Open Access Article
Open Access ArticleCationic conjugated polymer-based FRET aptasensor for label-free and ultrasensitive ractopamine detection†
Xuancheng Fuab,
Wei Dongab,
Chang Liuab,
Jing Han *c and
Chengzhi Huang
*c and
Chengzhi Huang *d
*d
aSchool of Sport Science, Beijing Sport University, Beijing 100084, China
bInstitute of Anti-Doping in China, Beijing Sport University, Beijing 100084, China
cBeijing National Laboratory for Molecular Sciences, Institute of Chemistry Chinese Academy of Sciences, Beijing 100190, P. R. China. E-mail: jinghan@iccas.ac.cn
dKey Laboratory of Luminescent and Real-Time Analytical Chemistry (Southwest University), Ministry of Education, College of Pharmaceutical Sciences, Southwest University, Chongqing 400715, China. E-mail: chengzhi@swu.edu.cn
First published on 7th April 2022
Abstract
A label-free fluorescence resonance energy transfer (FRET) platform based on cationic conjugated polymers and aptamers for ultrasensitive and specific ractopamine detection was constructed. This method exhibited a wide linear range from 0.05 to 500 μM and a low limit of detection of 47 nM, which make it an attractive assay platform for foodborne doping.
Ractopamine (RAC), as a typical type of beta-agonist (β-agonist), is widely utilized for curing human disease.1–3 Owing to its mechanism of promoting the efficiency of lean generation and its potential risks to human health, it has been identified as an S3-type dopant and is banned in most countries around the world.4 However, β-agonists are widely employed as illegal additives driven by the high profit of β-agonists in livestock, which could have serious consequences for athletes along with health risks.5 Hence, from a food safety standpoint, it is urgent to develop ultrasensitive technology to detect RAC in food.
To resolve this food safety dilemma, multiple analytical strategies have been employed to detect the abuse of RAC, including high-performance liquid chromatography (HPLC),6 liquid chromatography-mass spectrometry (LC-MS),7 and gas chromatography-mass spectrometry (GC-MS).8 However, their cost of equipment, highly demanding testing environment, and high time consumption limit their extensive application under limited experimental conditions.9 Therefore, a series of simple and rapid analytical methods for RAC detection, such as enzyme-linked immunosorbent assay (ELISA),10 chemiluminescence11 and electrochemistry,12 have been introduced for RAC analysis. It is worth noting that these technologies commonly require a labelling process, which not only increases the cost of analysis but might also reduce the specificity and affinity of the RAC-related probes.13 Therefore, developing a simple and ultrasensitive method for RAC detection that could avoid the labelling steps is still urgently needed.
Conjugated polymers (CPs) are a type of organic semiconductor material with large delocalized π-conjugated backbones.14 The ideal light-harvested and light-amplified properties of conjugated backbones result in the wide use of CPs in the biological field.15 For better application in a biological environment, cationic conjugated polymers (CCPs) are constructed through the introduction of a positive charge onto the side chain of CPs.16–18 The cation of CCPs provides a wonderful soluble environment in biological solutions, and can also bind with DNA via electrostatic interactions.19 Therefore, a fluorescence resonance energy transfer (FRET) aptasensor based on CCPs could sensitively detect various biological indicators, such as DNA methylation20 and disease-related proteins.21 Meanwhile, the label-free properties of an aptasensor based on genefinder could also decrease the cost of analysis, which might provide a strategy for simple and ultrasensitive RAC detection with relatively low cost.
Herein, we present a label-free FRET aptasensor platform using CCPs, aptamers and genefinder for detecting RAC. As shown in Scheme 1, PFP (poly[(9,9-bis(60-N,N,N-trimethylammonium) hexyl) fluorenylene phenylene dibromide]) is used as an energy donor. The RAC aptamer (A1, 5′-AGTGCGGGC-3′) and complementary DNA (C2, 5′-GCCCGCACT-3′) were designed and pre-formed as dsDNA before the detection process. In the absence of the target molecule, the aptamer still forms a stable dsDNA with C2. Then, the genefinder (GF) could insert into dsDNA and induce an over 100-fold fluorescence enhancement compared with that of the free state. The electrostatic interactions between the positively charged PFP and negative bone of dsDNA shorten the distance of CPs and GF, and thus the fluorescence of PFP can be significantly quenched due to FRET from PFP to GF, resulting in the enhancement of the fluorescence intensity of GF. In the presence of RAC, A1 formed a more specific and stable complex with RAC compared with C2, following the disassembly of dsDNA and the separation of GF from the DNA. Then, the free state of GF finally breaks the FRET of this system. Therefore, the ultrasensitive and quantitative detection of RAC could be realized by valuing the quenching degree of the FRET phenomenon.
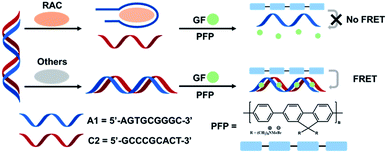 | ||
| Scheme 1 Schematic illustration of the assay for label-free ractopamine detection via the cationic conjugated polymers FRET strategy. | ||
The principle of the FRET phenomenon is that the fluorescence spectrum of the donor molecule overlaps with the absorption spectra of the acceptor molecule, and the molecular distance between the donor and acceptor need to be within the Förster radius.22–24 To match the absorption band of GF, a cationic conjugated polymer (PFP) was selected as the energy donor in this system. As shown in Fig. 1a, the fluorescence and absorption spectra of PFP and GF were first evaluated. A large overlap between the fluorescence spectrum of PFP and the absorption spectrum of GF of around 460 nm is observed. The cationic side chain of PFP could catch the dsDNA through electrostatic interactions, which satisfied the distance requirement of the FRET phenomenon. The spectral overlap and suitable distance provided a good possibility for us to construct an efficient FRET system. Then, the FRET phenomenon was confirmed in the mixed system of PFP, GF, and dsDNA. Under 380 nm excitation, the fluorescence intensity belonging to PFP was decreased, following the appearance of a fluorescence signal around 538 nm (Fig. 1b), which confirmed the FRET phenomenon from donor to acceptor. Compared with the control groups, the fluorescence signal belonging to PFP exhibited the sharpest decrement, following the strongest fluorescence signal enhancement of the GF dye in FRET groups. The fluorescence enhancement of GF before and after inserting into dsDNA was also demonstrated under 490 nm excitation, indicating that the fluorescence of GF could only be detected when GF was inserted into dsDNA. Therefore, the FRET phenomenon proved that PFP could bind to DNA and further act as an energy donor to transfer energy to GF in dsDNA, owning to the band gap overlap between PFP and GF.
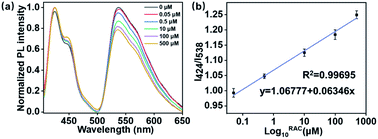
After successfully constructing this label-free FRET platform, its sensitivity during RAC detection was further evaluated. After incubating various amounts of RAC with dsDNA, the RAC competitively assembled with the aptamer and dissociated the dsDNA, which finally induced the disappearance of the FRET phenomenon. Therefore, the amount of RAC could be detected by the quenching degree of this FRET system. As displayed in Fig. 2a, on adding RAC, the emission of GF around 538 nm decreased. By increasing the concentration of RAC, the emission intensity of GF decreased step-wise owing to the competitive interaction between RAC and C2. The ratios of the intensities at 424 nm and 538 nm (I424/I538) were plotted to evaluate the linear response of the FRET system during RAC detection. As shown in Fig. 2b, the results exhibit an excellent logarithmic linear response from 0.05 to 500 μM. An extraordinary limit of detection (LOD) was further determined at 47 nM. This method was also compared with classical existing analytical methods of RAC detection, as shown in Table 1, which shows that our method has good sensitivity. Compared with other RAC detection methods, this FRET strategy exhibited a wider linear range and lower LOD value. Furthermore, it should be pointed out that some of the existing methods require the addition of organic solvents and surfactants, which may limit their biological application. The results of our study have clearly shown that this label-free FRET platform exhibits a relatively wide linear range and high sensitivity during detection.
| Method | Linear range (μM) | Limit of detection (μM) | Reference |
|---|---|---|---|
| Chemiluminescence | 0.059–0.296 | 0.014 | 11 |
| High-performance liquid chromatography | 200–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
50 | 22 |
| Electrochemistry | 0.1–380 | 50 | 12 |
| Gas chromatography-mass spectrometry | 0.073–0.591 | 0.059 | 6 |
| Fluorescence resonance energy transfer | 0.05–500 | 0.047 | This work |
The competency of resisting interference over possibly challenging species is a necessary requirement for wide application in food safety detection. Therefore, the selectivity of this strategy was further determined. Four structurally similar molecules, namely clenbuterol (CLB), salbutamol (SAL), glucose (Glu), and fructose (Fru), were used, whose structures are listed in Fig. 3a. As shown in Fig. 3b, the I424/I538 ratio of the RAC groups exhibited the highest value among all groups at the same concentration of 1 mM. The introduction of similar molecules did not influence the accuracy of the results. All these results clearly prove that this label-free FRET method exhibits high selectivity during RAC determination.
 | ||
| Fig. 3 (a) The structures of RAC and the control molecules. (b) The I424/I538 ratio with RAC and the control molecules. The concentrations of RAC and the control molecules are 500 μM. | ||
To further value the practical application and accuracy of this strategy, we employed this method to detect RAC in energy drinks. Three real samples containing different concentrations of RAC (0.05, 1, and 100 μM) were tested by this method. According to the results of these real samples (Table 2), this method exhibited a good average recovery of 104–112% with an RSD of less than 4.00% in real samples. Therefore, the application behavior demonstrates that this label-free FRET system based on CCPs and aptasensors can be utilized to detect RAC in real samples following a simple and cheap process. Furthermore, by replacing the RAC-targeting aptamer to other dopant-specific aptamers, this FRET strategy could be easily employed to detect various dopants.
| Sample | Added (μM) | Found (μM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 0.0500 | 0.0560 | 112 | 3.48 |
| 2 | 1.00 | 1.08 | 108 | 3.66 |
| 3 | 100 | 104 | 104 | 1.74 |
Conclusions
In summary, we built a label-free FRET aptasensor for RAC detection that could ultrasensitively detect RAC through a simple process. A dsDNA molecule, including a RAC-aptamer and complementary DNA, was pre-constructed, and the GF dye could insert into this dsDNA and further enhance its fluorescence signal. Then, a cationic conjugated polymer (PFP) was selected as a signal amplifier and added to this system to build a FRET platform via electrostatic interaction and a suitable band gap overlap. The introduction of RAC into this system could disassemble the dsDNA and form a more stable complex with the RAC-aptamer, which induces the separation of GF from DNA and ultimately results in the quenching of the FRET phenomenon. This method exhibited high sensitivity and selectivity during detection. Furthermore, a good average recovery of 104–112% and an RSD of less than 4.00% demonstrate the application potential of this method. Owing to the avoidance of any special labelling process, including modification, coating, immobilization, and separation, this method simplifies the detection process and reduces testing costs. Therefore, we believe that this label-free FRET aptasensor based on CCPs could be widely applied in RAC detection and could be further expanded for the on-site screening of harmful food additives due to its simple process, low cost as well as excellent sensitivity and selectivity.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, No. 22134005) and the Chongqing Talents Program for Outstanding Scientists (cstc2021ycjh-bgzxm0178).Notes and references
- L. Wang, Z. Zeng, X. Huang, Y. Su and L. He, RSC Adv., 2014, 4, 25719 RSC.
- G. Cao, F. Xu, S. Wang, K. Xu, X. Hou and P. Wu, Anal. Chem., 2017, 89, 4695 CrossRef CAS PubMed.
- M. Xu, X. Qian, K. Zhao, A. Deng and J. Li, Sens. Actuators, B, 2015, 215, 323 CrossRef CAS.
- S. Yan, Z. Wang, H. Yen, Y. Lee and M. Yin, Food Chem. Toxicol., 2016, 98, 119 CrossRef CAS PubMed.
- Z. Zhang, Y. Zhang, R. Song, M. Wang, F. Yan, L. He, X. Feng, S. Fang, J. Zhao and H. Zhang, Sens. Actuators, B, 2015, 211, 310 CrossRef CAS.
- J. P. Wang, X. W. Li, W. Zhang and J. Z. Shen, Chromatographia, 2006, 64, 613 CrossRef CAS.
- C. Li, Y.-L. Wu, T. Yang, Y. Zhang and W.-G. Huang-Fu, J. Chromatogr. A, 2010, 1217, 7873 CrossRef CAS PubMed.
- J. Cheng, S. Wang and X. O. Su, PLoS One, 2013, 8, e76400 CrossRef CAS PubMed.
- Q. Zhu, H. Liu, J. Zhang, K. Wu, A. Deng and J. Li, Sens. Actuators, B, 2017, 243, 121 CrossRef CAS.
- X. Li, W. Wang, L. Wang, Q. Wang, X. Pei and H. Jiang, Anal. Bioanal. Chem., 2015, 407, 7615 CrossRef CAS PubMed.
- G. Zhang, Y. Tang, J. Shang, Z. Wang, H. Yu, W. Du and Q. Fu, Luminescence, 2015, 30, 102 CrossRef CAS PubMed.
- Y. Orooji, P. N. Asrami, H. Beitollahi, S. Tajik, M. Alizadeh, S. Salmanpour, M. Baghayeri, J. Rouhi, A. L. Sanati and F. Karimi, J. Food Meas. Charact., 2021, 15, 4098 CrossRef.
- F. Pu, Z. Huang, D. Hu, J. Ren, S. Wang and X. Qu, Chem. Commun., 2009, 7357 RSC.
- Z. Li, W. Lu, S. Jia, H. Yuan and L.-H. Gao, ACS Appl. Bio Mater., 2021, 4, 370 CrossRef CAS PubMed.
- Y. Wang, L. Feng and S. Wang, Adv. Funct. Mater., 2019, 29, 1806818 CrossRef.
- H. Zhang, Y. Liang, H. Zhao, R. Qi, Z. Chen, H. Yuan, H. Liang and L. Wang, Macromol. Biosci., 2020, 20, 1900301 CrossRef CAS PubMed.
- Z. Chen, H. Yuan, H. Liang, C. Lu and X. Liu, Chinese Chem. Lett., 2018, 29, 339 CrossRef CAS.
- X. Fu, H. Bai, F. Lyu, L. Liu and S. Wang, Chem. Res. Chinese Univ., 2020, 36, 237 CrossRef CAS.
- C. Zhu, L. Liu, Q. Yang, F. Lv and S. Wang, Chem. Rev., 2012, 112, 4687 CrossRef CAS PubMed.
- L. Ma, Y. Huang, H. Zhang, W. Ning, R. Qi, H. Yuan, F. Lv, L. Liu, C. Yu and S. Wang, ACS Appl. Mater. Interfaces, 2021, 13, 9291 CrossRef CAS PubMed.
- R. Chai, C. Xing, J. Qi, Y. Fan, H. Yuan, R. Niu, Y. Zhan and J. Xu, Adv. Funct. Mater., 2016, 26, 9026 CrossRef CAS.
- A. J. Kell, L. Pagé, S. Tan, I. Charlebois, M. Boissinot, M. LeClercc and B. Simarda, Nanoscale, 2011, 3, 3747 RSC.
- P. Zeng, P. Hou, C. Jing and C. Huang, Talanta, 2018, 185, 118 CrossRef CAS PubMed.
- F. You, W. Tanga and L. L. Yun, Nanoscale, 2018, 10, 7726 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00574c |
| This journal is © The Royal Society of Chemistry 2022 |