various trajectory files i.e., root mean square deviations (RMSD), root mean square fluctuations (RMSF) and others were analyzed by using GROMACS scripts and VMD molecular visualization tool.54,55 The RMSF trajectory profile curves were calculated on the basis of Cα-atoms superimposition of proteins. The graphical tool XMGRACE was used for plotting various trajectories.56 Further, binding affinity analysis between inhibitor and receptor molecules was performed for all three simulated complexes. In the present study we have performed binding free energy calculation methods to analyze the binding affinity of our inhibitor with respect to three receptor molecules. For the binding energy calculation between the receptor and inhibitor molecule we performed MM-PBSA calculations method using the GROMACS tool. Python script MmPbSaStat.py and the graphical tool XMGRACE were used for the final statistical analysis of binding energy calculations and trajectory analysis, respectively.57,58Protein–protein interaction analysis
For the protein–protein interactions study, MBD1, MBD2 and MeCP2 were analyzed by using the STRING 10.5 online database (https://string-db.org/). Initially, MBD proteins of Homo sapiens origin were selected as the input for performing a protein–protein network study. Finally, the interacting protein partners of MBD proteins were predicted for further protein–protein interactions analysis.59Results and discussion
Sequential and structural analysis of MBD proteins
The three-dimensional structure of MBD1 (PDB ID: 6d1t), MBD2 (PDB ID: 6c1a) and MeCP2 (PDB ID: 5bt2) are retrieved from the Protein Data Bank database (https://www.rcsb.org/pdb/home/home.do) (Fig. S3B†). The 3D structures of MBD proteins are composed of a β-sheet superimposed over an α-helix and loop. The MBD1 protein is a monomer comprising chain A. The MBD1 primary structure comprising 79 amino acids and a conserved MBD motif, along with CXXC-type 1, type 2 and type 3. This protein is rich in proline and also comprises the TRD region. MBD2 is a homo tetramer protein comprising four chains A, B, E and F and each chain carries a sequence length of 79 amino acids residue. This protein is rich in glycine, arginine, MBD motifs (CXXC) and TRD regions. Additionally, we observed that MeCP2 is also a monomer protein comprising chain A containing a sequence length of 97 amino acids. MeCP2 has one MBD motif, a pro-rich region and a TRD region (Fig. S1†). The TRD domain includes a nuclear localization signal.These proteins have a motif region which interacts with other proteins in the nucleus and forms a complex with it and then binds to a specific region of the DNA sequence as well as interacting with histone proteins, due to its binding gene expression being repressed. MBD1 protein has chain A and a MBD motif, CXXC-type 1, 2 and 3, pro-rich and TRD sequence (Fig. S1†). Its repressive activity is reported to be mediated by lysine 9 (K9) of histone H3 methylation through SETDB1 histone methyl transferase (HMT) recruitment.60 It interacts with Suv39h, another HMT that methylates K9 of histone H3.61 Isoforms containing the CXXC3 are able to bind unmethylated DNA so these proteins not only repress the transcription of methylated sequences but also of unmethylated regions.62 MBD1 has been shown to be significantly associated with lung cancer and in human pancreatic carcinomas. Elevated expression of MBD1 showed association with lymph node metastasis.63 Loss of MBD1 function could affect the normal regulation of gene expression by lacking suppression of genes.12,64 The MBD1 role in gene regulation is confirmed by in situ hybridization and RNAi.64,65 Knockdown of MBD1 inhibited cell proliferation, invasion, and induces apoptosis in pancreatic cancer cells.63
MBD2 has A, B, E, F chains which contain glycine rich regions; the arginine-rich region also has one MBD and TRD motif in its sequence (Fig. S1†). Repressive activity of MBD2 is mediated by MeCP1, an ATP dependent chromatin remodeling complex formed by MBD2 and the Mi-2/NuRD complex.66 It is mainly associated with colorectal cancer, stomach cancer and breast cancer.67,68 MBD2 deficiency also dramatically reduced tumorigenesis and extended life span in the in vivo model.69,70 It has been shown that MBD2 can also bind to unmethylated DNA to cause changes in gene expression.63
MeCP2 has chain A and an MBD motif, a pro-rich region and a TRD region (Fig. S1†). The TRD domain includes a nuclear localization signal. Repression by MeCP2 is mediated by chromatin remodeling complexes recruitment to methylated DNA sequences. The TRD domain interacts with Sin3A, a complex containing histone deacetylase enzymes HDAC1 and HDAC2. Histone deacetylation is not only a way in which MeCP2 represses transcription and establishes heterochromatin formation; it is also known that MeCP2 interacts with a complex containing lysine 9 of histone H3 methyl transferase activity.71 MeCP2 is involved in cancer by binding to the hyper methylated regions of promoters of tumor suppressor genes and thereby causes their subsequent repression in breast cancer, prostate cancer, lung cancer, liver cancer, and colorectal cancer and the elevated expression of this gene has been reported in different cancers. Loss of MeCP2 function has been reported to inhibit cell proliferation and increase apoptosis of prostate cancer cells in vitro.72,73 In addition, treatment with several natural compounds has been shown to down regulate the elevated MeCP2 expression in prostate and breast cancer cells in vitro.74,75
Analysis of binding cavity
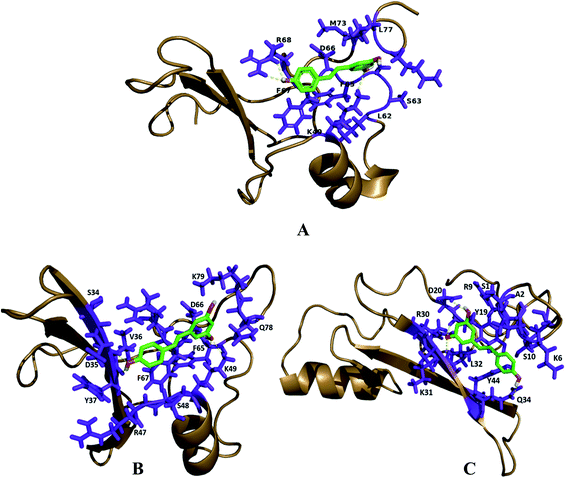
Prediction of the consensus ligand binding amino acid on MBD protein sequence was analyzed by the COACH online meta-server which predicts ligand binding amino acids (Table 1) based on comparative methods. Amino acids present on the ligand binding site of the protein have high affinity to bind the ligand as compared to other amino acids (Fig. 1 and 2). In MBD1 protein, 23 residues have been predicted to have ligand binding interaction, these amino acids are located on the MBD motif present on the early position of the protein structure. However, the TRD domain is located at the later position on the backbone of the protein. It makes the active site between the terminal loop and beta sheet of the protein. These MBD motifs interact with the methylated DNA and TRD domain attached to histone proteins which further regulate the transcription process. The MBD2 protein has been predicted to contain 26 residues which have ligand binding interaction; the MBD motif and TRD domain are jointly present within the middle of this protein backbone. The active site formed between the beta sheet, alpha helical and terminal loop of the proteins structure and these predicted amino acid falls within the early and middle position of this protein's structure. MeCP2 protein comprising 36 residues is predicted to have ligand binding interaction and is mostly present on the later sequence of the protein structure. The active site is formed between the beta sheet and terminal loop of the protein's structure (Fig. S1† and 2).| PDB ID | Protein | Affinity score ΔG (kcal mol−1) | Predicted consensus ligand binding amino acid residue in MBD proteins | Physical interaction | H bonds distance (A0) |
|---|---|---|---|---|---|
| 6d1t | MBD1 | −5.8 kcal mol−1 | GLY-6, GLU-21, ARG-25, LYS-26, SER-27, GLY28, LEU-29, SER-30, LYS-33, ASP-35, GLY-43, LYS-44, LYS-45, PHE-46 TYR-37, ARG-47, SER-48, LYS-49, PRO-50, GLN-51, PHE-67, ARG-68, THR-69 | ARG68(2HB) | 2.29 A0 |
| LYS49(1HB) | 2.18 A0 | ||||
| 2.42 A0 | |||||
| 6c1a | MBD2 | −5.9 kcal mol−1 | GLY-6, GLU-21, VAL-23, ARG-25, LYS-26, SER-27, GLY-28, LEU-29, SER-30, LYS-33, SER-34, ASP-35, VAL-36, TYR-37, GLY-43, LYS-44, LYS-45, PHE-46, ARG-47, SER-48, LYS-49, PRO-50, GLN-51, PHE-67, ARG-68, THR-69 | VAL36(2HB) | 2.03 A0 |
| LYS49(1HB) | 2.38 A0 | ||||
| PHE65(1HB) | 2.42 A0 | ||||
| 2.48 A0 | |||||
| 5bt2 | MeCP2 | −6.5 kcal mol−1 | GLY-16, PRO-25, GLU-26, GLY-27, TRP-28, THR-29, LYS-31, LYS-33, GLY-34, ARG-35, LYS-36, SER-37, GLY-38, ARG-39, SER-40, LYS-43, ASP-45, TYR-47 | LEU32(1HB) | 2.47 A0 |
| ILE-49, ASN-50, PRO-51 | TYR19(1HB) | 2.04 A0 | |||
| PHE-56, ARG-57, SER-58, LYS-59, VAL-60, GLU-61, LEU-62, TYR-65, PHE-66 | GLN34(1HB) | 2.15 A0 | |||
| ASP-71, LEU-74, ASP-80, PHE-81, THR-82, VAL-83 | ASP20(1HB) | 2.33 A0 |
 | ||
| Fig. 1 Prediction of ligand binding amino acids of MBD1, MBD2 and MeCP2 proteins by the COACH online tool. | ||
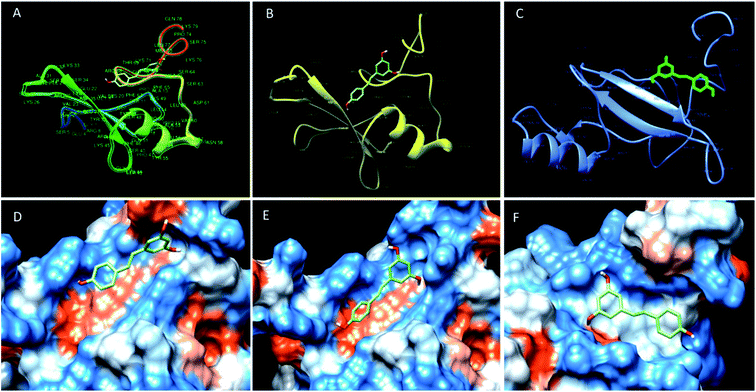 | ||
| Fig. 2 Binding mode of resveratrol in the deep cavity of MBD1 (A and D), MBD2 (B and E) and MeCP2 (C and F). | ||
Molecular docking analysis of resveratrol on MBD proteins
The molecular docking analysis of resveratrol with all three proteins MBD1, MBD2 and MeCP2 has been done to understand the binding mode of resveratrol with all three target molecules. Docking analysis helps us to calculate the relative binding affinity of resveratrol with respect to all 3 target proteins which should be directly proportional to the docking score. In this respect, we observed that the predicted binding affinity score of resveratrol with MBD1 is −5.8 along with 3 hydrogen bonds, where resveratrol binds to the Arg 68 residue by two hydrogen bonds to the 4′ position of the hydroxyl group along with one hydrogen bond between Lys 49 and 3rd carbon position of hydroxyl group of resveratrol. These two amino acids are present on the TRD domain of the protein and this domain is known to play a key role in gene transcription. The affinity score for resveratrol with MBD2 has been observed as −5.9. The resveratrol forms four hydrogen bonds with the MBD2 target. The amino acid residue Val 36 forms two hydrogen bonds at the 4′ position of the hydroxyl group whereas Phe 65 and Lys 49 form one hydrogen bond to each at the 5′ carbon hydroxyl group of resveratrol. These three amino acids Val 36, Lys 49 and Phe 65 are found to be allocated on the α-helix and β-sheet of the protein back bone, known for the methyl domain binding and TRD property. Binding of resveratrol to methyl domain binding will interfere with functioning of DNA binding and gene transcription. Further, we observed the affinity score of resveratrol for MeCP2 as −6.5 along with 4 hydrogen-bonds. Resveratrol forms hydrogen bonds with four residues of MeCP2, Leu 32, Tyr 19, Gln 34 and Asp 20 bind to the resveratrol hydroxyl group with one hydrogen bond to each. Amino acid residues Leu 32 and Gln 34 present on the beta sheet of the protein backbone bind to the 3′ carbon hydroxyl group and 4′ carbon hydroxyl group of resveratrol, respectively. Tyr 19 binds to the 3′ carbon hydroxyl group and Asp 20 bind to the 5′ carbon hydroxyl group of resveratrol and present on before the beta sheet of the protein backbone. Binding of resveratrol to these amino acids will inhibit the methyl domain binding and transcription of genes in chromosomes (Fig. 3) (Table 1). Further, using the structure analysis tool chimera, we found that there is no conformational change in MBD1, MBD2 and MeCP2 proteins after resveratrol binding and showing 0.00 RMSD and there is 100% similarity of the protein sequence between native and ligand bound proteins (Table 2).| Proteins–ligand complex | Similarity of sequence between native protein and ligand binding proteins after docking |
|---|---|
| MBD1 + resveratrol | >MBD1 docked.pdb/1-79GATESGKRMDCPALPPGWKKEEVIRKSGLSAGKSDVYYFSPSGKKFRSKPQLARYLGNTVDLSSFDFRTGKMMPSKLQK |
| MBD1 | >MBD1.receptor.pdb/1-79GATESGKRMDCPALPPGWKKEEVIRKSGLSAGKSDVYYFSPSGKKFRSKPQLARYLGNTVDLSSFDFRTGKMMPSKLQK |
| MBD2 + resveratrol | >MBD2 docked.pdb/1-79GATESGKRMDCPALPPGWKKEEVIRKSGLSAGKSDVYYFSPSGKKFRSKPQLARYLGNTVDLSSFDFRTGKMMPSKLQK |
| MBD2 | >MBD2.receptor.pdb/1-79GATESGKRMDCPALPPGWKKEEVIRKSGLSAGKSDVYYFSPSGKKFRSKPQLARYLGNTVDLSSFDFRTGKMMPSKLQK |
| MeCP2 + resveratrol | >MeCP2 docked.pdb/1-97ASASPKQRRSIIRDRGPMYDDPTLPEGWTRKLKQRKSGRSAGKYDVYLINPQGKAFRSKVELIVYFEKVGDTSLDPNDFDFTVTGRGSPSRHHHHHH |
| MeCP2 | >MeCP2.receptor.pdb/1-97ASASPKQRRSIIRDRGPMYDDPTLPEGWTRKLKQRKSGRSAGKYDVYLINPQGKAFRSKVELIVYFEKVGDTSLDPNDFDFTVTGRGSPSRHHHHHH |
Molecular dynamic simulations binding free energy calculations between protein–ligand complexes
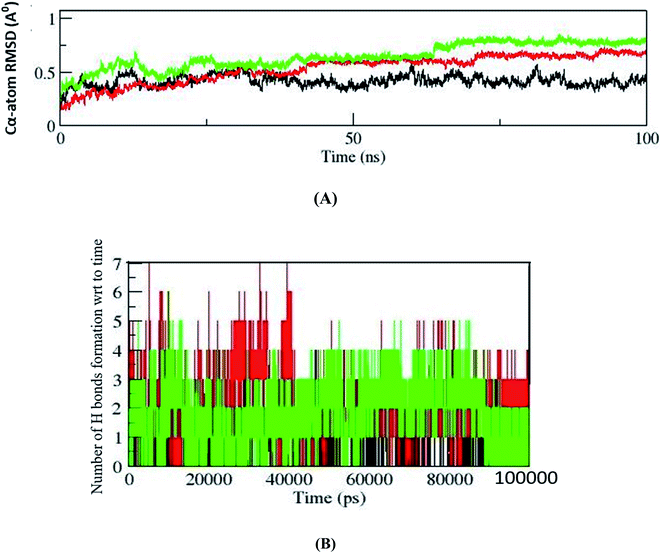
Molecular dynamic simulations of docked protein–ligand complexes have been done to understand the binding mode of resveratrol in a dynamic state. In the present study, the GROMACS 2019 package and GROMACS96 43a1 force field have been used to perform MD simulations of the three complexes. Altogether, the outcomes of the MD simulations studies have been analyzed by analyzing distinct trajectories. Comparative C-α atoms root mean square deviations (RMSD) trajectory curve analysis showed that all three complexes are stabilized after the simulation time of 75 ns, suggesting that these complexes stabilized before the end of 100 ns simulations (Fig. 4A). Further, the dynamic natures of hydrogen bond formation between protein–ligand complexes are analyzed to understand the comparative strength of the intermolecular hydrogen bonding between protein–ligand complexes during MD simulations of 100 ns. In this respect, we observed that the receptor molecule MBD1 forms up to four hydrogen bonds, MBD2 forms up to six hydrogen bonds and MeCP2 forms up to seven hydrogen bonds during simulations of 100 ns (Fig. 4B). The outcomes of dynamic state hydrogen bonding analysis suggest that the MeCP2–resveratrol complex forms a more stable hydrogen bond as compared to other two proteins. Further, it is also suggested that similar to the maximum number of hydrogen bond formation they might follow a similar order for their binding affinity.Today, binding free energy calculations between receptor and inhibitor is a popular method to calculate the binding affinity of an inhibitor for a particular receptor. Therefore, in the present study we have performed a binding energy calculation between the receptor molecules (MeCP2, MBD2 and MBD1) and resveratrol. The RMSD trajectory analysis outcomes of the MD simulations study suggested that all three systems have reached equilibrium after the time period of 75 ns and fluctuate around 0.4 Å, 0.5 Å and 0.7 Å for MBD1, MBD2 and MeCP2, respectively, with individually constant RMSD. Hence, all three systems achieved equilibrium after 75 ns; therefore, binding energy calculations for all three systems are performed for the trajectory of 10 ns between the simulation periods of 80 to 90 ns. After performing binding energy calculations for all three individual complexes we compared the final binding energies of all three cases (Table 3). Our binding energy analysis showed that the inhibitor molecule's resveratrol binds with the lowest binding energy of −94.76 kJ mol−1 for receptor molecule MeCP2 followed by the binding energy of −53.83 kJ mol−1 for the receptor molecule MBD2, followed by the binding energy of −36.73 kJ mol−1 for the receptor molecule MBD1 (Table 3). Our comparative binding energy analysis outcomes of resveratrol for all three receptor molecules showed that resveratrol showed the lowest binding energy with MeCP2 suggesting the higher binding affinity of resveratrol for MeCP2 and the binding affinity order following the order MBD2 and then MBD1. So, here our binding energy analysis results follow the same outcomes which we have seen in docking and binding interactions analysis where we have observed higher binding interactions of resveratrol in the case of MeCP2 followed by MBD2 and MBD1. Our binding energy analysis outcomes are strengthening our previous docking studies and binding interactions analysis results. Therefore, the combined outcomes of our docking studies, the binding interactions analysis and binding energy analysis suggest that resveratrol could be an effective and putative inhibitor of MeCP2 which could be used for further experimental validation studies.
| Complex (receptor & inhibitor) | van der Waal energy (kJ mol−1) | Electrostatic energy (kJ mol−1) | Polar solvation energy (kJ mol−1) | SASA energy (kJ mol−1) | Binding energy (kJ mol−1) |
|---|---|---|---|---|---|
| MeCP2 & resveratrol | −116.55 ± 20.50 | −42.59 ± 13.45 | 75.52 ± 18.76 | −11.14 ± 1.69 | −94.76 ± 22.69 |
| MBD2 & resveratrol | −66.25 ± 39.92 | −29.81 ± 19.27 | 49.39 ± 35.59 | −7.16 ± 3.56 | −53.83 ± 27.16 |
| MBD1 & resveratrol | −47.69 ± 29.43 | −10.04 ± 11.25 | 25.42 ± 25.74 | −4.42 ± 3.44 | −36.73 ± 18.37 |
Our result has clearly stated that MeCP2 has a greater number of intra-molecular hydrogen bonds and its backbone is more stable than MBD1 and MBD2 up to 100 ns of simulation (Fig. 4). Our binding energy calculation of protein ligand complexes and MD simulation studies also confirms that resveratrol has strong binding affinity with MeCP2 compared to MBD1 and MBD2 proteins (Table 3).
Protein–protein interaction analyses of MBD proteins with their neighboring metabolic pathway counterparts
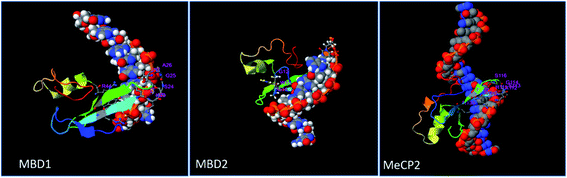
Protein–protein interaction analysis of MBD proteins with their neighboring proteins has been analyzed to identify the interacting partners of MBD proteins which are involved in cancer initiation and apoptotic pathways. The present study aims to understand the comparative binding affinity of MBD1, MBD2 and MeCP2 with resveratrol; therefore, we have identified the key interacting protein partners of MBD1, MBD2 and MeCP2 which directly and indirectly interact with these three proteins. Additionally, we assumed that by inhibiting these three proteins by the inhibitor molecule resveratrol we are able to block the corresponding metabolic pathways of these three proteins to which they belong. Accordingly, we observed that MBD1 protein interacts with ATF7IP, SETDB1, SUV39H1, CHAF1A, TENM1, CBX5, SUMO1, RARA, HDAC3 and CBX3 proteins. The MBD2 protein interacts with HDAC2, RBBP7, GATAD2A, MTA2, DNMT1, HDAC1, SIN3A, RBBP4, CHD3, PRMT5 proteins and MeCP2 protein interacts with HDAC2, RBBP7, GATAD2A, MTA2, DNMT1, HDAC1, SIN3A, RBBP4, CHD3, PRMT5 proteins (Fig. 5). These proteins are directly or indirectly involved in the regulation of gene expression, apoptosis, cell growth and proliferation, and signaling pathways in cancer cells. Our protein–protein interactions analysis suggests that proteins MBD1, MBD2 and MeCP2 are interacting with many of the other proteins which are crucial for many key metabolic pathways; therefore, inhibiting MBD1, MBD2 and MeCP2 with resveratrol could be a good approach to block the metabolic activity of the above discussed key pathways.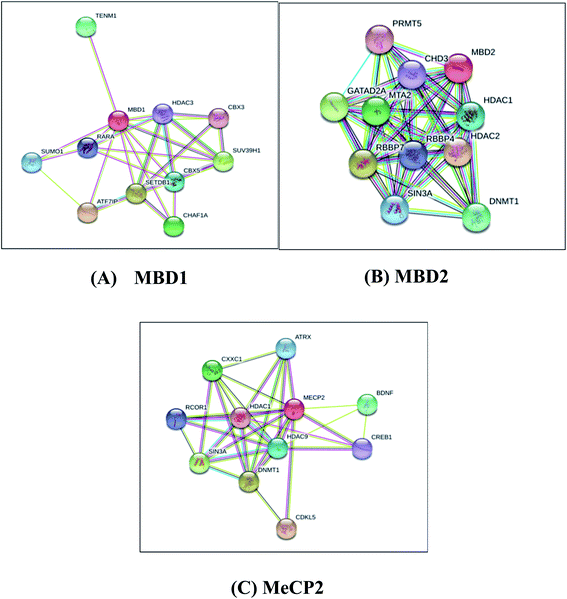 | ||
| Fig. 5 Protein–protein interaction network studies of (A) MBD1, (B) MBD2 and (C) MeCP2 methyl CpG binding proteins by STRING-10. | ||
MBD1 is a transcriptional repressor that binds to CpG islands in promoters where the DNA is methylated at position 5 of cytosine within CpG dinucleotides. MBD1 acts as a transcriptional repressor which plays a key role in gene silencing by recruiting AFT7IP, which in turn recruits factors such as the histone methyl transferase SETDB1. It probably forms a complex with SETDB1 and ATF7IP that represses transcription and couples DNA methylation and histone ‘Lys-9’ tri methylated (Fig. 5A).76 MBD2 binds to hemi methylated DNA as well, it recruits histone deacetylases and DNA methyl transferases and acts as a transcriptional repressor and plays a key role in gene silencing. It functions as a scaffold protein, targeting GATAD2A and GATAD2B to chromatin to promote repression. It may enhance the activation of some unmethylated cAMP-responsive promoters (Fig. 5B).77,78 The MeCP2 protein that binds to methylated DNA can bind specifically to a single methyl-CpG pair. It is not influenced by sequences flanking the methyl-CpG mediating transcriptional repression through interaction with histone deacetylase and the co-repressor SIN3A, which binds both 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC)-containing DNA, with a preference for 5-methylcytosine (5mC) (Fig. 5C).79 The above results explore the importance of these MBD proteins in the regulation of gene expression and their involvement in cancer initiation pathways.
Conclusions
In conclusion, we have found that MBD proteins are made up of an α-helix, β-sheet and MBD1 and MeCP2. These two proteins have a single chain whereas MBD2 is made up of four polypeptide chains. These proteins have an MBD motif, TRD and a CXXC region which bind to the histone protein and methylated DNA sequence which regulates the gene expression. Binding analysis revealed that MeCP2 has a maximum number of amino acids which interact with the ligand as compared to MBD1 and MBD2 proteins. After docking with resveratrol and the MD simulation of the protein ligand complex it is confirmed that resveratrol has high binding affinity toward MeCP2 protein compared to MBD1 and MBD2 proteins. These MBD proteins interact with the other signaling proteins which are directly or indirectly involved the cancer initiation pathways. The detailed mechanism and pathways of these MBD proteins in cancer development are still unclear. Resveratrol could be used as the inhibitor of these MBD proteins and further in vitro study should be done to explore the effects of MeCP2 protein in different cancer cells.Abbreviations
| CDD | Conserved domain database |
| HDAC | Histone deacetylase |
| HMT | Histone methyl transferases |
| MBD | Methyl-CpG binding proteins |
| MD | Molecular dynamic |
| MeCP2 | Methyl-CpG binding protein 2 |
| NuRD | Nucleosome remodeling complex |
Conflicts of interest
The authors declare that they have no conflicts or financial interests in the work reported in this paper.Acknowledgements
We thank the Science and Engineering Research Board, Department of Science and Technology, Govt. of India and Indian Council of Medical Research, India for providing fellowship.References
- Epigenetics, Research bulletin December, TOCRIS a biotechne brand, 2014.
- M. A. Dawson and T. Kouzarides, Cancer Epigenetics: From Mechanism to Therapy, Cell, 2012, 150, 12–27 CrossRef CAS PubMed.
- S. Sharma, T. K. Kelly and P. A. Jones, Epigenetics in cancer, Carcinogenesis, 2010, 31, 27–36 CrossRef CAS PubMed.
- J. L. Miller and P. A. Grant, The Role of DNA Methylation and Histone Modifications in Transcriptional Regulation in Humans, Subcell. Biochem., 2013, 61, 289–317 CAS.
- P. Karakaidos, D. Karagiannis and T. Rampias, Resolving DNA Damage: Epigenetic Regulation of DNA Repair, Molecules, 2020, 25, 2496 CrossRef CAS PubMed.
- R. Jaenisch and A. Bird, Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals, Nat. Genet., 2003, 33, 245–254 CrossRef CAS PubMed.
- P. A. Wade and A. P. Wolffe, Recognizing methylated DNA, Nat. Struct. Mol. Biol., 2001, 8, 575–577 CrossRef CAS PubMed.
- B. Hendrich and A. Bird, Identification and characterization of a family of mammalian methyl-CpG binding proteins, Mol. Cell. Biol., 1998, 18, 6538–6547 CrossRef CAS PubMed.
- Q. Du, P. L. Luu, C. Stirzaker and S. J. Clark, Methyl-CpG-binding domain proteins: readers of the epigenome, Epigenomics, 2015, 7(6), 1051–1073 CrossRef CAS PubMed.
- A. Marchler-Bauer, J. B. Anderson, F. Chitsaz, A. Marchler-Bauer, J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, S. Lu, G. H. Marchler, M. Mullokandov, J. S. Song, A. Tasneem, N. Thanki, R. A. Yamashita, D. Zhang and N. Zhang, CDD: specific functional annotation with the Conserved Domain Database, Nucleic Acids Res., 2009, 37, D205–D210 CrossRef CAS PubMed.
- A. Marchler-Bauer, S. Lu, J. B. Anderson, F. Chitsaz, M. K. Derbyshire, C. DeWeese-Scott, J. H. Fong, L. Y. Geer, R. C. Geer, N. R. Gonzales, M. Gwadz, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, F. Lu, G. H. Marchler, M. Mullokandov, M. V. Omelchenko, C. L. Robertson, J. S. Song, N. Thanki, R. A. Yamashita, D. Zhang, N. Zhang, C. Zheng and S. H. Bryant, CDD: a Conserved Domain Database for the functional annotation of proteins, Nucleic Acids Res., 2011, 39, D225–D229 CrossRef CAS PubMed.
- L. Parry and A. R. Clarke, The roles of the methyl-CpG binding proteins in cancer, Genes Cancer, 2011, 2, 618–630 CrossRef CAS PubMed.
- X. Zou, W. Ma, I. A. Solov'yov, C. Chipot and K. Schulten, Recognition of methylated DNA through methyl-CpG binding domain proteins, Nucleic Acids Res., 2012, 40, 2747–2758 CrossRef CAS PubMed.
- H. F. Jørgensen, I. Ben-Porath and A. P. Bird, Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains, Mol. Cell. Biol., 2004, 24, 3387–3395 CrossRef PubMed.
- M. Nakao, S. Matsui, S. Yamamoto, K. Okumura, M. Shirakawa and N. Fujita, Regulation of transcription and chromatin by methyl-CpG binding protein MBD1, Brain Dev., 2001, 23(suppl 1), S174–S176 CrossRef PubMed.
- N. Mahmood and S. A. Rabbani, DNA Methylation Readers and Cancer: Mechanistic and Therapeutic Applications, Front. Oncol., 2019, 9, 489 CrossRef PubMed.
- Q. Feng and Y. Zhang, The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes, Genes Dev., 2001, 15, 827–832 CAS.
- Q. Feng, R. Cao, L. Xia, H. Erdjument-Bromage, P. Tempst and Y. Zhang, Identification and functional characterization of the p66/p68 components of the MeCP1 complex, Mol. Cell. Biol., 2002, 22, 536–546 CrossRef CAS PubMed.
- X. Le Guezennec, M. Vermeulen, A. B. Brinkman, W. A. M. Hoeijmakers, A. Cohen, E. Lasonder and H. G. Stunnenberg, MBD2/NuRD and MBD3/NuRD, Two Distinct Complexes with Different Biochemical and Functional Properties, Mol. Cell. Biol., 2006, 26, 843–851 CrossRef CAS PubMed.
- H. Y. Park, Y. K. Jeon, H. J. Shin, I. J. Kim, H. C. Kang, S. J. Jeong, D. H. Chung and C. W. Lee, Differential promoter methylation may be a key molecular mechanism in regulating BubR1 expression in cancer cells, Exp. Mol. Med., 2007, 39, 195–204 CrossRef CAS PubMed.
- T. Shestakova, E. Zhuravel, L. Bolgova, O. Alekseenko, M. Soldatkina and P. Pogrebnoy, Expression of human beta-defensins-1, 2 and 4 mRNA in human lung tumor tissue: a pilot study, Exp. Oncol., 2008, 30, 153–156 CAS.
- S. M. Pulukuri and J. S. Rao, CpG island promoter methylation and silencing of 14-3-3σ gene expression in LNCaP and Tramp-C1 prostate cancer cell lines is associated with methyl-CpG-binding protein MBD2, Oncogene, 2006, 25, 4559–4572 CrossRef CAS PubMed.
- N. Shukeir, P. Pakneshan, G. Chen, M. Szyf and S. A. Rabbani, Alteration of the Methylation Status of Tumor-Promoting Genes Decreases Prostate Cancer Cell Invasiveness and Tumorigenesis In vitro and In vivo, Cancer Res., 2006, 66, 9202–9210 CrossRef CAS PubMed.
- X. Yu, A. Azzo, S. M. Bilinovich, X. Li, M. Dozmorov, R. Kurita, Y. Nakamura, D. C. Williams and G. D. Ginder, Disruption of the MBD2-NuRD complex but not MBD3-NuRD induces high level HbF expression in human adult erythroid cells, Haematologica, 2019, 104, 2361–2371 CrossRef CAS PubMed.
- G. Li, Y. Tian and W. G. Zhu, The Roles of Histone Deacetylases and Their Inhibitors in Cancer Therapy, Front. Cell Dev. Biol., 2020, 8, 2296–2634 Search PubMed.
- S. Majid, A. A. Dar, A. E. Ahmad, H. Hirata, K. Kawakami, V. Shahryari, S. Saini, Y. Tanaka, A. V. Dahiya, G. Khatri and R. Dahiya, BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer, Carcinogenesis, 2009, 30, 662–670 CrossRef CAS PubMed.
- B. Heitmann, T. Maurer, J. M. Weitzel, W. H. Stratling, H. R. Kalbitzer and E. Brunner, Solution structure of the matrix attachment region-binding domain of chicken MeCP2, Eur. J. Biochem., 2003, 270, 3263–3270 CrossRef CAS PubMed.
- I. Ohki, N. Shimotake, N. Fujita, M. Nakao and M. Shirakawa, Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1, EMBO J., 1999, 18, 6653–6661 CrossRef CAS PubMed.
- R. I. Wakefield, B. O. Smith, X. Nan, A. Free, A. Soteriou, D. Uhrin, A. P. Bird and P. N. Barlow, The solution structure of the domain from MeCP2 that binds to methylated DNA, J. Mol. Biol., 1999, 291, 1055–1065 CrossRef CAS PubMed.
- Y. Imaizumi and R. Feil, Emerging chromatin structural roles of the methyl-CpG binding protein MeCP2, Epigenomics, 2021, 13, 405–409 CrossRef CAS PubMed.
- I. Ohki, N. Shimotake, N. Fujita, J. Jee, T. Ikegami, M. Nakao and M. Shirakawa, Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA, Cell, 2001, 105, 487–497 CrossRef CAS PubMed.
- P. A. Wade and A. P. Wolffe, Recognizing methylated DNA, Nat. Struct. Mol. Biol., 2001, 8, 575–577 CrossRef CAS PubMed.
- B. B. Aggarwal, A. Bhardwaj, R. S. Aggarwal, N. P. Seeram, S. Shishodia and Y. Takada, Role of resveratrol in prevention and therapy of cancer: preclinical and clinical studies, Anticancer Res., 2004, 24(5A), 2783–2840 CAS.
- L. Frémont, Biological effects of resveratrol, Life Sci., 2000, 66, 663–673 CrossRef.
- M. Jasiński, L. Jasińska and M. Ogrodowczyk, Resveratrol in prostate diseases – a short review, Cent. Eur. J. Urol., 2013, 66, 144–149 Search PubMed.
- D. Sato, N. Shimizu, Y. Shimizu, M. Akagi, Y. Eshita, S. Ozaki, N. Nakajima, K. Ishihara, N. Masuoka, H. Hamada, K. Shimoda and N. Kubota, Synthesis of glycosides of resveratrol, pterostilbene, and piceatannol, and their anti-oxidant, anti-allergic, and neuroprotective activities, Biosci., Biotechnol., Biochem., 2014, 78, 1123–1128 CrossRef CAS PubMed.
- Y. Shukla and R. Singh, Resveratrol and cellular mechanisms of cancer prevention, Ann. N. Y. Acad. Sci., 2011, 1215, 1–8 CrossRef CAS PubMed.
- G. J. Soleas, E. P. Diamandis and D. M. Goldberg, The world of resveratrol, Adv. Exp. Med. Biol., 2001, 492, 159–182 CrossRef CAS PubMed.
- M. Sovak, Grape Extract, Resveratrol, and Its Analogs: A Review, J. Med. Food, 2001, 4, 93–105 CrossRef CAS PubMed.
- S. Pervaiz, Resveratrol: from grapevines to mammalian biology, FASEB J., 2003, 17, 1975–1985 CrossRef CAS PubMed.
- B. C. Trela and A. L. Waterhouse, Resveratrol: Isomeric Molar Absorptivities and Stability, J. Agric. Food Chem., 1996, 44(5), 1253–1257 CrossRef CAS.
- E. Auriol, L. M. Billard, F. Magdinier and R. Dante, Specific binding of the methyl binding domain protein 2 at the BRCA1-NBR2 locus, Nucleic Acids Res., 2005, 13, 4243–4254 CrossRef PubMed.
- P. Fustier, L. Le Corre, N. Chalabi, C. Vissac-Sabatier, Y. Communal, Y. J. Bignon and D. J. Bernard-Gallon, Resveratrol increases BRCA1 and BRCA2 mRNA expression in breast tumour cell lines, Br. J. Cancer, 2003, 89, 168–172 CrossRef CAS PubMed.
- P. S. Reddy, K. B. Lokhande, S. Nagar, V. D. Reddy, P. S. Murthy and K. V. Swamy, Molecular Modeling, Docking, Dynamics and Simulation of Gefitinib and its Derivatives with EGFR in Non-small Cell Lung Cancer, Curr. Comput.-Aided Drug Des., 2018, 14, 246–252 CrossRef CAS PubMed.
- S. C. Lovell, I. W. Davis, W. B. Adrendall, P. I. W. De Bakker, J. M. Word, M. G. Prisant, J. S. Richardson and D. C. Richardson, Structure validation by C alpha geometry: phi, psi and C beta deviation, Proteins: Struct., Funct., Bioinf., 2003, 50, 437–450 CrossRef CAS PubMed.
- J. Yang, A. Roy and Y. Zhang, Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment, Bioinformatics, 2013, 29, 2588–2595 CrossRef CAS PubMed.
- M. Fatemi and P. A. Wade, MBD family proteins: reading the epigenetic code, J. Cell Sci., 2006, 119, 3033–3037 CrossRef CAS PubMed.
- F. Fuks, P. J. Hurd, R. Deplus and T. Kouzarides, The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase, Nucleic Acids Res., 2003, 31, 2305–2312 CrossRef CAS PubMed.
- I. V. Leontyev and A. A. Stuchebrukhov, Polarizable mean-field model of water for biological simulations with AMBER and CHARMM force fields, J. Chem. Theory Comput., 2012, 8, 3207–3216 CrossRef CAS PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. Van Gunsteren and J. Hermans, in Interaction Models for Water in Relation to Protein Hydration, Springer, Dordrecht, 1981, pp. 331–342 Search PubMed.
- M. N. Vrahatis, G. S. Androulakis, J. N. Lambrinos and G. D. Magoulas, A class of gradient unconstrained minimization algorithms with adaptive step size, J. Comput. Appl. Math., 2000, 114, 367–386 CrossRef.
- B. Hess, P-LINCS: a parallel linear constraint solver for molecular simulation, J. Chem. Theory Comput., 2008, 4, 116–122 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, Particle mesh ewald-an n
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(n) method for ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
log(n) method for ewald sums in large systems, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS. - H. J. C. Berendsen, D. Van der Spoel and R. Van Drunen, GROMACS: A message-passing parallel molecular dynamics implementation, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- K. Lindorff-Larsen, S. Piana, K. Palmo, P. Maragakis, J. L. Klepeis, R. O. Dror and D. E. Shaw, Improved side-chain torsion potentials for the Amber ff99SB protein force field, Protein, 2010, 78, 1950–1958 CrossRef CAS PubMed.
- S Dinesh, S. Sudharsana, A Mohanapriya and T. Itami, Molecular docking and simulation studies of Phyllanthus amarus phytocompounds against structural and nucleocapsid proteins of white spot syndrome virus, 3 Biotech, 2017, 7, 1–12 CrossRef PubMed.
- R. Kumari and R. Kumar, OSDD Consortium and A. Lynn. g_mmpbsa - A GROMACS tool for high-throughput MM-PBSA calculations, J. Chem. Inf. Model., 2014, 54, 1951–1962 CrossRef CAS PubMed.
- N. A. Baker, D. Sept, S. Joseph, M. J. Holst and J. A. McCammon, Electrostatics of nanosystems: Application to microtubules and the ribosome, Proc. Natl. Acad. Sci., 2001, 98, 10037–10041 CrossRef CAS PubMed.
- J. Yang, A. Roy and Y. Zhang, Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment, Bioinformatics, 2013, 29, 2588–2595 CrossRef CAS PubMed.
- D. Szklarczyk, A. Franceschini, M. Kuhn, M. Simonovic, A. Roth, P. Minguez, T. Doerks, M. Stark, J. Muller, P. Bork, L. J. Jensen and C. Von Mering, The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored, Nucleic Acids Res., 2011, 39, D561–D568 CrossRef CAS PubMed.
- N. V. Sankaranarayanan, How to Study Protein–ligand Interaction through Molecular Docking Today's exercise: Molecular Docking of Serine Protease with its inhibitors, 2016, 1–2 Search PubMed.
- R. J. Klose and A. P. Bird, Genomic DNA methylation: the mark and its mediators, Trends Biochem. Sci., 2006, 31, 89–97 CrossRef CAS PubMed.
- N. Mahmood and S. A. Rabbani, DNA methylation readers and cancer: Mechanistic and therapeutic applications, Front. Oncol., 2019, 9, 489 CrossRef PubMed.
- C. Liu, Y. Chen, X. Yu, C. Jin, J. Xu, J. Long, Q. Ni, D. Fu, H. Jin and C. Bai, Proteomic analysis of differential proteins in pancreatic carcinomas: Effects of MBD1 knock-down by stable RNA interference, BMC Cancer, 2008, 8, 1–10 CrossRef PubMed.
- S. Bader, M. Walker, H. A. McQueen, R. Sellar, E. Oei, S. Wopereis, Y. Zhu, A. Peter, A. P. Bird and D. J. Harrison, MBD1, MBD2 and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers, Oncogene, 2003, 22, 3506–3510 CrossRef CAS PubMed.
- N. Fujita, D. L. Jaye, M. Kajita, C. Geigerman, C. S. Moreno and P. A. Wade, MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer, Cell, 2003, 113, 207–219 CrossRef CAS PubMed.
- Y. Kanai, S. Ushijima, Y. Nakanishi and S. Hirohashi, Reduced mRNA expression of the DNA demethylase, MBD2, in human colorectal and stomach cancers, Biochem. Biophys. Res. Commun., 1999, 264, 962–966 CrossRef CAS PubMed.
- L. M. Billard, F. Magdinier, G. M. Lenoir, L. Frappart and R. Dante, MeCP2 and MBD2 expression during normal and pathological growth of the human mammary gland, Oncogene, 2002, 21, 2704–2712 CrossRef CAS PubMed.
- J. Berger and A. Bird, Role of MBD2 in gene regulation and tumorigenesis, Biochem. Soc. Trans., 2005, 33, 1537–1540 CrossRef CAS PubMed.
- O. J. Sansom, J. Berger, S. M. Bishop, B. Hendrich, A. Bird and A. R. Clarke, Deficiency of Mbd2 suppresses intestinal tumorigenesis, Nat. Genet., 2003, 34, 145–147 CrossRef CAS PubMed.
- N. Fujita, N. Shimotake, I. Ohki, T. Chiba, H. Saya, M. Shirakawa and M. Nakao, Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1, Mol. Cell. Biol., 2000, 20, 5107–5118 CrossRef CAS PubMed.
- D. Bernard, J. Gil, P. Dumont, S. Rizzo, D. Monté, B. Quatannens, D. Hudson, T. Visakorpi, F. Fuks and Y. De Launoit, The methyl-CpG-binding protein MECP2 is required for prostate cancer cell growth, Oncogene, 2006, 25, 1358–1366 CrossRef CAS PubMed.
- A. Yaqinuddin, F. Abbas, S. Z. Naqvi, M. U. Bashir, R. Qazi and S. A. Qureshi, Silencing of MBD1 and MeCP2 in prostatecancer-derived PC3 cells produces differential gene expression profiles and cellular phenotypes, Biosci. Rep., 2008, 28, 319–326 CrossRef CAS PubMed.
- M. Pandey, S. Shukla and S. Gupta, Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells, Int. J. Cancer, 2010, 126, 2520–2533 CAS.
- S. Mirza, G. Sharma, R. Parshad, S. D. Gupta, P. Pandya and R. Ralhan, Expression of DNA methyltransferases in breast cancer patients and to analyze the effect of natural compounds on DNA methyltransferases and associated proteins, J. Breast Cancer, 2013, 16, 23–31 CrossRef PubMed.
- N. Fujita, S. Takebayashi, K. Okumura, S. Kudo, T. Chiba, H. Saya and M. Nakao, Methylation-Mediated Transcriptional Silencing in Euchromatin by Methyl-CpG Binding Protein MBD1 Isoforms, Mol. Cell. Biol., 1999, 19, 6415–6426 CrossRef CAS PubMed.
- B. Hendrich and A. BIRD, Identification and characterization of a family of mammalian methyl CpG-binding proteins, Genet. Res., 1998, 72, 59–72 CrossRef.
- H. H. Ng, Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg and A. Bird, MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex, Nat. Genet., 1999, 23, 58–61 CrossRef CAS PubMed.
- C. G. Spruijt, F. Gnerlich, A. H. Smits, T. Pfaffeneder, P. W. T. C. Jansen, C. Bauer, M. Münzel, M. Wagner, M. Müller, F. Khan, H. C. Eberl, A. Mensinga, A. B. Brinkman, K. Lephikov, U. Müller, J. Walter, R. Boelens, H. Van Ingen, H. Leonhardt, T. Carell and M. Vermeulen, Dynamic readers for 5-(Hydroxy)methylcytosine and its oxidized derivatives, Cell, 2013, 152, 1146–1159 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra00432a |
| This journal is © The Royal Society of Chemistry 2022 |