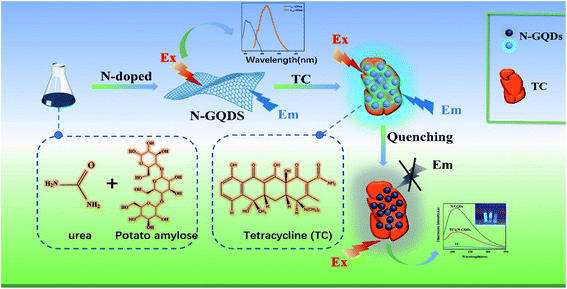
Open Access Article
Open Access ArticleGreen synthetic nitrogen-doped graphene quantum dot fluorescent probe for the highly sensitive and selective detection of tetracycline in food samples
Huanyu Xiea,
Yudong Lu *a,
Ruiyun Youa,
Wei Qian*b and
Shan Linc
*a,
Ruiyun Youa,
Wei Qian*b and
Shan Linc
aCollege of Chemistry and Materials Science, Fujian Provincial Key Laboratory of Advanced Oriented Chemical Engineer, Fujian Key Laboratory of Polymer Materials, Fujian Normal University, Fuzhou, Fujian 350007, China. E-mail: luyd@fjnu.edu.cn
bResearch Centre of Wetlands in Subtropical Region, School of Geographical Sciences, Fujian Normal University, Fuzhou, Fujian 350007, China. E-mail: fjnuqw@fjnu.edu.cn
cJiangxi Province Key Laboratory of Polymer Micro/Nano Manufacturing and Devices, East China University of Technology, Nanchang, Jiangxi 330013, China
First published on 14th March 2022
Abstract
Tetracycline (TC) is a broad-spectrum antibiotic. When humans consume too much food containing tetracycline residues, it can be a serious health hazard. Therefore, it is essential to develop a strategy to detect TC. In this study, we prepared light blue-green luminescent nitrogen-doped graphene quantum dots (N-GQDs) by a hydrothermal method using the natural products potato straight-chain starch and urea as precursors; the fluorescence quantum yield of the prepared N-GQDs was 5.2%. We investigated the detection of tetracycline (TC) by this N-GQD fluorescent sensor based on the internal filtration effect (IFE) of TC on N-GQDs. The reaction is green, simple and no other contaminating products are present. A good linear relationship was established between the relative fluorescence intensity ratio of the system and the logarithm of the TC concentration of 2.5 × 10−10 to 5 × 10−6 M (R2 = 0.9930), with a detection limit of 9.735 × 10−13 M. The method has been used to analyze TC in three real food samples (whole milk, skim milk, honey) with low detection limits (3.750 × 10−11 to 2.075 × 10−9 M), wide linear range, and satisfactory recoveries of 93.80–109.20% were obtained. In conclusion, the proposed method is a green, rapid, highly sensitive and selective method for the detection of tetracycline in real food samples, demonstrating the potential application of N-GQDs in food detection.
1. Introduction
Tetracycline (TC) is a broad-spectrum antibiotic commonly used in the treatment of infections caused by different pathogenic bacteria, and is commonly used to prevent bacterial infections and treat infections.1 However, the excessive consumption of human food containing TC or improper handling of residues can be harmful to human health.2,3 Therefore, we urgently need a method to quickly detect TC in different samples. Currently, scientists have developed several conventional methods for detecting TC residues. The routine detection techniques for TC include liquid chromatography (HPLC),4 surface enhanced Raman spectroscopy (SERS),5 flow injection analysis,6 capillary electrophoresis (CE),7,8 colorimetric analysis9 and enzyme-linked immunosorbent assay (ELISA).10 These detection methods have the disadvantages of complex pre-processing, time-consuming and expensive, low sensitivity, and long detection time, which limit its application and make it challenging in practical applications. Therefore, there is an urgent need to develop a simple, economical and highly sensitive TC sensor. Compared with the above methods, fluorescence spectrometry has the advantages of simple operation, high sensitivity, low cost and quantitative detection.Graphene quantum dots (GQDs) are fluorescent materials less than 100 nm in size.11–14 Scientists can bring fluorescence to graphene by converting 1D graphene into 0D GQDs or by chemical surface modification methods such as graphene oxide (GO) modification, reduced graphene (r-GO), doped r-GO, etc15. Compared to organic dyes and fluorescent proteins, nanomaterials fluorescent quantum dots (GQDs) have significant advantages due to their special physical and chemical properties, such as high emission quantum yield, anti-light bleaching, tunable photoluminescence (PL),16 molar extinction high coefficients, good photostability and good biocompatibility.17 The raw materials for many synthetic quantum dots are biomass with the advantages of being environmentally friendly, green, low cost, high carbon content, and rich in functional groups, which are excellent carbon sources for synthesizing GQDs.18,19 So far, scientists have used different biomass-based feedstocks such as graphite waste,20 green tea extract,21 orange juice,22 rice husk,23 honey,24 and many other biomass-based materials for the preparation and production of GQDs. Many studies have shown that the domain of GQDs can be adjusted by doping heteroatoms to adjust the energy band gap of GQDs. For example, nitrogen, chlorine, fluorine and sulfur can be reacted with GQDs at the early stage of synthesis. Probably owing to the interaction of defective clusters and different dopants.25,26 Nitrogen-doped graphene quantum dots (N-GQDs) can control their electronic properties and their fluorescence properties due to the doping of chemically bonded N atoms.27 N-GQDs with their special fluorescence and chemical properties have a broad range of applications in the detection of toxins and antibiotics, bioimaging, photocatalysts, photodetectors, electrochemiluminescence, optical sensors;28–33 GQDs doped with nitrogen exhibit superior fluorescence properties, with pyrrolic-N and pyridinic-N being the main contributors to their enhanced electrocatalytic activity, and graphitic-N also having strong electrocatalytic activity due to its inherent strong electrical conductivity However, recent reports suggest that the enhanced fluorescence is largely due to the formation of pyrrolic-N the fluorescence enhancement has been recently reported to be due in large part to the formation of nitrogen.34,35 In recent years, Qu et al. proposed a blue fluorescent carbon quantum dot for the indirect detection of tetracycline in foods;36 Yang and coworkers prepared N-GQDs with blue fluorescence emission for the detection of tetracycline.37 However, the lack of sensitivity of the above detection methods remains to be addressed. Ju et al. reported a method for the synthesis of N-GQDs from citric acid and dicyandiamide, which requires a complex post-treatment process;38 Fan and co-workers proposed to obtain graphene quantum dots by oxidizing graphene flakes with concentrated nitric acid for 20 h and then reducing them with NaBH4 for 12 h. The acid residue of the product in this method is not easy to handle and the long experimental time is still to be solved.39 Therefore, despite the efforts of scientists, there is a need to further develop a highly sensitive, simple and green low-cost method for the synthesis of N-GQDs in order to overcome these drawbacks.
In this article, we have developed an N-GQD fluorescent sensor utilizing the natural products potato straight-chain starch and urea as the carbon and nitrogen sources, respectively, and used for rapid, accurate and highly sensitive TC detection (Scheme 1). The structure and size of the prepared N-GQDs are promising, and the fluorescence intensity of N-GQDs depends on the excitation wavelength with wavelength dependence. And N-GQDs exhibited intense blue fluorescence and stable photoluminescence with a quantum yield of 10.2%, good water solubility and stability with high PL emission when irradiated with UV lamp in aqueous solution. In addition, the fluorescence of N-GQDs was effectively quenched based on the internal filtering effect of TC on N-GQDs (IFE). The proposed N-GQDs can be used as a fluorescent sensor for rapid detection of TC with a possible detection limit of 9.375 × 10−13 M. Compared with the reported methods, this detection method exhibits more excellent characteristics with higher sensitivity and high selectivity for the target. It provides a new and effective method for the simple, rapid and economical detection of TC.
2. Experimental section
2.1. Chemicals and materials
Potato starch (s112495) was purchased from Shanghai Aladdin Biochemistry (Shanghai, China), urea from Shanghai Aladdin Biochemistry Co. (Shanghai, China), tetracycline was purchased from Shanghai Aladdin Biochemical Co., Ltd, milk and honey (within shelf life) was purchased from supermarkets (Fuzhou, China). Cysteine (Cys), tyrosine (Tyr), aspartic acid (Asp), lysine (Lys), threonine (Thr), glutamic acid (Glu) were purchased from Aladdin Chemical Reagents (Shanghai, China). Tetracycline, hydrochloride, tobramycin, oxytetracycline, kanamycin sulphate, chloramphenicol, amikacin, streptomycin, gentamicin, metronidazole were purchased from Shanghai Maclean's Biochemical Technology Co Ltd (Shanghai, China). NaCl, KCl, CuCl2, FeCl2, CaCl2, MgCl2 were purchased from Aladdin Chemical Reagents (Shanghai, China). Analytical grade chemicals and reagents were used throughout the experiment, and the solutions were prepared with distilled water (Guozhiyuan Y1/2-10UV Kaitong, Changsha, China).2.2. Instruments
The UV1902 spectrophotometer was used to record the UV-Vis absorption spectra at room temperature (Shanghai cold light technology, China), Transmission electron microscopy (TEM) observations was purchased from Hitachi (Tokyo, Japan), Thermo Scientific K-Alpha was used to conduct X-ray photoelectron spectroscopy (XPS) investigations (Shimadzu, UK); Hitachi F-7000 Fluorescence Spectrophotometer was purchased from Hitachi (Tokyo, Japan), PHC-3C type pH meter purchased from YOKE Instruments (Shanghai, China), Edinburgh – Steady state/transient fluorescence spectrometer FLS1000 was purchased from United Kingdom (London, England), The Zeta-sizer nanometer equipment was used to examine the particle size distribution and zeta potential of nanoparticles dispersed in water (Malvern panalytical, England), Hamamatsu Q-Tau fluorescence life meter purchased from Japan (Tokyo, Japan); 365 nm UV lamp model ZF-5 was purchased from Wenzhou Mingren Electronics Instrument Co. Ltd. (Wenzhou, China). A FLS1000 spectrometer (London, England) was used to detect time-resolved fluorescence decays, and the resulting lifetime decay curves were fitted using a double-exponential function Y(t) (eq (1)) and the average lifetime was estimated using eqn (2).
Y(t) = A1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τ1) + A2 exp(−t/τ1) + A2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τ2) exp(−t/τ2)
| (1) |
| τ3 = (A1τ12 + A2τ22)/(A1τ1 + A2τ2) | (2) |
2.3. Preparation of N-GQDs
Using starch as the carbon source, weigh 0.3 g of potato amylose with a balance, weigh 25 ml of deionized water with a graduated cylinder, transfer all the starch and deionized water to a three-necked flask and stir to dissolve, then add 0.18 g of urea to In a three-necked flask, the solution was stirred for 15 min and then heated to 65 °C and stirred for 20 min, then transferred to the PTFE autoclave after natural cooling to room temperature, transferred to a high-temperature oven, heated to 180 °C for 8 h, cooled freely to room temperature and transferred in the centrifuge tube, centrifuge at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min in the centrifuge, then filter the N-GQDs using a 220 nm filter membrane and transfer the light yellow liquid to a centrifuge tube after filtration. Use a 1000 DA dialysis bag for dialysis for 48 h, the deionized water was changed every 4 h. Finally, the light-yellow nitrogen-doped graphene quantum dot solution was obtained and stored in a refrigerator at 4 °C for the next experiments. The reaction mechanism for the synthesis of GQDs from starch is as follows: potato starch is hydrolyzed to produce glucose, the glucose molecule is dehydrated to become C
000 rpm for 30 min in the centrifuge, then filter the N-GQDs using a 220 nm filter membrane and transfer the light yellow liquid to a centrifuge tube after filtration. Use a 1000 DA dialysis bag for dialysis for 48 h, the deionized water was changed every 4 h. Finally, the light-yellow nitrogen-doped graphene quantum dot solution was obtained and stored in a refrigerator at 4 °C for the next experiments. The reaction mechanism for the synthesis of GQDs from starch is as follows: potato starch is hydrolyzed to produce glucose, the glucose molecule is dehydrated to become C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, which is the fundamental unit of graphene's structure, and then the hydrogen atom of the glucose molecule interacts with the adjacent hydroxyl group and is dehydrated under hydrothermal conditions. Finally, the C atoms are covalently bonded to each other thus generating GQDs.40–43
C, which is the fundamental unit of graphene's structure, and then the hydrogen atom of the glucose molecule interacts with the adjacent hydroxyl group and is dehydrated under hydrothermal conditions. Finally, the C atoms are covalently bonded to each other thus generating GQDs.40–43
2.4. Fluorescence quantum yield determination
The quantum yields (Q) of fluorescence were calculated using quinine sulphate in water (Q = 0.54) as a benchmark. The following eqn (3) was used to compute the fluorescence quantum yields (Q) of N-GQDs:44| Qx = QR × (Ix/IR) × (AR/Ax) × (ηx/ηR)2 | (3) |
2.5. Optimization of experimental conditions parameters
In order to obtain N-GQDs with better fluorescence effect, the experimental conditions of the reaction process were optimized. Other conditions were kept constant, and the reaction temperature, reaction time and pH were changed during the experiment. The solution pH was adjusted using 0.1 M NaOH and HCl solution using a rubber-tipped dropper into a beaker containing 2 ml (1 mg ml−1) of N-GQDs solution, and the solution pH was measured using a PHC-3C type pH meter to obtain different pH values of the N-GQDs solution. The experimental condition parameters with the best fluorescence effect were selected to prepare N-GQDs for the next experiments.2.6. Detection of TC with N-GQDs fluorescent sensors
Different concentrations of TC standard solutions by dilution method, placed them in a centrifuge tube, add 20 μl of different concentrations of TC standard solutions and 2 ml of the above diluted N-GQDs solution in pH 7.0 dilution buffer, dilute to 5.0 ml, and shake well. The fluorescence emission test was performed with a fluorescence spectrophotometer at an excitation wavelength of 325 nm with both excitation and emission slits of 10 nm after the reaction had been at room temperature for 2 min, and the fluorescence intensity (F) of the system after the addition of TC and the fluorescence intensity (F0) of the system without TC were measured, and the error curves were plotted.2.7. Selective evaluation of TC assays
The selectivity of the method was assessed by measuring the relative fluorescence intensity ratio (F/F0) of the system with the addition of different substances to the N-GQDs solution. Then, in 1000 μl of diluted N-GQDs solution by adding 50 μl (10−5 M) tobramycin, oxytetracycline, kanamycin sulphate, chloramphenicol, amikacin, streptomycin, gentamicin, metronidazole, cysteine (Cys), tyrosine (Tyr), aspartic acid (Asp), lysine (Lys), threonine (Thr), glutamic acid (Glu) and Na+, Ca2+, Cu2+, Mg2+, Fe2+, K+ (10 times the TC concentration). Finally, the relative fluorescence emission intensity (F/F0) was measured using a fluorescence spectrophotometer.2.8. Determination of TC in real food samples
To investigate the feasibility of using N-GQDs to detect TC in real samples, the suitability of this fluorescent sensor was studied in different food samples (whole milk, skim milk, honey). Shelf-stable skim milk (0% fat), whole milk (6% fat) and honey purchased from a local supermarket were used in the experiments. The main components of both types of milk are raw cow's milk, protein and carbohydrates; the main components of honey are glucose, fructose, protein and carbohydrates. After adding 1% trichloroacetic acid (TCA) to the whole milk samples and sonicating for 30 min, the proteins in the milk were precipitated and centrifuged at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 min to facilitate separation. Finally, the proteins in the whole milk samples were removed using a 220 nm filter membrane. The real sample solution after the pre-treatment of the appeal was placed in a 4 °C environment to be used. Subsequently different concentrations (50 μl) of TC solution (5 × 10−6, 2.5 × 10−6, 5 × 10−7 M) were taken and added to the appeal pretreated whole milk sample mix and fully reacted for 8 min at room temperature. Skim milk sample and honey sample were pre-treated in the same way as whole milk. After pretreatment, different concentrations (50 μl) of TC solutions (5 × 10−7, 2.5 × 10−7 and 5 × 10−8 M) were mixed with the above pretreated skim milk samples and the reaction was carried out for 8 min at room temperature. The obtained mixed solutions were subjected to fluorescence emission measurement experiments with a fluorescence spectrophotometer.
000 rpm min−1 for 15 min to facilitate separation. Finally, the proteins in the whole milk samples were removed using a 220 nm filter membrane. The real sample solution after the pre-treatment of the appeal was placed in a 4 °C environment to be used. Subsequently different concentrations (50 μl) of TC solution (5 × 10−6, 2.5 × 10−6, 5 × 10−7 M) were taken and added to the appeal pretreated whole milk sample mix and fully reacted for 8 min at room temperature. Skim milk sample and honey sample were pre-treated in the same way as whole milk. After pretreatment, different concentrations (50 μl) of TC solutions (5 × 10−7, 2.5 × 10−7 and 5 × 10−8 M) were mixed with the above pretreated skim milk samples and the reaction was carried out for 8 min at room temperature. The obtained mixed solutions were subjected to fluorescence emission measurement experiments with a fluorescence spectrophotometer.
3. Results and discussion
3.1. Characterization of N-GQDs
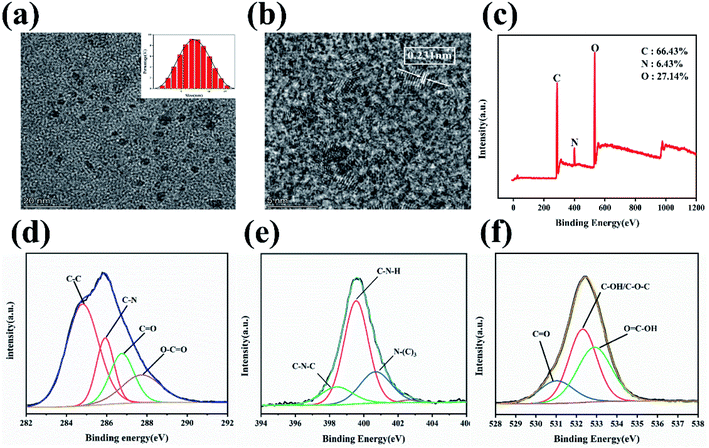
Transmission electron microscopy (TEM) was used to investigate the morphological characteristics and size distribution of N-GQDs. A typical plane view of a TEM image is shown in Fig. 1(a). The N-GQDs that have been synthesized are evenly distributed and well scattered. High-resolution transmission electron microscopy (HRTEM) images of N-GQDs (Fig. 1(b)) show a clear lattice structure with a lattice spacing of 0.231 nm, which corresponds to the (100) plan of graphene.45 The size distribution of the statistical N-GQDs (Fig. 1(a) inset) was in the range of 3.5–5.5 nm, with an average diameter of 3.8 nm. It confirms that N-GQD is well crystallized. X-ray photoelectron spectroscopy (XPS) is used to analyze the chemical compositions of N-GQDs. As demonstrated in Fig. 1(c), C, N, and O with atomic percentages of 66.43%, 6.43% and 27.14% are given, which proves that amination took place during the synthesis process. And the relating C 1s, N 1s, and O 1s. The peak is at ca. 284.6, 399.8 and 532.6 eV respectively.46 The C1s spectra of N-GQDs is shown in Fig. 1(d), confirming the presence of C–C (284.6 eV), C–N (285.8 eV), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (286.8 eV) and O–C
O (286.8 eV) and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (287.6 eV) functional groups; Fig. 1(e) shows the spectra of N1s, indicating the presence of N–H (399.7 eV), N–C3 (400.6 eV) and C–N–C (398.6 eV) on the surface of N-GQDs;47,48 Fig. 1(f) demonstrates the N-GQD surface O 1s spectra of C
O (287.6 eV) functional groups; Fig. 1(e) shows the spectra of N1s, indicating the presence of N–H (399.7 eV), N–C3 (400.6 eV) and C–N–C (398.6 eV) on the surface of N-GQDs;47,48 Fig. 1(f) demonstrates the N-GQD surface O 1s spectra of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (531.0 eV) and C–OH/C–O–C (532.5 eV) and O
O (531.0 eV) and C–OH/C–O–C (532.5 eV) and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH (533.0 eV);49,50 N atoms have been effectively doped into N-GQDs by hydrothermal reaction and functional groups such as amino groups have been generated on the original N-GQDs during the synthesis process, according to XPS analysis.
C–OH (533.0 eV);49,50 N atoms have been effectively doped into N-GQDs by hydrothermal reaction and functional groups such as amino groups have been generated on the original N-GQDs during the synthesis process, according to XPS analysis.
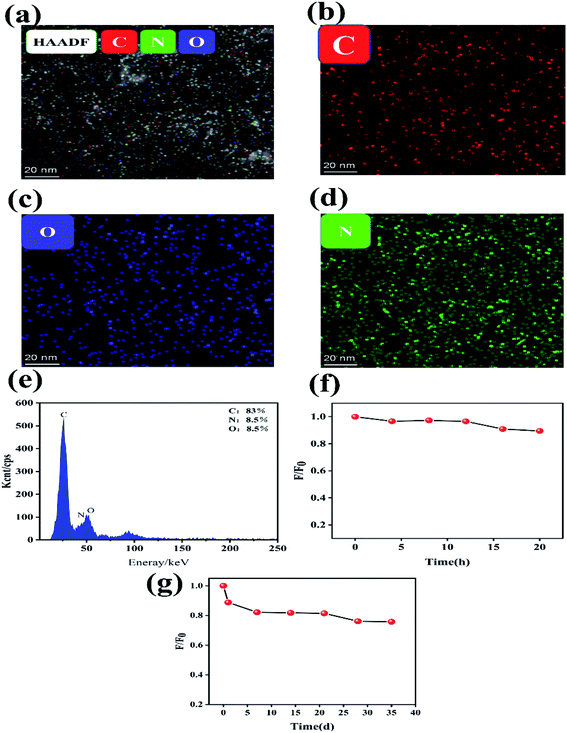
Energy Dispersive Spectrometer (EDS) is used to determine the type and amount of elements in material micro-region components, we also studied the chemical composition analysis of N-GQDs using EDS. Fig. 2(a–d) shows the EDS element mapping test of N-GQDs. From the EDS elemental mapping plots, it can be seen that the synthesized N-GQDs fluorescent probes contain C, N and O elements, which demonstrates that the doping of N elements is successful. According to Fig. 2(e), it can be seen that the content of C, N, O elements in N-GQDs was analyzed by EDS instrument, and the percentage of C, N and O elements were 83%,8.5% and 8.5%, respectively. All the above characterizations prove that N elements are successfully doped into GQDs to form N-GQDs fluorescent probes.
For the purpose of investigating the stability of N-GQDs, we tested the fluorescence stability of the prepared N-GQDs solution. Fig. 2(f) reveals that the fluorescence intensity of N-GQDs was not significantly changed after 4 h of 365 nm UV irradiation, indicating strong photostability; after 10 h, the fluorescence intensity gradually decreased. Fig. 2(g) shows that the fluorescence intensity of N-GQDs changed with time, and its fluorescence intensity slightly decreased in the first 5 days. Even though 35 days, the fluorescence of N-GQDs showed good stability with negligible changes.
3.2. Optimization of parameters for the synthesis conditions of N-GQDs
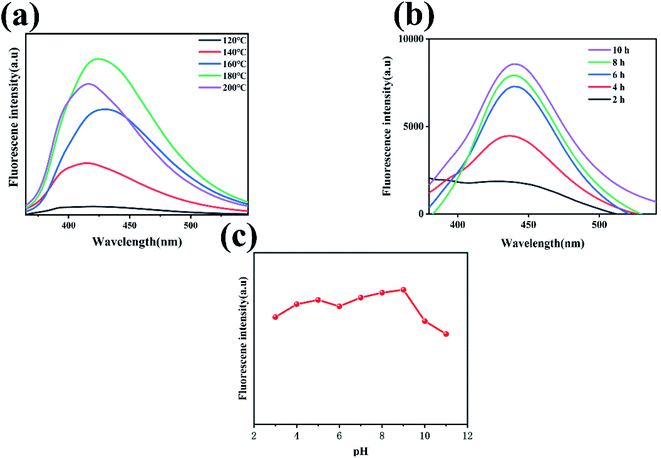
To obtain a quantum dot fluorescent sensor with good fluorescence intensity and high sensitivity, the condition parameters (reaction temperature, reaction time and pH value) in the synthesis reaction of N-GQDs were optimized to obtain N-GQDs with better fluorescence effect. Other experimental conditions were controlled consistently, and N-GQDs were prepared by changing the reaction temperature. The fluorescence emission curves of the produced N-GQDs at various reaction temperatures are given in Fig. 3(a). The intensity of the fluorescence increased when the reaction temperature was raised, as shown in Fig. 3(a), and the fluorescence intensity peaked at 180 °C. When the reaction temperature was higher than 180 °C, the fluorescence intensity began to decrease, and with the change in reaction temperature, the overall fluorescence intensity showed a trend of increasing and subsequently declining. The possible mechanism is due to the dehydration reaction of starch to form glucose and the dehydration of oligosaccharides at higher temperatures to undergo intermolecular cross-linking. Formyl groups react with hydrocarbon groups to dehydrate under hydrothermal conditions. Finally, the cyclic condensation produces GQDs.40 The other experimental conditions were controlled consistently, and different N-GQDs were prepared by changing the reaction time, as shown in Fig. 3(b), the excitation wavelength was 325 nm, and the fluorescence emission of the prepared N-GQDs was tested at different reaction times. Furthermore, as seen in Fig. 3(b), the fluorescence intensity of the quantum dots increased when the reaction time was increased, and the better fluorescence effect was achieved at the reaction time of 10 h. We optimized the optimal pH of the solution for the product quantum dots reacted at 180 °C for 10 h. As shown in Fig. 3(c). The fluorescence intensity starts to increase in the pH range of 2–5, and then decreases when the pH is in the range of 5–6; when the pH is in the range of 6–9, the fluorescence intensity starts to increase again and remains relatively stable; when the pH is greater than 9, the fluorescence intensity starts to show a significant decrease, and the N-GQDs solution has a better fluorescence emission intensity and is relatively stable in a neutral environment at around pH = 7. The results show that the fluorescence behaviour of N-GQDs is attenuated by strong acidic and alkaline environments, due to the change in fluorescence intensity of N-GQDs with pH associated with the protonation and deprotonation of a large number of groups on its surface such as amide and carboxyl groups.51 Moreover, the presence of many charge ions in a strongly acidic and alkaline environment leads to the occurrence of surface effects thereby causing the N-GQDs radiative leap efficiency to be affected and the N-GQDs fluorescence emission intensity to change accordingly. Therefore, we chose pH = 7, reaction temperature of 180 °C and reaction time of 10 h as the optimal parameters for the reaction process to prepare N-GQDs with better fluorescence effect for the next study.3.3. Optical characteristics of N-GQDs
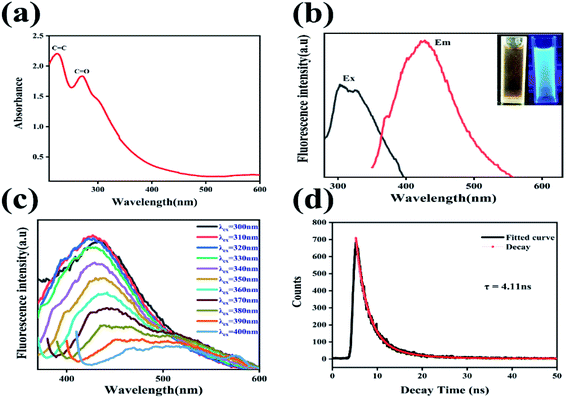
To examine the optical characteristics of N-GQDs, UV-vis absorption and PL activities were recorded. At 225 nm and 271 nm, there are two strong absorption bands, as well as a weak absorption band at 302 nm, are clearly visible in Fig. 4(a). The π–π* transition is responsible for the absorption bands at 225 nm, while the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bonds is responsible for the weak absorption peak at 273 nm.49 In addition, as is shown in Fig. 4(b), excitation and emission spectra of N-GQDs are displayed. The excitation and emission wavelengths are 325 nm and 425 nm respectively. As can be seen from the inset of Fig. 4(b), the N-GQDs solution appears yellow under daylight and blue-green under UV lamp irradiation at an excitation wavelength of 365 nm, indicating that the fluorescence of this N-GQDs solution has better photoluminescence. The emission wavelength of the synthesized N-GQDs is found to be highly dependent on the excitation wavelength, as shown in Fig. 4(c). As the excitation wavelength is steadily increased from 300 to 400 nm (with a 10 nm step length), the emission wavelength shifts from 435 to 503 nm. It suggests that this N-GQDs fluorescent probe has excitation wavelength-dependent fluorescence properties. We also performed the fluorescence lifetime test. Fluorescence decay curves of N-GQDs measured by Edinburgh FLS 1000 lifetime and steady-state spectrometer. As shown in Fig. 4(d), we performed the second order fluorescence lifetime test, the fluorescence lifetime of N-GQDs τ1 = 2.18 ns, τ2 = 5.84 ns; The fluorescence lifetime of the synthesized N-GQDs was calculated to be 4.11 ns on average. The above characterization shows that the synthesized N-GQDs have great photoluminescence and fluorescence lifetime, while the N-GQD shave wavelength-dependent characteristics.
N bonds is responsible for the weak absorption peak at 273 nm.49 In addition, as is shown in Fig. 4(b), excitation and emission spectra of N-GQDs are displayed. The excitation and emission wavelengths are 325 nm and 425 nm respectively. As can be seen from the inset of Fig. 4(b), the N-GQDs solution appears yellow under daylight and blue-green under UV lamp irradiation at an excitation wavelength of 365 nm, indicating that the fluorescence of this N-GQDs solution has better photoluminescence. The emission wavelength of the synthesized N-GQDs is found to be highly dependent on the excitation wavelength, as shown in Fig. 4(c). As the excitation wavelength is steadily increased from 300 to 400 nm (with a 10 nm step length), the emission wavelength shifts from 435 to 503 nm. It suggests that this N-GQDs fluorescent probe has excitation wavelength-dependent fluorescence properties. We also performed the fluorescence lifetime test. Fluorescence decay curves of N-GQDs measured by Edinburgh FLS 1000 lifetime and steady-state spectrometer. As shown in Fig. 4(d), we performed the second order fluorescence lifetime test, the fluorescence lifetime of N-GQDs τ1 = 2.18 ns, τ2 = 5.84 ns; The fluorescence lifetime of the synthesized N-GQDs was calculated to be 4.11 ns on average. The above characterization shows that the synthesized N-GQDs have great photoluminescence and fluorescence lifetime, while the N-GQD shave wavelength-dependent characteristics.
3.4. Fluorescence response of N-GQDs to TC
In this research, the fluorescence of N-GQDs could be quenched by TC. To obtain the best response between N-GQDs and TC, the pH of the reaction system was investigated. In general, pH plays an important role in fluorescence testing, which can sometimes impact not only the fluorescence of N-GQDs, but also how they react with the target. The protonation and deprotonation of oxygen-containing groups on the surface of N-GQDs could explain the fluctuation in fluorescence intensity with pH.52 We tested the stability of the fluorescence signal at different pH values. Fig. 5(a) shows the variation of the relative fluorescence intensity of the system with pH before and after the addition of TC. The value of the system F/F0 varies with pH; at pH 2.0 to 4.0, F/F0 increases steadily; at pH 4.0 to 8.0, F/F0 remains basically constant, while at pH 8.0, the system F/F0 starts to decrease. The fluorescence intensity diminishes dramatically, if the pH value reaches this range, as the surface bound carboxyl groups are protonated in an acidic environment and the hydroxide ions are protonated in alkaline media, resulting in a decrease in the fluorescence intensity of the system.53 Therefore, we used the N-GQDs/TC solution system with pH = 7.0 in a neutral environment for the next experimental exploration.Fig. 5(b) shows the fluorescence emission spectra of N-GQDs in the presence of TC, and the fluorescence spectra were analyzed directly on the TC solution, in which there was no essentially fluorescence emission intensity; the addition of TC to the N-GQDs solution resulted in a significant quenching of the fluorescence emission intensity of the N-GQDs, and the fluorescence response of N-GQDs underwent a sharp quenching after the addition of 50 μl TC (10−5 M), The computed quenching efficiency was around 67%, demonstrating that TC could efficiently quenching N-GQD fluorescence. The fluorescence intensity of N-doped GQDs decreased with the addition of TC (Fig. 5(e)). The relative fluorescence intensity F/F0 versus TC concentration (0–10−4 M) is shown in Fig. 5(e) (F and F0 are the fluorescence emission intensity in the presence or absence of TC in the N-GQDs solution, respectively). And TC concentration for the N-GQDs-based system at the optimized pH. The results reveal that the relative fluorescence intensity F/F0 in the fluorescent system of N-GQDs decreases steadily with increasing TC concentration, indicating that TC can effectively quench the fluorescence of N-GQDs. Fig. 5(f) shows that over the concentration range (2.5 × 10−10 to 5 × 10−6 M), there is a good linear relationship between F/F0 and TC concentration. The linear regression equation was F/F0 = −0.0383(log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C) + 0.6234 with a correlation coefficient R2 = 0.993, where C is the TC concentration. The limit of detection (LOD) was 9.375 × 10−13 M according to the rule of 3σ/s (where s is the slope of the linear calibration plot and σ is the standard deviation of the blank signal). Thus, these results suggest that the synthesized N-GQDs have great practical applications for the detection of TC.
C) + 0.6234 with a correlation coefficient R2 = 0.993, where C is the TC concentration. The limit of detection (LOD) was 9.375 × 10−13 M according to the rule of 3σ/s (where s is the slope of the linear calibration plot and σ is the standard deviation of the blank signal). Thus, these results suggest that the synthesized N-GQDs have great practical applications for the detection of TC.
3.5. Selective determination of the system
The fluorescence system has a good sensitivity for TC, and in order to study the selectivity of N-GQDs in different antibiotic (tetracycline, tobramycin, oxytetracycline, kanamycin sulphate, chloramphenicol, amikacin, streptomycin, gentamicin, metronidazole), amino acids (cysteine (Cys), tyrosine (Tyr), aspartic acid (Asp), lysine (Lys), threonine (Thr), glutamic acid (Glu)) and metal ions (Na+, Ca2+, Cu2+, Mg2+, Fe2+, K+) in coexistence conditions for interference experiments. Determination of the relative fluorescence intensity of N-GQDs solution with 50 μl (10−4 M) of different antibiotics, amino acids and metal ions (10 times the TC concentration) to perform the selectivity study of N-GQDs for TC. As shown in Fig. 6, the results showed that the fluorescence intensity of the system changed very little with the addition of other antibiotics, amino acids and their different metal ions under coexistence conditions, while TC had a significant fluorescence quenching effect on the fluorescent system of N-GQDs. The results indicate that N-GQDs have a good selectivity for TC.3.6. Detection for TC content in real food samples
To validate the applicability and feasibility of the N-GQDs fluorescent sensor of this work for the detection of TC in real food samples (including: whole milk, skim milk, honey). In this work, we calculated the limit of detection (LOD) and limit of quantification (LOQ) using fluorescence emission spectra and linearity plots of real food samples at multiple spiked concentrations. Real food samples (whole milk, skim milk and honey) were spiked at 1.0 × 10−8 to 1.0 × 10−6 M. The spiked samples were pre-treated and analyzed using the method N-GQDs fluorescent probes. The results show that in Fig. 7(a), (c) and (e). In the three real food samples, the fluorescence intensity in the system diminishes as the TC concentration increases, suggesting that the technology is applicable to real food samples. Fig. 7(b), (d) and (f) show good linearity (R2 = 0.9924–0.9985) in the range of 1.0 × 10−8 M to 1.0 × 10−6 M, with a limit of detection (LOD) of 3.75 × 10−11 to 2.075 × 10−9 M (3σ/s, where s is the slope of the linear calibration plot and σ is the standard deviation of the blank signal). Therefore, the method has good applicability with low detection limits for different actual food samples. To assess the reliability of the N-GQDs fluorescent probes for TC analysis in real food samples (whole milk, skim milk and honey), recovery experiments were performed on TC in real samples using the standard addition method. Different concentrations of TC (5 × 10−8 to 2.5 × 10−6 M) were added to treated real food samples (whole milk, skim milk and honey) for the experiments. Then, the fluorescence emission intensity of these samples was systematically tested. According to the results in Table 1, the recoveries ranged from 93.80% to 109.20%. The results showed good recoveries for this real sample assay, indicating the promising application of N-GQDs fluorescent probes in real food sample detection. Comparing the method proposed in this study with some of the other traditional methods reported (Table 2), the method in this work has a low detection limit and a good linear range. The results show that the method in this work has a low detection limit and a wide linear range for the detection of tetracycline and real food samples. The proposed strategy is feasible and applicable and has potential applications in the sensitive, rapid and highly sensitive detection of TC in real food samples.| TC Added(M) | TC found (M) | Recovery (%) | RSD (%, N = 3) | |
|---|---|---|---|---|
| a ND means not detected. | ||||
| Full fat milk | 0 | ND | ND | ND |
| 5.00 × 10−6 | 4.83 × 10−6 | 96.60 | 11.17 | |
| 2.50 × 10−6 | 2.73 × 10−6 | 109.20 | 10.09 | |
| 5.00 × 10−7 | 5.44 × 10−7 | 108.80 | 9.19 | |
| Skim milk | 0 | ND | ND | ND |
| 5.00 × 10−7 | 5.46 × 10−7 | 109.20 | 5.50 | |
| 2.50 × 10−7 | 2.62 × 10−7 | 104.80 | 10.32 | |
| 5.00 × 10−8 | 4.76 × 10−8 | 95.20 | 7.15 | |
| Honey | 0 | ND | ND | ND |
| 5.00 × 10−7 | 4.69 × 10−7 | 93.80 | 8.26 | |
| 2.50 × 10−7 | 2.49 × 10−7 | 99.60 | 6.33 | |
| 5.00 × 10−8 | 5.06 × 10−8 | 101.20 | 9.58 | |
| Analytical method | Target | Linear range (nM) | LOD (M) | Ref. |
|---|---|---|---|---|
| Capillary electrophoresis | TC | 2.00 × 10−8 | 8 | |
| HPLC | TC | 10−6 | 54 | |
| Colorimetric analysis | TC | 0.3–10 | 2.660 × 10−10 | 9 |
| ELISA | TC | 0.225–2250 | 2.200 × 10−10 | 55 |
| Fluorescence | TC | 0.001–0.4 | 6.000 × 10−6 | 56 |
| Fluorescence | TC | 3000–3.5 × 10−4 | 1.200 × 10−8 | 57 |
| Fluorescence | TC | 0.25–5000 | 9.375 × 10−13 | This work |
3.7. Mechanism of quenching of TC by N-GQDs
With the addition of TC, fluorescence quenching of N-GQDs occurs, and this phenomenon suggests that there is a certain interaction between TC quenching of N-GQDs fluorescence intensity. To investigate the possible fluorescence quenching mechanism, we tested the UV-vis spectra of TC and the fluorescence emission and excitation spectra of N-GQDs, as shown in Fig. 5(c), TC has obvious UV absorption bands at wavelengths of 274 nm and 360 nm, and the excitation spectra of N-GQDs overlap with the absorption spectra of TC. Therefore, the quenching mechanism of fluorescence is generally caused by fluorescence resonance energy transfer (FRET) or internal filtration effect (IFE).58 As a result, we tested the fluorescence lifetime to further distinguish and fluorescence resonance energy transfer (FRET) and internal filtering effect (IFE) quenching mechanisms. As illustrated in Fig. 5(d), the fluorescence lifetime of the N-GQDs was essentially unchanged by the addition of TC, the fluorescence lifetimes of N-GQDs were 4.11 ns (τ1 = 2.18 ns, τ2 = 5.84 ns) and 4.03 ns (τ1 = 2.12 ns, τ2 = 5.52 ns) after the addition of TC, where the concentration of TC was 5 × 10−6 M. Thus, IFE was shown to be TC induced fluorescence quenching of N-GQDs as the main mechanism.594. Conclusions
In summary, we developed a green method to prepare N-GQDs fluorescent sensors, synthesized N-GQDs for TC detection using the natural product potato starch as raw material, and the internal filtering effect (IFE) quenching mechanism of TC on N-GQDs was used to detect TC in food samples. Firstly, the strategy achieves a reduction in the preparation cost of nanomaterials (using green, low-cost natural products as raw materials), eliminating complicated steps and expensive materials. Secondly, the strategy overcomes the drawbacks of existing methods, and the proposed strategy has the advantages of high sensitivity, good selectivity and stability. Finally, the method has a low detection limit of 9.735 × 10−13 M, a wide linear range and good recoveries for TC and the low detection limits in the range of 3.750 × 10−11 to 2.075 × 10−9 M for the detection of TC in real food samples. N-GQDs fluorescence sensor has good linearity (R2 = 0.993) in the concentration range of TC (2.5 × 10−10 to 5 × 10−6 M), in addition, a low concentration of TC was detected in three real food samples with good recoveries of 93.80% to 109.20%, which indicates that the N-GQDs quantum dot sensor has a good prospect for the practical analysis of food samples. Therefore, the results suggest that the N-GQDs quantum dot fluorescence sensor is a promising tool for TC residue detection, which is important in food safety testing.Conflicts of interest
The authors declare that there are no conflicts of interest.Acknowledgements
We are grateful for the support by Fujian Provincial Department of Science and Technology guiding Project (2020Y0019); Industry-university Cooperation Project of Fujian Provincial Department of Science and Technology (2020N5006); The Opening Project of Jiangxi Province Key Laboratory of Polymer Micro/Nano Manufacturing and Devices (PMND202007); Fushimei agricultural and rural maker space (Minke Xing (2019) No. 2); Innovative Research Team in Science and Technology in Fujian Province University.References
- C. Lu, Q. Su and X. Yang, Nanoscale, 2019, 11, 16036–16042 RSC.
- L. A. Goncharova, N. G. Kobylinska, M. Díaz-Garcia and V. N. Zaitsev, J. Anal. Chem., 2017, 72, 724–733 CrossRef CAS.
- S. Jahanbani and A. Benvidi, Biosens. Bioelectron., 2016, 85, 553–562 CrossRef CAS PubMed.
- M. X. Feng, G. N. Wang, K. Yang, H. Z. Liu and J. P. Wang, Food Control, 2016, 69, 171–176 CrossRef CAS.
- D. Jin, Y. Bai, H. Chen, S. Liu, N. Chen, J. Huang, S. Huang and Z. Chen, Anal. Methods, 2015, 7, 1307–1312 RSC.
- M. P. Rodríguez, H. R. Pezza and L. Pezza, Spectrochim. Acta, Part A, 2016, 153, 386–392 CrossRef PubMed.
- D. Moreno-González, F. J. Lara, L. Gámiz-Gracia and A. M. García-Campaña, J. Chromatogr. A, 2014, 1360, 1–8 CrossRef PubMed.
- I. S. Ibarra, J. A. Rodriguez, J. M. Miranda, M. Vega and E. Barrado, J. Chromatogr. A, 2011, 1218, 2196–2202 CrossRef CAS PubMed.
- M. Ramezani, N. M. Danesh, P. Lavaee, K. Abnous and S. M. Taghdisi, Biosens. Bioelectron., 2015, 70, 181–187 CrossRef CAS PubMed.
- Y. Chen, D. Kong, L. Liu, S. Song, H. Kuang and C. Xu, Food Analytical Methods, 2016, 9, 905–914 CrossRef.
- J. Shen, Y. Zhu, C. Chen, X. Yang and C. Li, Chem. Commun., 2011, 47, 2580–2582 RSC.
- Q. Xu, J. Wei, J. Wang, Y. Liu, N. Li, Y. Chen, C. Gao, W. Zhang and T. S. Sreeprased, RSC Adv., 2016, 6, 28745–28750 RSC.
- Y. Yan, J. Gong, J. Chen, Z. Zeng, W. Huang, K. Pu, J. Liu and P. Chen, Adv. Mater., 2019, 31, 1808283 CrossRef PubMed.
- Z. Zhang, J. Zhang, N. Chen and L. Qu, Energy Environ. Sci., 2012, 5, 8869–8890 RSC.
- L. Li, G. Wu, G. Yang, J. Peng, J. Zhao and J.-J. Zhu, Nanoscale, 2013, 5, 4015–4039 RSC.
- S. S. Arumugam, J. Xuing, A. Viswadevarayalu, Y. Rong, D. Sabarinathan, S. Ali, A. A. Agyekum, H. Li and Q. Chen, J. Photochem. Photobiol., A, 2020, 401, 112788 CrossRef CAS.
- X. Zhou, S. Guo and J. Zhang, ChemPhysChem, 2013, 14, 2627–2640 CrossRef CAS PubMed.
- R. Das, R. Bandyopadhyay and P. Pramanik, Mater. Today Chem., 2018, 8, 96–109 CrossRef CAS.
- M. Pirsaheb, S. Moradi, M. Shahlaei and N. Farhadian, J. Hazard. Mater., 2018, 353, 444–453 CrossRef CAS PubMed.
- K. Jlassi, S. Mallick, A. Eribi, M. M. Chehimi, Z. Ahmad, F. Touati and I. Krupa, Sens. Actuators, B, 2021, 328, 129058 CrossRef CAS.
- T. Ye, C. Peng, I. Ali and J. Liu, Journal of Materials and Applications, 2018, 7, 20–24 Search PubMed.
- S. Sahu, B. Behera, T. K. Maiti and S. Mohapatra, Chem. Commun., 2012, 48, 8835–8837 RSC.
- Z. Wang, J. Yu, X. Zhang, N. Li, B. Liu, Y. Li, Y. Wang, W. Wang, Y. Li and L. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 1434–1439 CrossRef CAS PubMed.
- S. Mahesh, C. L. Lekshmi, K. D. Renuka and K. Joseph, Part. Part. Syst. Charact., 2016, 33, 70–74 CrossRef CAS.
- S. Kundu, R. M. Yadav, T. Narayanan, M. V. Shelke, R. Vajtai, P. M. Ajayan and V. K. Pillai, Nanoscale, 2015, 7, 11515–11519 RSC.
- A. Pramanik, S. Biswas, C. S. Tiwary, R. Sarkar and P. Kumbhakar, ACS Omega, 2018, 3, 16260–16270 CrossRef CAS PubMed.
- H. W. Kim, D. J. Lee, H. Lee, J. Song, H. T. Kim and J. K. Park, J. Mater. Chem. A, 2014, 2, 14557–14562 RSC.
- S. Gupta, T. Smith, A. Banaszak and J. Boeckl, Nanomaterials, 2017, 7, 301 CrossRef PubMed.
- D. Jiang, Y. Chen, N. Li, W. Li, Z. Wang, J. Zhu, H. Zhang, B. Liu and S. Xu, PLoS One, 2015, 10, e0144906 CrossRef PubMed.
- S. Paulo, E. Palomares and E. Martinez-Ferrero, Nanomaterials, 2016, 6, 157 CrossRef PubMed.
- L. Tong, F. Qiu, T. Zeng, J. Long, J. Yang, R. Wang, J. Zhang, C. Wang, T. Sun and Y. Yang, RSC Adv., 2017, 7, 47999–48018 RSC.
- M.-L. Tsai, W.-C. Tu, L. Tang, T.-C. Wei, W.-R. Wei, S. P. Lau, L.-J. Chen and J.-H. He, Nano Lett., 2016, 16, 309–313 CrossRef CAS PubMed.
- B. Zhang, Y. He and Z. Fan, J. Photochem. Photobiol., A, 2018, 367, 452–457 CrossRef CAS.
- D. Qu, M. Zheng, L. Zhang, H. Zhao, Z. Xie, X. Jing, R. E. Haddad, H. Fan and Z. Sun, Sci. Rep., 2014, 4, 1–11 Search PubMed.
- Y. Li, Y. Zhao, H. Cheng, Y. Hu, G. Shi, L. Dai and L. Qu, J. Am. Chem. Soc., 2012, 134, 15–18 CrossRef CAS PubMed.
- F. Qu, Z. Sun, D. Liu, X. Zhao and J. You, Microchim. Acta, 2016, 183, 2547–2553 CrossRef CAS.
- Y. Yang, Z. Liu, D. Chen, B. Gu, B. Gao, Z. Wang, Q. Guo and G. Wang, J. Photochem. Photobiol., A, 2021, 405, 112977 CrossRef CAS.
- J. Ju, R. Zhang, S. He and W. Chen, RSC Adv., 2014, 4, 52583–52589 RSC.
- L. Fan, Y. Hu, X. Wang, L. Zhang, F. Li, D. Han, Z. Li, Q. Zhang, Z. Wang and L. Niu, Talanta, 2012, 101, 192–197 CrossRef CAS PubMed.
- A. Bayat and E. Saievar-Iranizad, J. Lumin., 2017, 192, 180–183 CrossRef CAS.
- X. H. Li, S. Kurasch, U. Kaiser and M. Antonietti, Angew. Chem., Int. Ed., 2012, 51, 9689–9692 CrossRef CAS PubMed.
- T. Miyazawa and T. Funazukuri, in Hydrothermal Reactions and Techniques, World Scientific, 2003, pp. 75–81 Search PubMed.
- L. Tang, R. Ji, X. Cao, J. Lin, H. Jiang, X. Li, K. S. Teng, C. M. Luk, S. Zeng and J. Hao, ACS Nano, 2012, 6, 5102–5110 CrossRef CAS PubMed.
- Y. Song, C. Zhu, J. Song, H. Li, D. Du and Y. Lin, ACS Appl. Mater. Interfaces, 2017, 9, 7399–7405 CrossRef CAS PubMed.
- S. H. Jin, D. H. Kim, G. H. Jun, S. H. Hong and S. Jeon, ACS Nano, 2013, 7, 1239–1245 CrossRef CAS PubMed.
- S. Li, Y. Li, J. Cao, J. Zhu, L. Fan and X. Li, Anal. Chem., 2014, 86, 10201–10207 CrossRef CAS PubMed.
- J. Ju and W. Chen, Biosens. Bioelectron., 2014, 58, 219–225 CrossRef CAS PubMed.
- S. Liu, J. Tian, L. Wang, Y. Zhang, X. Qin, Y. Luo, A. M. Asiri, A. O. Al-Youbi and X. Sun, Adv. Mater., 2012, 24, 2037–2041 CrossRef CAS PubMed.
- Y.-X. Chen, D. Lu, G.-G. Wang, J. Huangfu, Q.-B. Wu, X.-F. Wang, L.-F. Liu, D.-M. Ye, B. Yan and J. Han, ACS Sustainable Chem. Eng., 2020, 8, 6657–6666 CrossRef CAS.
- X. Hai, Z. Guo, X. Lin, X. Chen and J. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 5853–5861 CrossRef CAS PubMed.
- X. Jia, J. Li and E. Wang, Nanoscale, 2012, 4, 5572–5575 RSC.
- Y. Hao, Z. Gan, X. Zhu, T. Li, X. Wu and P. K. Chu, J. Phys. Chem. C, 2015, 119, 2956–2962 CrossRef CAS.
- X. Wang, L. Zhang, A. Hao, Z. Shi, C. Dai, Y. Yang and H. Huang, ACS Appl. Nano Mater., 2020, 3, 9796–9803 CrossRef CAS.
- K. Ng and S. W. Linder, J. Chromatogr. Sci., 2003, 41, 460–466 CAS.
- S. Wang, J. Liu, W. Yong, Q. Chen, L. Zhang, Y. Dong, H. Su and T. Tan, Talanta, 2015, 131, 562–569 CrossRef CAS PubMed.
- Y. Yan, J. H. Liu, R. S. Li, Y. F. Li, C. Z. Huang and S. J. Zhen, Anal. Chim. Acta, 2019, 1063, 144–151 CrossRef CAS PubMed.
- L. Li, L. Shi, J. Jia, O. Eltayeb, W. Lu, Y. Tang, C. Dong and S. Shuang, Sens. Actuators, B, 2021, 332, 129513 CrossRef CAS.
- H. Yang, L. Yang, Y. Yuan, S. Pan, J. Yang, J. Yan, H. Zhang, Q. Sun and X. Hu, Spectrochim. Acta, Part A, 2018, 189, 139–146 CrossRef CAS PubMed.
- B. Chen, S. Chai, J. Liu, C. Liu, Y. Li, J. He, Z. Yu, T. Yang, C. Feng and C. Huang, Anal. Bioanal. Chem., 2019, 411, 2291–2300 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |