Open Access Article
Open Access ArticleA gold nanoparticle-based visual aptasensor for rapid detection of acetamiprid residues in agricultural products using a smartphone†
Chengnan Xu‡
 *a,
Mei Lin‡a,
Chaonan Songb,
Danli Chenb and
Caimiao Bian*b
*a,
Mei Lin‡a,
Chaonan Songb,
Danli Chenb and
Caimiao Bian*b
aZhejiang Citrus Research Institute, Taizhou, 318026, China. E-mail: xcnpub@163.com
bSchool of Life Science, Taizhou University, Taizhou, 318001, China. E-mail: blancaimiao@tzc.edu.cn
First published on 16th February 2022
Abstract
Based on the colorimetric analysis of gold nanoparticles and a smartphone readable strategy, a stable, sensitive, and visual method was established for rapid detection of acetamiprid residues in agricultural products. By optimizing the key parameters, the detection process only took 40 minutes with good specificity. The acetamiprid aptamer can help AuNPs to resist salt-induced aggregation. Conversely, in the presence of acetamiprid, the anti-protection is weakened and the AuNPs aggregated with the color change of the solution. The photographs of the solution are recorded by the smartphone and analyzed through image processing. In the range from 25 to 300 μM the method can realize a quantitative analysis of acetamiprid, and the detection limit is about 3.81 μM. Excellent recoveries are taken in samples of cucumber, cabbage, and river water, ranging from 96.78% to 129.95%. These results show no significant difference from the results obtained by the microplate reader. What's more, the method employs a smartphone to read without the assistance of professional equipment, which greatly reduces the cost of detection, and shows a promising application prospect for on-site rapid detection of acetamiprid.
1. Introduction
Acetamiprid is a kind of broad-spectrum systemic pesticide that acts on the nicotinic acetylcholine receptor; it interferes with the transmission of a stimulus and causes accumulation of neurotransmitters, finally resulting in the paralysis and death of pests.1,2 In this way, insects are efficiently killed, which is of low toxicity to mammals.3 Thus, acetamiprid is widely used to control various pests but a residue of acetamiprid has occurred in agricultural products and the environment due to the overuse in modern agriculture.4 The accumulated acetamiprid not only has high toxicity to organisms but also poses potential risks to human health through food.5,6 Therefore, it has been a serious concern and it is urgent to develop sensitive and rapid acetamiprid detection methods for agricultural safety and environmental monitoring. For this reason, numerous methods have been established for the quantitation of acetamiprid. Chromatographic techniques are widely applied as a conventional detection method to determine acetamiprid residues due to their sensitivity and accuracy. However, these methods are time-consuming, tedious, and require expensive instruments and professional operators, which makes them unsuitable for on-site detection. Hence, more innovative and reliable sensing methods including electrochemical method,7 fluorescence method,8 immunoassay,9 and colorimetric method10 have been developed, which greatly reduce the dependence on large instruments through assistant equipment is still needed. Among them, the colorimetric method has great potential in the rapid detection of pesticide residues due to its unique characteristics such as simple operation, cost-effectiveness, fast response, and so on.The colorimetric sensing method is normally based on the absorbance change and is sometimes accompanied with color changes. The step of signal conversion plays a key role in the colorimetric assay, normally it is nanomaterials or chromogenic reactions. Due to its unique optical properties and high extinction coefficient,11,12 gold nanoparticles (AuNPs) are commonly used as the signal conversion element during the colorimetric sensing process. The AuNPs-based colorimetric assay relies on the absorbance change and color variation caused by the state change of AuNPs, which is reliable and easy to operate, thus it is popularly employed in pesticide residue detection.13,14 Based on the above colorimetric method, researchers have applied intelligent strategies to achieve rapid detection, such as naked eyes visual inspection, test strip, and digital recognition.15–18 However, these colorimetric detections mostly are qualitative/semi-quantitative or still in need of professional instruments.
As a daily essential electronic device, the smartphone has gained greatly attractive attention in the fields of medical health, environmental monitoring, and food safety.19 The smartphone is equipped with numerous modules that can be utilized for measuring, for example, high-efficiency processor, high-resolution camera, open software platform, and so on. In the past decade, lots of works have been done to push the development of smartphone-based colorimetric assays in sensitive detection of various species including small molecules,20,21 pesticides,22 proteins,23,24 and pathogens.25 By adding the necessary components, the smartphone is expected to become a portable detection platform, especially showing great application prospects for on-site detection.
Herein, we proposed an instrument-free visual assay for acetamiprid detection with the smartphone readout strategy. As well known, AuNPs synthesized by sodium citrate reduction can remain stable and not aggregate under the protection of surface citrate electrostatic repulsion. However, under high salt concentration condition, the repulsion between particles is weakened and aggregation occurs, meanwhile the color of the solution changes. The sensing mechanism is displayed in Scheme 1, when acetamiprid aptamer is introduced to the above solution, the aptamer can help AuNPs to resist salt-induced aggregation. On the contrary, when acetamiprid presents in the solution, the anti-protection will be weakened due to the specific binding between aptamer and acetamiprid. Eventually, AuNPs aggregated causing the color of the solution to change from red to blue-purple. The photographs of the solution are obtained by the smartphone, then the acetamiprid can be quantitatively detected through image processing.
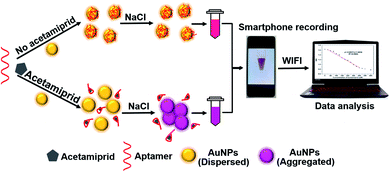 | ||
| Scheme 1 Schematic illustration of instrument-free visual method for acetamiprid detection with smartphone readout strategy. | ||
2. Materials and methods
2.1 Materials
Hydrogen tetrachlorocuprate hydrate (HAuCl4·3H2O), and sodium citrate tribasic dihydrate (Na3C6H5O7·2H2O) were purchased from Sigma-Aldrich (St Louis, MO, USA). Acetamiprid was purchased from Aladdin Chemistry Co., Ltd. (Shanghai, China). Tris–HCl buffer (pH = 7.5) was purchased from Sangon Biotech Company, Ltd. (Shanghai, China). Glyphosate, diquat, paraquat, chloramphenicol, and tetracycline were purchased from Tan-Mo Technology Co., Ltd. (Changzhou, China). The acetamiprid aptamer with the sequence of 5′-CTGACACCATATTATGAAGA-3′ was purchased from Sangon Biotech Company, Ltd. (Shanghai, China). The polypropylene centrifuge tubes were purchased from KIRGEN Biotechnology Co., Ltd. (Shanghai, China). Tris-buffer solution (20 mM Tris–HCl, 50 mM NaCl) was used as working buffer. All other analytical grade reagents and ultrapure water were used in the experiments. Cabbage and cucumber were procured from a local supermarket, and the river water was taken from YongNing River.2.2 Instruments
The UV-vis absorption spectra were recorded by a ReadMax 1900 Microplate reader (Flash Spectrum, Shanghai, China). For all measurements, the absorption spectrum for 280 μL of the sample was scanned from 300 to 999 nm. All the photographs were taken with a smartphone of Honor V40. For transmission electron microscopy (TEM) characterization, the images of nanoparticles were obtained by a JEM-1200EX microscopy (JEOL, Tokyo, Japan). The TEM sample was carried out by dropping 5 μL of the sample solution on a carbon-coated copper grid fixed with a tweezer and then drying out at room temperature.2.3 Synthesis of AuNPs
AuNPs were chemically synthesized via a citrate reduction method according to the previous report and the concentration was estimated to be 13 nM.262.4 Colorimetric detection of acetamiprid
The typical acetamiprid detection process was as follows, acetamiprid aptamer was heated at 95 °C for 10 min and quickly cooled down to 25 °C. Then, the aptamer mixed with different concentrations of acetamiprid were incubated at 37 °C for 30 min. After that, 60 μL of these incubated solutions were mixed with 220 μL of diluted AuNPs solution and incubated at 37 °C for 5 min. Then, 20 μL of NaCl solution (0.6 M) was added to these above solutions ensuring that the final concentration of NaCl was 50 mM and incubated for another 5 min at 37 °C. Finally, the photographs of these reacted solutions were quickly recorded, and the 280 μL of above solutions were transferred to 96-well plates for UV-vis characterization.2.5 Signal recording with a smartphone
For simple operation, the sample solutions were held in the polypropylene centrifuge tubes. And the tubes were placed in a 3D printed black box, illuminated with two LED lamps (shown in Fig. S1†). The lamps with a power of 1.0 W were fixed on bottom of the box to ensure homogeneous illumination. Photographs were taken with the built-in camera of the smartphone zoomed at 2.0×. Each sample was individually photographed and repeated at three times for whole experiment. Then the photographs were sent to the computer via WIFI and analyzed on the software Photoshop to read the RGB values for calculation. In order to ensure the accuracy of the points, the surrounding light-transmitting area and the middle reflective part were discarded. Out of these areas, 10 points with one pixel were evenly taken from the left and right areas of each sample by an experimentalist. And the average RGB values of total 10 points were set as the RGB values of each sample.2.6 Visual detection of acetamiprid in real samples
To verify the reliability of the method, real environmental and agricultural products samples including cabbage, cucumber, and river water samples were pretreated and spiked with the quantitatively standard acetamiprid solution according to the previous report.27,28 Briefly, filter the river water after standing still, and then pass it through a 0.22 μM membrane. The cucumber and cabbage samples were made into a paste with a homogenizer, added with an appropriate amount of buffer, and centrifuged at 4000 rpm for 20 minutes. The precipitate was removed and the supernatant was filtered with a membrane. Finally, a certain amount of acetamiprid was spiked into the pretreated samples, then the photographs and absorption spectra of the solutions were carried out.3. Results and discussions
3.1 Characterizations of the visual acetamiprid detection assay
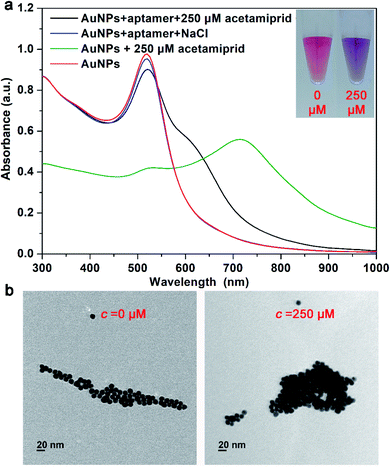
To verify the detection principle was based on the existing state changes of AuNPs caused by acetamiprid and aptamer, the corresponding absorption spectrum of AuNPs was recorded. The result is shown in Fig. 1a, the absorption peak of the synthesized AuNPs solution (red line) is at 520 nm. When 250 μM acetamiprid is added to the AuNPs solution (green line), the absorption peak at 520 nm significantly decreases with an obvious aggregation peak appearing at 730 nm. It showed that acetamiprid has no protection ability to AuNPs, but to accelerate the AuNPs aggregation. In the case of aptamer and NaCl (blue line) are added at the same time when no acetamiprid exists, the absorption peak at 520 nm only slightly decreases with no aggregation peak shows, indicating that the aptamer can effectively protect AuNPs against the aggregation caused by high salt concentration. Adding 250 μM acetamiprid to the above solution (black line), its absorption peak at 520 nm decreases, and an aggregation peak appears at 630 nm. Thus, we define the intensity ratio of the absorption peak of AuNPs at 520 nm and 630 nm (A520/A630) as the degree of AuNPs aggregation, and the value is negatively correlated to the aggregation degree. It can be inferred that when the aptamer specifically binds to acetamiprid, AuNPs lose the protection of aptamer and are aggregated under high salt concentration conditions. The color of the solution changes from red to blue-purple, which proves this hypothesis (as shown in the embedded photo). And the TEM results further prove the assay is based on the existing state changes of AuNPs (shown in Fig. 1b).3.2 Optimization of the visual assay
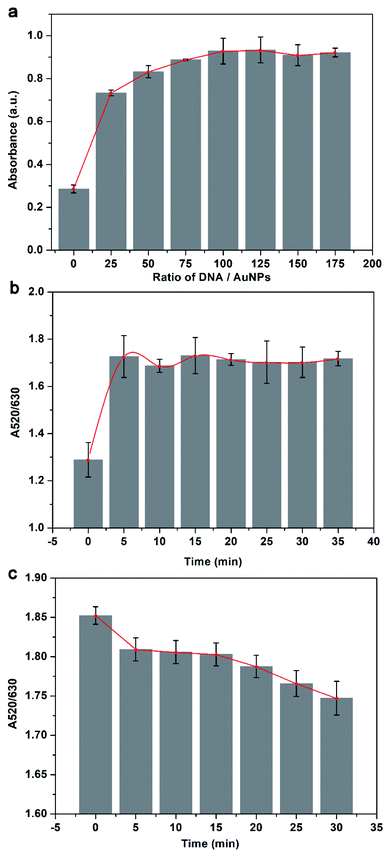
To obtain the optimal detection performance, the crucial experimental parameters of the visual acetamiprid detection assay were optimized. In this work, the main factors that affect the assay are the concentration ratio of aptamer probe and AuNPs, the reaction time between AuNPs and aptamer, and the detection time after salting. As shown in Fig. 2a, at the beginning the intensity of the absorption peak at 520 nm increases as the concentration ratio increases, but when the concentration ratio exceeds 100, the intensity value remains unchanged. It suggests that the protection of the aptamer is saturated when the concentration ratio reaches 100, so the concentration ratio is applied to be 100. For nanoparticle aggregation-based assay, false positive results and low sensitivity may cause by insufficient conjugation between AuNPs and aptamer. The result gives in Fig. 2b, the process of AuNPs conjugates with the aptamer is fast and full conjugation takes only 5 min, thus the incubation time is chosen to be 5 min for saving assay time. Besides, it is essential to determine the measuring time after salting since the salt-induced aggregation goes on with time. It can be seen from Fig. 2c that the A520/A630 continues to decline overall but keeps stable from 5 to 15 min, indicating that the salt-induced aggregation achieves short-lived dynamic equilibrium in 5 to 15 min. Therefore, the best detection time is 5 minutes after salting.3.3 Analytical performance
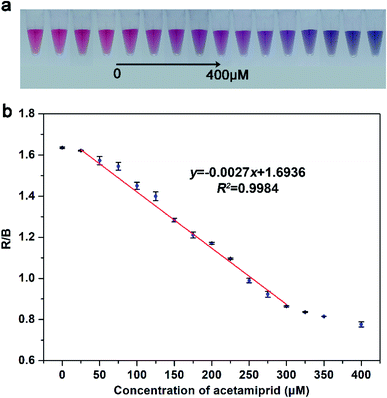
To investigate the actual detection performance, the developed visual assay method was used to quantitatively analyze different concentrations of acetamiprid. Based on the above-optimized conditions, a smartphone was employed to record photographs of the solution after adding with acetamiprid, then the data were sent to the computer via WIFI and analyzed on the software Photoshop. Fig. 3a displays the photograph of the AuNP solutions after adding with different concentrations target acetamiprid ranging from 0 to 400 μM. With the increment of target acetamiprid increases, the color of AuNPs solution gradually changes from pink to light purple, and finally to blue-violet. The software Photoshop was applied to analyze the three-channel changes (respectively red (R), green (G), and blue (B)) of the photograph. Among them, the R and B channels respond significantly to the concentration change of acetamiprid, so the R/B value is used as the detection index (ESI Fig. S2†). As Fig. 3b shows that a good linear correlation between the R/B value and concentration of target acetamiprid is obtained in the range of 25–300 μM. The linear regression equation is fitted as y = −0.0027x + 1.6936, with R2 = 0.9984, where y and x represent the R/B value and the concentration of target acetamiprid, respectively, and R2 is the correlation coefficient. The error bars represent the standard deviation of RGB values for three times repeat of each sample. The estimated limit of detection (LOD) is evaluated to 3.81 μM (3 times determinations of the blank solution). These results are similar to the results obtained by a microplate reader (Fig. S3†). The remarkable aggregation phenomenon of AuNPs is emerging for that the absorption peak at 630 nm increasingly grows and reduces at 520 nm upon addition of acetamiprid. Furthermore, a good linear correlation exists in the range from 0 to 220 μM and the linear regression equation obtained from absorption spectrum is fitted as z = −0.0183c + 5.8191, with R2 = 0.9924, where z and c represent the A520/A630 value and the concentration of target acetamiprid, respectively, and the calculated LOD is about 2.61 μM. Compare with results obtained by the microplate reader, the visual assay method recording by the smartphone can achieve the sensitivity in the same concentration level, meanwhile, it has a wider detection range compared with reported methods (shown in Table S1†).3.4 Specificity of the visual sensing and practical performance in real samples
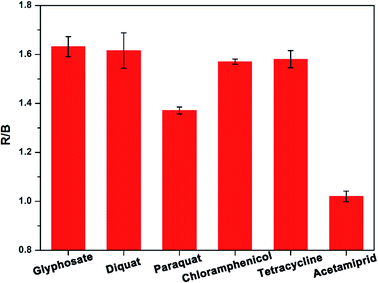
To explore the specificity of detection, we employed the established visual sensing method to detect acetamiprid and five other substances (i.e. glyphosate, diquat, paraquat, tetracycline, and chloramphenicol). The experimental results are shown in Fig. 4, it can be observed that the R/B values obtained from acetamiprid and the other five substances are significantly different, and only acetamiprid can cause a greater response, which indicates that the visual assay method detects acetamiprid has good selectivity.Cabbage, cucumber, and river water were taken as actual environmental samples to carry out the recovery experiments, those three blank samples were spiked with two concentration levels of acetamiprid for assay to evaluate the practical detection performance of our method. As summarized in Table 1, the recoveries of our developed visual assay for acetamiprid range from 96.78% to 129.95%, and an acceptable relative standard deviation (RSD) also is received. Besides, the results were compared with those gained by the microplate reader, and no significant difference was found between the two methods. The results suggest that the developed visual assay method is reliable and stable for acetamiprid residues detection in the real environmental sample. Worthwhile, the smartphone replaces the large instrument microplate reader, which greatly reduces the detection cost and restriction, and it is expected to achieve on-site real-time detection of pesticide residue in agricultural products.
| Samples | Spiked (μM) | Detected (μM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Cabbage | 0 | Undetected | ||
| 50 | 48.39 | 96.78 | 1.05 | |
| 200 | 254.23 | 127.12 | 1.71 | |
| Cucumber | 0 | Undetected | ||
| 50 | 50.21 | 100.42 | 1.33 | |
| 200 | 248.71 | 124.35 | 0.51 | |
| River water | 0 | Undetected | ||
| 50 | 53.26 | 106.53 | 2.03 | |
| 200 | 259.91 | 129.95 | 1.33 | |
4. Conclusion
In summary, we developed a visual sensing method for the detection of acetamiprid residues in agricultural products, which is convenient to operate and time-saving (only takes 40 minutes). The smartphone replaces the large instrument microplate reader to achieve quantitative determination of acetamiprid, which greatly reduces the detection cost and restriction. At the same time, this method is stable and reliable, with good sensitivity and high specificity. Compare with the microplate reader, the visual assay method recording by the smartphone can achieve the sensitivity in the same concentration level, meanwhile, it has a wider detection range. Furthermore, the successful application for acetamiprid detection in real environmental samples demonstrates visual sensing method established in this paper has a good field application prospect in on-site real-time detection. What's more, the visual sensing method can provide technical support for the rapid detection of pesticide residues in agricultural products and it is practical for the quality and safety evaluation of agricultural products.Conflicts of interest
There are no conflict of interest to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 31801642), the National Agricultural Product Quality and Safety Risk Assessment Plan (No. GJFP2019012) and the Taizhou Municipal Science and Technology Bureau (No. 20ny20).References
- Y. Qi, Y. Chen, F.-R. Xiu and J. Hou, Sens. Actuators, B, 2020, 304, 127359 CrossRef CAS.
- Z. Jiao, H. Zhang, S. Jiao, Z. Guo, D. Zhu and X. Zhao, Food Anal. Methods, 2019, 12, 668–676 CrossRef.
- Z. Yang, J. Qian, X. Yang, D. Jiang, X. Du, K. Wang, H. Mao and K. Wang, Biosens. Bioelectron., 2015, 65, 39–46 CrossRef CAS PubMed.
- L. Zeng, D. Zhou, J. Wu, C. Liu and J. Chen, Anal. Methods, 2019, 11, 1168–1173 RSC.
- K. Fan, W. Kang, S. Qu, L. Li, B. Qu and L. Lu, Talanta, 2019, 197, 645–652 CrossRef CAS.
- Y. Guo, F. Yang, Y. Yao, J. Li, S. Cheng, H. Dong, H. Zhang, Y. Xiang and X. Sun, J. Hazard. Mater., 2021, 401, 123794 CrossRef CAS PubMed.
- J. Yi, Z. Liu, J. Liu, H. Liu, F. Xia, D. Tian and C. Zhou, Biosens. Bioelectron., 2020, 148, 111827 CrossRef CAS PubMed.
- Q. Yu, C. He, Q. Li, Y. Zhou, N. Duan and S. Wu, Microchim. Acta, 2020, 187, 1–10 CrossRef PubMed.
- J. Li, W. Sun, Y. Qin, P. Cui, G. Song, X. Hua, L. Wang and M. Wang, Food Agric. Immunol., 2021, 32, 740–753 CrossRef CAS.
- L. Yang, X. Wang, H. Sun, W. Yao, Z. Liu and L. Jiang, Anal. Chim. Acta, 2021, 1150, 238118 CrossRef CAS PubMed.
- S. Zeng, K.-T. Yong, I. Roy, X.-Q. Dinh, X. Yu and F. Luan, Plasmonics, 2011, 6, 491–506 CrossRef CAS.
- C.-W. Liu, Y.-T. Hsieh, C.-C. Huang, Z.-H. Lin and H.-T. Chang, Chem. Commun., 2008, 2242–2244 RSC.
- I. C. Sulaiman, B. Chieng, M. Osman, K. Ong, J. Rashid, W. W. Yunus, S. Noor, N. Kasim, N. Halim and A. Mohamad, Microchim. Acta, 2020, 187, 1–22 CrossRef PubMed.
- R. Singh, P. Thakur, A. Thakur, H. Kumar, P. Chawla, J. V. Rohit, R. Kaushik and N. Kumar, Int. J. Environ. Anal. Chem., 2021, 101, 3006–3022 CrossRef CAS.
- Y. Zhou, C. Li, R. Liu, Z. Chen, L. Li, W. Li, Y. He and L. Yuan, ACS Biomater. Sci. Eng., 2020, 6, 2805–2811 CrossRef CAS PubMed.
- W. Wang and H. Ouyang, Microchem. J., 2019, 149, 104055 CrossRef CAS.
- H. Ouyang, M. Wang, W. Wang and Z. Fu, Sens. Actuators, B, 2018, 266, 318–322 CrossRef CAS.
- Y. Li, L. Liu, S. Song and H. Kuang, Food Agric. Immunol., 2018, 29, 14–26 CrossRef CAS.
- S. Kanchi, M. I. Sabela, P. S. Mdluli and K. Bisetty, Biosens. Bioelectron., 2018, 102, 136–149 CrossRef CAS PubMed.
- C. Dong, Z. Wang, Y. Zhang, X. Ma, M. Z. Iqbal, L. Miao, Z. Zhou, Z. Shen and A. Wu, ACS Sens., 2017, 2, 1152–1159 CrossRef CAS PubMed.
- A. Amirjani and D. H. Fatmehsari, Talanta, 2018, 176, 242–246 CrossRef CAS PubMed.
- Y. Chen, Q. Fu, D. Li, J. Xie, D. Ke, Q. Song, Y. Tang and H. Wang, Anal. Bioanal. Chem., 2017, 409, 6567–6574 CrossRef CAS.
- F. Wang, Y. Lu, J. Yang, Y. Chen, W. Jing, L. He and Y. Liu, Analyst, 2017, 142, 3177–3182 RSC.
- H. Li, M. Yang, D. Kong, R. Jin, X. Zhao, F. Liu, X. Yan, Y. Lin and G. Lu, Sens. Actuators, B, 2019, 282, 366–372 CrossRef CAS.
- L. Zheng, G. Cai, S. Wang, M. Liao, Y. Li and J. Lin, Biosens. Bioelectron., 2019, 124, 143–149 CrossRef PubMed.
- J. Liu and Y. Lu, Nat. Protoc., 2006, 1, 246–252 CrossRef CAS PubMed.
- Y. Sun, Z. Li, X. Huang, D. Zhang, X. Zou, J. Shi, X. Zhai, C. Jiang, X. Wei and T. Liu, Biosens. Bioelectron., 2019, 145, 111672 CrossRef CAS PubMed.
- M. P. Shirani, B. Rezaei and A. A. Ensafi, Spectrochim. Acta, Part A, 2019, 210, 36–43 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00348a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |