Open Access Article
Open Access ArticleEnzymatic hydrolysis of lignocellulosic biomass using a novel, thermotolerant recombinant xylosidase enzyme from Clostridium clariflavum: a potential addition for biofuel industry
Asma Zafar†
 a,
Attia Hamid
a,
Attia Hamid b,
Liangcai Pengc,
Yanting Wang
b,
Liangcai Pengc,
Yanting Wang c and
Muhammad Nauman Aftab†
c and
Muhammad Nauman Aftab† *b
*b
aFaculty of Life Sciences, University of Central Punjab, Lahore, Pakistan
bInstitute of Industrial Biotechnology, Government College University, Lahore, 54000, Pakistan. E-mail: nauman535@yahoo.com; Fax: +92 99213341; Tel: +92 3444704190
cBiomass and Bioenergy Research Center, Huazhong Agriculture University, Wuhan, China
First published on 18th May 2022
Abstract
The present study describes the cloning, expression, purification and characterization of the xylosidase gene (1650 bp) from a thermophilic bacterium Clostridium clariflavum into E. coli BL21 (DE3) using the expression vector pET-21a(+) for utilization in biofuel production. The recombinant xylosidase enzyme was purified to homogeneity by heat treatment and immobilized metal ion affinity chromatography. SDS-PAGE determined that the molecular weight of purified xylosidase was 60 kDa. This purified recombinant xylosidase showed its maximum activity at a temperature of 37 °C and pH 6.0. The purified recombinant xylosidase enzyme remains stable up to 90 °C for 4 h and retained 54.6% relative activity as compared to the control. The presence of metal ions such as Ca2+ and Mg2+ showed a positive impact on xylosidase enzyme activity whereas Cu2+ and Hg2+ inhibit its activity. Organic solvents did not considerably affect the stability of the purified xylosidase enzyme while DMSO and SDS cause the inhibition of enzyme activity. Pretreatment experiments were run in triplicate for 72 h at 30 °C using 10% NaOH. Saccharification experiment was performed by using 1% substrate (pretreated plant biomass) in citrate phosphate buffer of pH 6.5 loaded with 150 U mL−1 of purified recombinant xylosidase enzyme along with ampicillin (10 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg mL−1). Subsequent incubation was carried out at 50 °C and 100 rpm in a shaking incubator for 24 h. Saccharification potential of the recombinant xylosidase enzyme was calculated against both pretreated and untreated sugarcane bagasse and wheat straw as 9.63% and 8.91% respectively. All these characteristics of the recombinant thermotolerant xylosidase enzyme recommended it as a potential candidate for biofuel industry.
μg mL−1). Subsequent incubation was carried out at 50 °C and 100 rpm in a shaking incubator for 24 h. Saccharification potential of the recombinant xylosidase enzyme was calculated against both pretreated and untreated sugarcane bagasse and wheat straw as 9.63% and 8.91% respectively. All these characteristics of the recombinant thermotolerant xylosidase enzyme recommended it as a potential candidate for biofuel industry.
Introduction
Due to the increased prices of crude oils and depletion of fossil fuels, access to renewable energy sources has become essential to maintain the equilibrium of modern economies.1 The development of alternative fuels is important to reduce the carbon dioxide emission and use of fossil based fuels, as well as to face the anxiety of future petroleum sources. At present, bioethanol is obtained mostly from hydrolysis and fermentation of corn and sugar cane. However, facing the calculated rise in biofuels consumption worldwide, it has become obligatory to design new sources of biofuels and sustainable technologies. Therefore, there is a rising interest in increasing second and third generation biofuels, mainly based on ligno-cellulosic biomasses, which include agricultural residues, grasses cultivated as particular energy crops, wood (from softwood) and solid urban waste (cardboard, paper etc.).2,3 Lignocellulose being the most abundant renewable energy reservoir on earth, has the great potential to be utilized as a substrate for biofuel production in the biofuel industry. But the main limitation occurs in its conversion to fermentable sugars due to its rigid and compact structure.4Agricultural waste materials like sugarcane bagasse, wheat straw, rice bran and corn cobs are being utilized in many countries for the production of biofuels. Wheat straw and sugarcane bagasse are among the most generated agricultural waste in the world. The main constituents of wheat straw and sugarcane bagasse are cellulose, hemicellulose and lignin whose concentrations vary from region to region but typically they contain more than 20% hemicellulose in them.5,6 Hemicellulose contents of these agro waste materials can be efficiently converted into fermentable sugars by the microbial enzymes.
In nature, cellulose is one of the most plentiful renewable energy sources; its usage for biofuels has been hindered by the structural complexity of plant cell walls.7,8 The composition of the lignocellulosic biomass varies depending on the species to species that was being considered, but it is mainly composed by cellulose (35–50%), hemicellullose (20–35%), lignin (10–25%) and other less represented components.9 Every cellulose molecule is a linear polymer of β1 → 4d-glucose unit. Hemicellullose is a group of pentoses (arabinose and xylose) and hexoses (glucose, mannose, and galactose). Hemicellulose is mainly composed of D-xylose, L-arabinose, D-galactose, D-mannose as well as, 4-O-methyl-D-glucuronic acid.10
Hemicellulose is widely used as substrate in various industrial applications including biofuel, food, feed, cosmetics as well as paper industry due to the presence of various types of pentose sugars in it.11–15 But the degradation of hemicellulose is the most difficult process due to its high molecular weight and insoluble nature because of its linkage with cellulose and lignin. Xylan is the most commonly available hemicellulose found plant cell walls of angiosperms and herbaceous plants while in gymnosperm's cell wall, the hemicellulose portion is comprised of xylans as well as glucomannan.16 Xylans are composed of 1,4-linked xylosyl residues having a side chain of various acids like D-glucuronic, arabonic, p-coumaric, ferulic and acetic at 2,3-OH positions.17,18 Hemicellulases are a group of enzymes which are classified according to their substrates. The synergistic action of various enzymes is necessary to hydrolyze cellulose and hemicelluloses.19–22
A pretreatment step is also required in the enzymatic hydrolysis of lignocellulosic biomasses because of the recalcitrance nature of cellulose, hemicellulose, and lignin.23 Over the years, different methods of pretreatments are developed, counting physical (grinding, milling, and irradiation),24 physical–chemical (hydrothermolysis, steam pretreatment/autohydrolysis and wet oxidation),25 chemical methods (dilute acid, organic solvents, oxidizing agents and alkali)26 and biological, or a grouping of these methods.27 All above mentioned processes have some limitations like production of inhibitors etc. which leads to raise the cost of the processes.28 So there is a need to develop a pretreatment processes which reduce the energy requirement as well minimize the production of inhibitors during the processes of pretreatment.29
Clostridium clariflavum is anaerobic, Gram-positive, rod shaped, non-motile, moderately thermophilic, cellulose degrading, and chemo-organotrophic bacterium.30,31 The sequence of C. clariflavum revealed that its genome carries different genes that encode cellulosomal proteins as well as polysaccharide-degrading enzymes.32 The thermophilic features of C. clariflavum are of great interest to the research community of biomass degradation, since it is one of the few thermophilic cellulosome-producing bacteria known today. Moreover, C. clariflavum is capable of utilizing cellobiose, cellulose and natural substrates, for example switchgrass. C. clariflavum has got the potential to utilize xylose and xylan as sole sources of carbon.33
In this study cloning and expression of a novel xylosidase gene from C. clariflavum was performed into E. coli BL21 (DE3). The purification, optimization and characterization of recombinant xylosidase enzyme were carried out in order to explore it for utilization in plant biomass hydrolysis. Moreover, the saccharification potential of recombinant xylosidase enzyme was also investigated and reported in this study.
Methodology
Material
The chemicals and reagents utilized in this work were acquired from Merck (Germany) and Sigma (USA). Primers were manufactured from Gene LinkTM (USA) through World Wide Scientific Corp, Lahore, Pakistan. InsTAclone PCR cloning kit, QIAquick Gel Extraction kit, Plasmid isolation kit (Expin plasmid SV Mini, 50p) by GeneAll was used, restriction enzymes, T4 DNA ligase, DNA ladder, ColorPlus prestained protein marker, extensive ranging from (10–180 kDa) and Protino® Ni-TED kit were obtained from Thermo Fisher Scientific. Plant biomass (sugarcane bagasse & wheat straw) were collected from the fields of Lahore, Pakistan. PCR thermocycler (Applied Biosystem, Veriti, UK), rotatory shaking incubator (Ecocell). UV-spectrophotometer (Cecil-CE7200.0, Aquarius, UK) and SEM analysis was performed by MAIA3 TESCAN (Bartin University, Turkey). Other chemicals used in this work were of analytical grade.Selection of bacterial strain and plasmid
The genomic DNA of C. clariflavum was purchased from German collection of microorganisms and cell culture, DSMZ. The expression vector pET-21a(+) and the host organism E. coli BL21 (DE3) were obtained from culture collection of Institute of Industrial Biotechnology, GC University, Lahore Pakistan. Both strains were maintained in Lauria Bertani (1% tryptone, 0.5% yeast extract and 1% NaCl) for further use.Cloning of xylosidase gene of C. clariflavum in pET-21a(+)
Specific pair of primers was designed against the nucleotide sequence of the xylosidase gene of C. clariflavum to amplify the xylosidase gene. Vector NTI software was used to designed the primers. The amplified PCR product of xylosidase gene and expression vector pET-21a(+) were double digested with NdeI and HindIII in the presence of their suitable buffers using Qiaquick DNA purification kit. T4 DNA ligase was used for the ligation of xylosidase gene and pET-21a(+) vector in the presence of its specific buffer (10×). The ligated xylosidase gene was transformed into the competent cells of E. coli BL21 (DE3) that were freshly prepared. The positive clones were screened by colony PCR and restriction analysis of the isolated recombinant plasmid.Expression of recombinant gene in E. coli BL21 (DE3)
Fresh LB medium was inoculated (1%) by overnight grown culture of transformed E. coli BL21 (DE3) and incubated in a shaking incubator at 37 °C until the optical density of the medium reaches between 0.4-0.6. The cells were induced using IPTG and incubated for 4 h in a shaking incubator at 37 °C. The medium was then centrifuged for 10 min at 4 °C, 9000 rpm. The supernatant was separated and pellet was dissolved into Tris–Cl buffer (pH 6) for sonication. SDS-PAGE was used to examine the expression of the cloned xylosidase gene in extracellular and intracellular fractions.Enzyme activity assay
In the activity assay, the amount of p-nitrophenol emitted by the enzyme was measured using p-nitrophenyl-D-xylopyranoside as a substrate by following the protocol given by de Oliveira Rodrigues et al.34 The amount of p-nitrophenol released during the reaction, was determined by spectrophotometer at 405 nm. One unit of enzyme was defined as volume of enzyme that was needed to liberate 1 μmol of p-nitrophenol under standard enzyme assay conditions.Estimation of total protein
Total proteins were determined by Bradford method.35 Bovine serum albumin was used as a standard. In a 5 mL Bradford and 900 μl buffer and 100 μl of protein sample were added. A blank was run in parallel which contained 900 μl buffer and protein sample was replaced with 100 μl dH2O. The absorbance was taken at 595 nm in a spectrophotometer to determine the total protein contents.Optimization of xylosidase gene expression
The optimization of different parameters like pH of medium (4.0–9.0), incubation temperature of recombinant culture (16–42 °C), IPTG concentration for induction (0.1–0.6 mM), time of induction with IPTG (1–6 h) and optical density at 600 nm at the time of induction (0.2–0.8), was done to observe the maximum expression of cloned xylosidase gene in E. coli BL21 (DE3).Xylosidase enzyme purification
Recombinant xylosidase enzyme was purified by utilizing the methods mentioned below:Heat treatment
In order to denature host (E. coli BL21 DE3) proteins, recombinant enzyme was incubated at 70 °C for 30 min. After placing the enzyme for 30 min on ice, the host proteins were isolated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min.
000 rpm for 20 min.
Immobilized metal ion affinity chromatography
The Protino® Ni-TED kit was used to further purify the partially purified recombinant xylosidase enzyme to homogeneity. Four volumes of 1× LEW buffer were added to Protino column, and the column was held at room temperature to allow the buffer to travel through it via gravitational force. Partially purified xylosidase enzyme (3 mL) was transferred to pre-equilibrated column and allowed the enzyme to move through the column by the gravitational force. Eight bed volume of 1× LEW buffer was used to wash the column. Xylosidase enzyme was eluted with 3× elution buffer and collected in separate eppendorf tubes. Purified recombinant xylosidase protein fractions were analyzed by SDS-PAGE and protein determination analysis.Determination of molecular mass
SDS-PAGE was used to determine the molecular mass of the recombinant purified xylosidase protein by following the Laemmli method.36 A color plus pre-stained ladder of 10–250 kDa was run in parallel to the protein fraction samples. The gel was then stained in staining solution and destained in destaining solution for visualization of protein bands.Characterization of recombinant xylosidase
Various characters (pH stability, thermostability, effect of organic solvents on enzyme activity and stability, effect of different inhibitors on enzyme activity and stability and effect of various metal ions on enzyme activity and stability) of the purified recombinant xylosidase enzyme were studied for its potential use in biofuel industries. Purified enzyme was incubated for 1–3 h at room temperature with buffers of different pH (4.0–9.0) to assess pH stability, then residual activity was measured under standard assay conditions. To estimate thermostability, purified xylosidase enzyme was pre-incubated at different temperatures (50–90 °C) for 1–4 h and then residual activity of enzyme was calculated under standard enzyme assay conditions. The effect of organic solvents on the stability of purified recombinant xylosidase enzyme was determined by pre-incubating the enzyme for 1 hour at room temperature with various concentrations (10–30%) of different organic solvents (methanol, absolute ethanol, n-butanol, isopropanol) and then estimating its residual activity. The effect of metal ions and inhibitors on enzyme activity and stability was also investigated by incubating the enzyme with different concentrations (1–10 mM) of different metal ions (Mg2+, Ca2+, Ni2+, Mn2+, Zn2+, Hg2+, Cu2+, K1+, Co1+, Na1+) as well as with various concentrations (1–3%) of different inhibitors/surfactants (tween-20, DMSO, SDS, tween-80 and urea), respectively, for one hour at room temperature and then residual activities of purified recombinant enzyme was calculated under standards enzyme assay conditions.Collection, characterization and pretreatment of lignocellulosic biomass
Two types of plant biomass (sugarcane bagasse & wheat straw) were tested in this study to check the saccharification efficiency of purified recombinant xylosidase enzyme of C. clariflavum. Both plant biomasses were collected from local fields and dried at 60 °C for 48 h after thoroughly washing with tape water. After complete drying, the straws of sugarcane bagasse and wheat straw were ground in a milling machine and passed through a sieve to obtain a uniform particle size of plant biomass. Prior to the saccharification experiment, pretreatment of both plant biomass (sugarcane bagasse & wheat straw) was carried out by low temperature alkali method under optimized condition (temperature, NaOH concentrations, time duration). Pretreatment experiments were run in triplicates for 72 h at 30 °C using 10% NaOH. After pretreatment, samples were filtered and solid fractions were observed after air drying using scanning electron microscope for further use in enzymatic saccharification. The compositional analysis of the plant biomass used was performed using standard method of NREL for structural analysis of plant biomass Sluiter et al.37Saccharification activity of xylosidase enzyme
Hydrolytic activity of purified recombinant xylosidase enzyme from C. clariflavum was checked against pretreated as well as untreated plant biomass (sugarcane bagasse & wheat straw). Saccharification experiment was performed in a 250 mL Erlenmeyer flask using 1% substrate (pretreated plant biomass) in citrate phosphate buffer of pH 6.5 loaded with 150 U mL−1 of purified recombinant xylosidase enzyme. Subsequent incubation was carried out at 50 °C and 100 rpm in a shaking incubator for 24 h. To avoid the contamination risk, ampicillin (10 μg mL−1) was added in each experiment. Experimental controls were run in parallel which contained untreated plant biomass. The optimization of different experimental conditions (pH, temperature of incubation, time of incubation, substrate concentration, enzyme concentration, agitation speed) were carried to get the maximum saccharification efficiency of recombinant xylosidase enzyme of C. clariflavum. The untreated and pretreated plant biomass contents were determined by following the protocol of Sluiter et al.37 The structural morphologies of pretreated and untreated plant biomass (sugarcane bagasse & wheat straw) after enzymatic hydrolysis were determined by imaged using scanning electron microscopy. Reducing sugar residues in the hydro-lysate samples were estimated as described by Zafar et al.38 The saccharification percentage was calculated by using the following formula:| Saccharification (%) = xylose yield (mg mL−1) × 0.9 × total reaction volume × 100 |
Results
Cloning and expression of xylosidase gene into pET-21a(+)
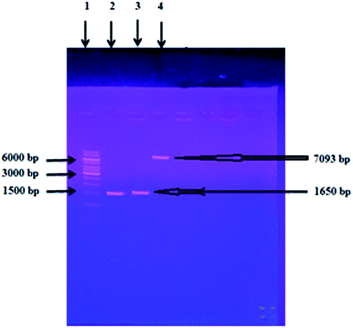
The concentration of the genomic DNA of C. clariflavum purchased from DSMZ was estimated to be 30 ng μl−1. Xylosidase gene of C. clariflavum was amplified by polymerase chain reaction (PCR). A band of 1650 bp of xylosidase gene was observed on agarose gel (Fig. 1). The purified, amplified xylosidase gene product was ligated into the pET-21a(+) vector and successfully transformed into E. coli BL21 (DE3). Positive clones were screened using colony PCR and single restriction analysis of the recombinant plasmid with HindIII in the presence of R buffer. A band of about 7093 bp was observed on agarose gel that determined the successful ligation of the xylosidase gene into the pET21a (+) (Fig. 1). Enzyme activity was estimated in both extracellular and intracellular fractions. The cell pellet was resuspended in 50 mM Tris–Cl buffer (pH: 8.0) and lyzed with a sonicator to obtain the intracellular fractions. The substrate for the enzyme activity assay was p-nitrophenyl-D-xylopyranoside. Enzyme units calculated in intracellular sample were 5.33 U mL−1.Optimization of enzyme expression
The following parameters were studies in order to determine the ideal conditions for maximum expression of the recombinant xylosidase gene.Effect of inducer concentration, time of induction and incubation temperature
IPTG is an inducer used in the production of recombinant proteins that are produced under lac operon. Periodic increase in IPTG concentration resulted in steady increase in the enzyme production. When 0.5 mM IPTG induction was provided to recombinant E. coli BL21 (DE3) cells, maximum xylosidase production (2.06 ± 0.47 U mL−1 0.89 ± 0.25 mg mL−1) was calculated. However, a decrease in the recombinant xylosidase production (1.35 ± 0.47 U mL−1) was observed at higher IPTG concentration (0.6 mM) as shown in Fig. 2A. The xylosidase enzyme activity was calculated after performing the activity assay in triplicate. Effect of time duration of IPTG induction on the xylosidase production as well as on total protein production was determined and optimum time of induction for maximum expression of recombinant xylosidase gene was 4 h incubation at 37 °C. Maximum xylosidase activity (2.24 ± 0.21 U mL−1) with maximum total protein contents (1.14 ± 0.11 mg mL−1) was observed after 4 h of IPTG induction as shown in Fig. 2B. After 6 h of incubation, however, considerable reduction in xylosidase enzyme activity (1.35 ± 0.17 U mL−1) was observed. After incubating the recombinant E. coli BL21 (DE3) culture at different temperatures (16–42 °C) for 4 h along with inducer, the effect of incubation temperature on the xylosidase gene expression was determined. Maximum xylosidase enzyme activity (2.6 ± 0.14 U mL−1) and total protein estimation (1.61 ± 0.04 mg mL−1) was observed when the recombinant E. coli BL21 (DE3) culture was incubated at 37 °C. However, enzyme activity was decreased when temperature fluctuated from 37 °C. At 16 °C, 22 °C, 30 °C, and 42 °C, low enzyme activity like 0.53 ± 0.07 U mL−1, 1.36 ± 0.09 U mL−1, 1.93 ± 0.09 U mL−1, and 1.75 ± 0.06 U mL−1 were observed as shown in Fig. 2C.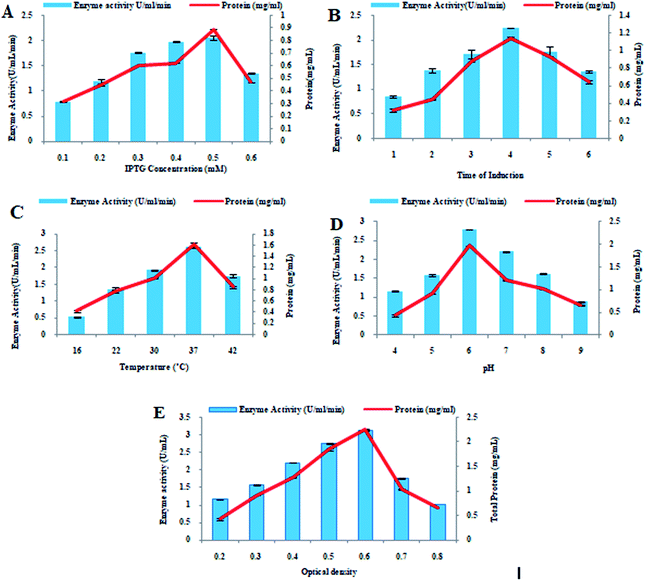 | ||
| Fig. 2 Optimization of xylosidase gene expression. (A) IPTG concentration, (B) induction duration, (C) incubation temperature (D) pH and (E) optical density. | ||
Effect of pH of culture medium and optical density
By changing the pH of medium ranging from 4.0 to 9.0, the effect of initial pH on xylosidase gene expression was calculated. Maximum enzyme production (2.8 ± 0.09 U mL−1) and total protein production (1.97 ± 0.01 mg mL−1) was calculated when the pH of media was 6.0. At pH 9.0, enzyme activity was lowered to 0.88 ± 0.08 U mL−1 as indicated in Fig. 2D. The activity of recombinant xylosidase was 1.16 ± 0.09 U mL−1, 1.58 ± 0.06 U mL−1, 2.21 ± 0.07 U mL−1, and 1.62 ± 0.09 U mL−1, at pH values of 4.0, 5.0, 7.0, and 8.0 respectively as shown in Fig. 2D. The effect of optical density of medium on xylosidase gene expression was determined by calculating enzyme activity at various optical density values ranging from 0.2 to 0.8. When the optical density of the medium was 0.6 at 600 nm at the time of induction, the maximum xylosidase enzyme activity (3.12 ± 07 U mL−1) and total amount of protein (2.25 ± 01 mg mL−1) were calculated. However, less enzyme units (1.03 ± 0.04 U mL−1) were obtained at optical density 0.8 as shown in Fig. 2E.Purification of recombinant xylosidase enzyme
The recombinant xylosidase enzyme was partially purified by incubating it at 70 °C for 30 min to denature the non-specific host proteins. The denatured host proteins were separated by centrifuge for 20 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Recombinant xylosidase enzyme was further purified by immobilized metal ion affinity chromatography. The final eluted fraction was used for the total protein estimation and activity assay. The purification fold and specific activity of the recombinant purified xylosidase fraction was estimated to be 41.96 fold and 141.85 U mg−1 respectively with 54% recovery as shown in Table 1.
000 rpm. Recombinant xylosidase enzyme was further purified by immobilized metal ion affinity chromatography. The final eluted fraction was used for the total protein estimation and activity assay. The purification fold and specific activity of the recombinant purified xylosidase fraction was estimated to be 41.96 fold and 141.85 U mg−1 respectively with 54% recovery as shown in Table 1.
| Purification steps | Xylosidase Unit (U) | Total protein (mg) | Specific activity (U/mg) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude xylosidase | 930 | 275 | 3.38 | 1 | 100 |
| Heat-treated sample | 625 | 12.5 | 50.0 | 14.79 | 67.20 |
| IMAC sample | 505 | 3.56 | 141.85 | 41.96 | 54.30 |
Molecular weight determination
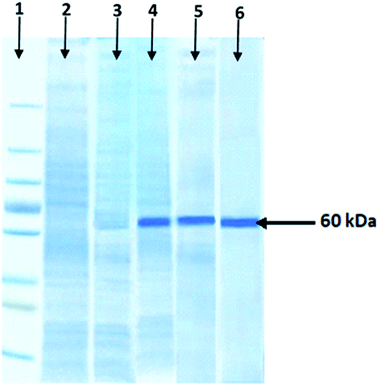
The molecular weight of purified xylosidase protein was determined by SDS-PAGE. An obvious single band of recombinant xylosidase protein was observed near 60 kDa position. However, there was no band at this position in the control; wild culture of E. coli BL21 (DE3) and non-induced pET-21a(+) vector having xylosidase gene as shown in Fig. 3.Characterization of recombinant purified xylosidase enzyme
To characterize the enzyme for industrial utilization, following parameters were studied.| S no. | Plant biomass | Sugar released (mg ml−1) | Saccharification (%) | Hemicellulose concentration in biomass (%) |
|---|---|---|---|---|
| 1 | Untreated sugarcane bagasse | 0.74 | 3.33 | 32 |
| 2 | Pretreated sugarcane bagasse | 2.14 | 9.63 | 21 |
| 3 | Untreated wheat straw | 0.57 | 2.56 | 28 |
| 4 | Pretreated wheat straw | 1.98 | 8.91 | 20 |
Discussion
For the complete conversion of lignocellulosic materials into monomeric units, a cocktail of cellulases and hemicellulases is required. Xylan being the major component of hemicelluloses especially in agro-industrial residues is converted into xylose sugar by the action of two major enzymes (xylanase and xylosidase). Xylosidase is an important enzyme for the affective hydrolysis of xylan into monomeric sugars for the application in various industrial processes as it acts upon non-reducing ends of the hemicellulose fragment generated as the result of xylanase enzyme action.39 Xylosidases are absent in most of the commercial enzyme cocktails and are obtained from microorganisms for industrial applications.40 So cocktail of commercially available biomass degrading enzymes must be supplemented with microbial enzymes in order to attain the full potential of plant biomass in industrial application.41 The aim of this study was to purify and characterize the xylosidase gene of C. clariflavum expressed in E. coli BL21 (DE3) using the pET-21a(+) expression vector for use in industrial applications.A 1650 bp gene of xylosidase from C. clariflavum was successfully amplified and cloned in pET-21a (+) vector and transformed into competent cells of E. coli BL21 (DE3). Colony PCR and restriction analysis of an isolated recombinant vector were used to screen positive clones. A 7093 bp band of single restricted recombinant vector confirmed the successful integration of xylosidase gene in pET-21a (+) as shown in Fig. 1. Other reports42–46 are present on the cloning and expression of xylosidase gene in E. coli from various sources but till now no report is present on the cloning and expression of xylosidase gene from C. clariflavum.
The expression of cloned gene was evaluated by SDS-PAGE analysis and a very clear band of almost 60 kDa was observed (Fig. 2). Enzyme production and yield from recombinant microorganisms is greatly influenced by various parameters like pH, temperature, time of incubation, etc.47 Metabolic burden on the recombinant cell is generated due to the expression of foreign gene which results in low expression of gene, reduced cell biomass due to the reduced growth rates as well as plasmid instability.48–50 Various reports are present on the optimization of production conditions from recombinant protein.51–54 Optimization of various parameters was carried out in order to get the maximum recombinant xylosidase enzyme. To induce the expression of recombinant gene under lac promoter, IPTG is a very commonly used inducer which offers number of advantages in lab scale experiments.55 Optimization of inducer concentration is very important factor because it directly affects the production of protein per cell as well as its folding. On the other hand, IPTG also showed inhibitory effect to the cell growth.56,57 Maximum xylosidase production (2.06 ± 0.89 U mL−1) as found with 0.5 mM IPTG induction. Various studies51,58 represent the effect of inducer concentration on the expression of cloned genes.
Induction time is another parameter which directly affects the cell biomass production and ultimately influences the production of enzyme yield. Moreover, it has been reported that prolonged induction phase of cloned genes in E. coli resulted in secretion of periplasmic recombinant proteins in the surrounding environment.59 Maximum expression of cloned xylosidase gene in this study was obtained after 4 h of induction with 0.5 mM IPTG (Fig. 2B). After 4 h of IPTG induction, Hamid and Aftab.58 observed the maximum expression of the xylosidase gene from Thermotoga naphthophila in E. coli BL21 (DE3). In another report Whitehead and Cotta.60 reported the maximum expression of xylosidase and arabinosidase genes after 2 h of induction.
Optimum growth temperature of E. coli ranges from 37 °C to 39 °C but this temperature coincides the maximal expression activity of many promoter systems61 and in turns affects the yield of recombinant protein. Similarly, pH of the culture medium can affect the yield of recombinant protein, secretion of protein from the cell as well as proteolytic activity.62 Optimal temperature and pH of recombinant E. coli BL21 for recombinant xylosidase production were observed as 37 °C and 6.0, respectively in this study. These are in agreement with the previous results for the growth of recombinant E. coli BL21 (DE3) cells at 37 °C38,58 but the optimized pH for high enzyme production was 7.0 in these studies. To induce the maximum expression of foreign genes in host cells in order to get the maximum yield of foreign proteins, the right time of inducer implementation during growth cycle of cell is very important because in many strains the growth and viability of the cells is affected by the inducer type and concentration.59 Maximum enzyme production in this study was observed when recombinant E. coli BL21 culture having optical density 0.6 nm, was induced with IPTG (Fig. 2E).
Heat denaturation and immobilized metal affinity chromatography were used to purify recombinant xylosidase enzyme, which yielded a recovery of 54.30 percent (Table 1). Xu et al.63 reported β-xylosidase enzyme from Thermoanaerobacterium aotearoense showing recovery yield less than 55% after the heat treatment followed by affinity chromatography. Sumarsih et al.64 reported the purification of recombinant β-xylosidase from Bacillus megaterium by affinity chromatographic method using agarose comprising Ni-NTA (Nickel-Nitrilotriacetic acid). For industrial applications of recombinant proteins, their chemical and physical instabilities are taken as bottleneck. Thermostable enzymes are attractive for industrial applications. Advantages of the thermostable enzymes in industrial usage include, higher rate of reaction, high yield of product, increased stability, low contamination problems and reduce viscosity.65 Similarly, pH stable enzymes have huge application because they can retain their specificity as well as catalytic activity.66 In biofuel industry for the production of biofuel from plant biomass, the thermostable and pH stable enzymes are considered as more advantageous due to their cost-effective and efficient process of bioconversion.67 An important aspect of this study is the thermostability of recombinant xylosidase at 80 °C up to 4 h (Fig. 4A) and pH stability at pH 6.0 up to 3 h (Fig. 4B). Previously various reports are present on thermostable xylosidases enzymes from thermophiles and hyperthermophiles.68–71 pH stability of enzyme is also important factor to characterize recombinant purified xylosidase. The results obtained in this study are similar to previous reports.63,72,73
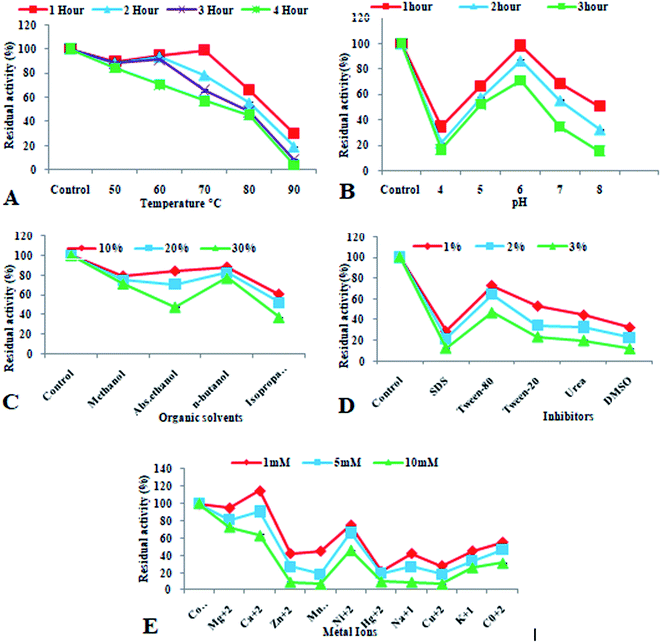 | ||
| Fig. 4 Characterization of purified recombinant xylosidase enzyme. (A) Thermostability, (B) pH stability, (C) effect of organic solvents, (D) effect of inhibitors, (E) effect of metal ions. | ||
Organic solvents influence the activity of enzyme in industrial applications. In biofuel production from plant biomass, presence of lignin causes the hindrance in release of fermentable sugars from plant biomass. So pretreatment of plant biomass is a very important step in biofuel industry in order to get the full potential of plant biomass. Efficient pretreatment of plant in the presence of organic solvents (methanol, ethanol, acetone etc.) in varied concentrations has been reported in previous studies.74–76 In order to characterize the enzyme under study for industrial applications, the effect of different solvents was checked on its activity and stability. The purified recombinant xylosidase enzyme showed almost 100% residual activity with various concentrations (10–30%) of organic solvents like methanol and ethanol (Fig. 4C) that was similar to the β-xylosidase from Thermotoga petrophila and β-xylosidase Xln-DT from Dictyoglomus thermophilum.68
In enzymatic hydrolysis of cellulose, the use of surfactants in the reaction is known to enhance the mechanism of reaction by facilitating the enzyme–substrate adsorption.77 However, some ionic agents are known to inhibit the enzyme activity in industrial applications as they interact with different hydrophobic and hydrophilic regions of the enzymes and change their tertiary structures.78 In this study, different inhibitors were used to observe the effect on residual activity of enzyme and the results showed the inhibition of activity of enzyme with SDS and DMSO (Fig. 4D) that are similar to Shin et al.79 Various other reports showed that β-xylosidase from Aspergillus oryzae and Paecilomyces thermophila, was inhibited by SDS.80,81 The effect of metal ions on purified recombinant xylosidase enzyme was different for various metal ions. It showed improved activity in the presence of Mg2+ and Ca2+, while inhibited in the presence of Cu2+ and Hg2+ (Fig. 4E). This inhibition was similar to β-xylosidase Dt-xyl3 activities that was strongly inhibited up to 66.3% and 9.9% in the presence of 1 mM and 5 mM Cu2+, individually. Cu2+ is known to catalyze the auto-oxidation of cysteine molecules, which cause the formation of inter and intra-molecular di-sulfide bonds or sulfenic acid and led to decreased enzyme production.82 Zhou et al.83 characterized recombinant β-xylosidase enzyme from Bacteroides ovatus and showed its activation in the presence of Mg2+ and Mn2+. Marcolongo et al.84 also reported β-xylosidase from Anoxybacillus sp. that was inhibited in the presence of Cu2+ and Hg2+.
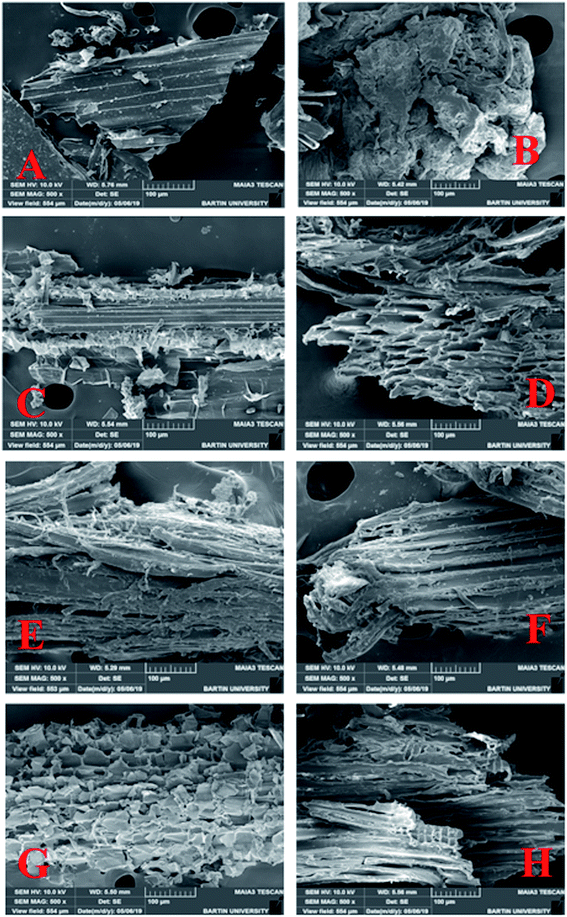
For efficient production of biofuel, the complete degradation of lignocellulosic biomass is a key factor, and the removal of impurities before enzymatic hydrolysis of biomass is very much important in order to make the plant biomass accessible to the enzymes working on it. In present study, the efficiency of recombinant xylosidase enzyme to hydrolyse the lignocellulosic biomass was checked for its final use in biofuel industry. Pretreatment of plant biomass (sugarcane bagasse & wheat straw) was carried out and resulting product was analyzed by SEM analysis as shown in Fig. 5. Untreated plant biomass samples (Fig. 5A and B) showed more compact and arranged surface morphology as compared to pretreated plant biomass whose surface area was increased as well as microfibrils were splitted and abrasive due to alkali treatment. Similar results were reported in previous reports.43,85–89 This pretreatment facilitated the enzymatic hydrolysis of biomass by recombinant xylosidase enzyme. The enzymatic hydrolysis of pre-treated plant biomass is mainly affected by the enzyme loadings, pH of the reaction, temperature of incubation, substrate concentration, reaction time as well as the applied pre-treatment method.90–95 A through study, under optimized conditions (pH, temperature, time of incubation, enzyme concentration, substrate concentration) was carried out to check the hydrolytic efficiency of recombinant xylosidase and results are represented in Table 2. Maximum hydrolytic activity (9.63%) of recombinant xylosidase enzyme was observed against pretreated sugarcane bagasse with 2.14 mg mL−1 sugar released. Enzyme was also found to be able to hydrolyse untreated plant biomass to some extent and 3.33% saccharification was observed against untreated sugarcane bagasse. Lesser hydrolytic activity of enzyme against untreated plant biomass might be due to compact and rigid structure of sugarcane bagasse as shown in Fig. 5E. These results are consistent with previous reports on saccharification of sugarcane bagasse.43,89,96,97 Effect of recombinant xylosidase on the hydrolysis of wheat straw was observed in similar pattern like higher percentage hydrolysis (8.91%) again pretreated wheat straw biomass. Many reports are present on the plant biomass hydrolysis by xylosidase enzyme from various microorganisms20,45,84,98–100 but not any report on hydrolysis of recombinant xylosidase enzyme from C. clariflavum yet present.
Conclusions
The demand of biofuel production has increased for the environmental preservation and the energy supply problems. In this study, an extensive research work was carried out on novel recombinant thermostable xylosidase enzyme for utilization in biofuel industry. The low temperature alkali method was used for the pretreatment of plant biomass because it requires relatively lower energy. Efficient hydrolytic activity of enzyme reported in this study against pretreated sugarcane bagasse and wheat straw (9.63% and 8.91%). Based on this work, it is concluded that the use of plant biomass can meet the energy needs. It is environment friendly and less polluting, representing it a more appealing power source. The present study also highlighted the importance of biofuel for the developing Asian countries, like Pakistan, facing an energy disaster because of exponential growth in population. According to literature study, the lignocellulosic plant biomass present in abundant amounts in Pakistan including wheat straw, sugarcane bagasse, rice straw and corn cob etc. For this intention, the government of Pakistan must endorse collaboration between research institutes and the industries for successful implementation of biofuel programs in the Pakistan.Author contributions
Asma Zafar: original manuscript writing, investigation. Attia Hamid: investigation, conceptualization. Muhammad Nauman Aftab: supervision (writing – reviewing and editing), project administration.Funding
This research was funded by the Higher Education Commission of Pakistan for Biofuel project No. 5535 and the Pak-Turk Researchers Mobility Program.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
Sincere gratitude is expressed to the Higher Education Commission of Pakistan and the Pak-Turk Researchers Mobility Program.References
- A. Rafandadi and I. Ozturk, Impacts of renewable energy consumption on the German economic growth: evidence from combined cointegration test, Renewable Sustainable Energy Rev., 2017, 75, 1130–1141, DOI:10.1016/j.rser.2016.11.093.
- B. Gabrielle, Significance and limitations of first generation biofuels, J. Soc. Biol., 2008, 202, 161–165, DOI:10.1051/jbio:2008028.
- E. M. Rubin, Genomics of cellulosic biofuels, Nature, 2008, 454, 841–845, DOI:10.1038/nature07190.
- Z. Wang, G. Cao, C. Jiang, J. Song, J. Zheng and Q. Yang, Butanol production from wheat straw by combining crude enzymatic hydrolysis and anaerobic fermentation using Clostridium acetobutylicum ATCC824, Energy Fuels, 2013, 27(10), 5900–5906, DOI:10.1021/ef4010658.
- E. de Morais Teixeira, T. J. Bondancia, K. B. R. Teodoro, A. C. Corrêa, J. M. Marconcini and L. H. C. Mattoso, Sugarcane bagasse whiskers: extraction and characterizations, Ind. Crops Prod., 2011, 33(1), 63–66, DOI:10.1016/j.indcrop.2010.08.009.
- R. R. C. Bakker, Availability of lignocellulosic feedstocks for lactic acid production-feedstock availability, lactic acid production potential and selection criteria, Wageningen UR-Food & Biobased Research. 2013, pp. 1–62 Search PubMed.
- W. Zhao, L. Glavas, K. Odelius, U. Edlund and A. C. Albertsson, Facile and green approach towards electrically conductive hemicellulose hydrogels with tunable conductivity and swelling behavior, Chem. Mater., 2014, 26(14), 4265–4273, DOI:10.1021/cm501852w.
- K. Y. Yoon, E. E. Woodams and Y. D. Hang, Enzymatic production of pentoses from the hemicellulose fraction of corn residues, Food Sci. Technol., 2006, 39(4), 388–392, DOI:10.1016/j.lwt.2005.02.005.
- L. Z. Liu, S. C. Badal, and J. Slininger, Lignocellulosic biomass conversion to ethanol by sacharomyces, Bioenergy, ed. J. D. Wall, C. S. Harwood, and A. Demain, ASM Press, Washington DC, 2008, pp. 17–36, DOI:10.1128/9781555815547.ch2.
- L. Z. Huang, M. G. Ma, X. X. Ji, S. E. Choi and C. Si, Recent developments and applications of hemicellulose from wheat straw: a review, Front. Bioeng. Biotechnol., 2021, 9, 440, DOI:10.3389/fbioe.2021.690773.
- J. Falco, M. Sieben, N. Brun, M. Sevilla Solís, T. V. D. Mauelen, E. Morallón, D. Cazorla Amoros and M. M. Titirici, Hydrothermal carbons from hemicellulose-derived aqueous hydrolysis products as electrode materials for supercapacitors, ChemSusChem, 2013, 6, 374–382, DOI:10.1002/cssc.201200817.
- L. Hu, M. Du, J. Zhang, L. Hu, M. Du and J. Zhang, Hemicellulose-based hydrogels present status and application prospects: A brief review, Open J. For., 2017, 8, 15–28, DOI:10.4236/ojf.2018.81002.
- K. Y. Yoon, E. E. Woodams and Y. D. Hang, Enzymatic production of pentoses from the hemicellulose fraction of corn residues, Food Sci. Technol., 2006, 39(4), 388–392, DOI:10.1016/j.lwt.2005.02.005.
- H. Du, W. Liu, M. Zhang, C. Si, X. Zhang and B. Li, Cellulose nanocrystals and cellulose nanofibrils based hydrogels for biomedical applications, Carbohydr. Polym., 2019, 209, 130–144, DOI:10.1016/j.carbpol.2019.01.020.
- C. Liu, G. Zou, X. Yan and X. Zhou, Screening of multimeric β-xylosidases from the gut microbiome of a higher termite, Globitermes brachycerastes, Int. J. Biol. Sci., 2018, 14(6), 608, DOI:10.7150/ijbs.22763.
- A. Kumagai and T. Endo, Effects of hemicellulose composition and content on the interaction between cellulose nanofibers, Cellulose, 2021, 28(1), 259–271, DOI:10.1007/s10570-020-03530-x.
- A. Rennie and H. V. Scheller, Xylan biosynthesis, Curr. Opin. Biotechnol., 2014, 26, 100–107, DOI:10.1016/j.copbio.2013.11.013.
- Y. Yang, J. Li, Q. Yu, J. Hou, C. Gao, D. Li, Y. Liu, C. Ran and Z. Zhou, Conformational determinants necessary for secretion of Paecilomyces thermophila β-1, 4-xylosidase that lacks a signal peptide, AMB Express, 2018, 8(1), 1–10, DOI:10.1186/s13568-018-0542-2.
- Q. Beg, M. Kapoor, L. Mahajan and G. S. Hoondal, Microbial xylanases and their industrial applications: a review, Appl. Microbiol. Biotechnol., 2001, 56(3), 326–338, DOI:10.1007/s002530100704.
- A. Zafar, M. N. Aftab and M. A. Saleem, Pilot scale production of recombinant hemicellulases and their saccharification potential, Prep. Biochem. Biotechnol., 2020, 50(10), 1063–1075, DOI:10.1080/10826068.2020.1783679.
- A. Martínez, A. R. Chaves and P. M. Civello, β-Xylosidase activity and expression of a β-xylosidase gene during strawberry fruit ripening, Plant Physiol. Biochem., 2004, 42(2), 89–96, DOI:10.1016/j.plaphy.2003.12.001.
- M. I. Rajoka and S. Khan, Hyper-production of a thermotolerant β-xylosidase by a deoxy-D-glucose and cycloheximide resistant mutant derivative of Kluyveromyces marxianus PPY 125, Electron. J. Biotechnol., 2005, 8(2), 58–65, DOI:10.2225/vol8-issue2-fulltext-9.
- S. Ethaib, R. Omar, M. K. S. Mazlina, A. B. D. Radiah and S. Syafiie, Development of a hybrid PSO–ANN model for estimating glucose and xylose yields for microwave-assisted pretreatment and the enzymatic hydrolysis of lignocellulosic biomass, Neural Comput. Appl., 2018, 30(4), 1111–1121, DOI:10.1007/s00521-016-2755-0.
- D. Kumari and R. Singh, Pretreatment of lignocellulosic wastes for biofuel production: A critical review, Renewable Sustainable Energy Rev., 2018, 90, 877–891, DOI:10.1016/j.rser.2018.03.111.
- S. R. Paudel, S. P. Banjara, O. K. Choi, K. Y. Park, Y. M. Kim and J. W. Lee, Pretreatment of agricultural biomass for anaerobic digestion: Current state and challenges, Bioresour. Technol., 2017, 245, 1194–1205, DOI:10.1016/j.biortech.2017.08.182.
- K. Malik, E. S. Salama, T. H. Kim and X. Li, Enhanced ethanol production by Saccharomyces cerevisiae fermentation post acidic and alkali chemical pretreatments of cotton stalk lignocellulose, Int. Biodeterior. Biodegrad., 2020, 147, 104869, DOI:10.1016/j.ibiod.2019.104869.
- A. Kumar, S. Singh, V. Rajulapati and A. Goyal, Evaluation of pre-treatment methods for Lantana camara stem for enhanced enzymatic saccharification, 3 Biotech, 2020, 10(2), 1–11, DOI:10.1007/s13205-019-2029-5.
- S. Ethaib, R. Omar, S. M. Kamal and D. R. Awang Biak, Comparison of sodium hydroxide and sodium bicarbonate pretreatment methods for characteristic and enzymatic hydrolysis of sago palm bark, Energy Sources, Part A, 2020, 2020, 1–11, DOI:10.1080/15567036.2020.1753857.
- S. Ethaib, R. Omar, M. S. Mazlina, A. D. Radiah and S. L. Zubaidi, Toward sustainable processes of pretreatment technologies of lignocellulosic biomass for enzymatic production of biofuels and chemicals: a review, BioResources, 2020, 15, 10063, DOI:10.15376/biores.15.4.10063-10088.
- H. Shiratori, S. Ikeno, N. Ayame, A. Kataoka, K. Miya, T. Hosono, T. Beppu and K. Ueda, Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor, Appl. Environ. Microbiol., 2006, 72(5), 3702–3709, DOI:10.1128/AEM.72.5.3702-3709.2006.
- K. Shiratori, H. Sasaya, H. Ohiwa, S. Ikeno, N. Ayame, A. Kataoka, T. Miya, T. Beppu and K. Ueda, Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge, Int. J. Syst. Evol. Microbiol., 2009, 59(7), 1764–1770, DOI:10.1099/ijs.0.003483-0.
- A. Izquierdo, L. Goodwin, K. W. Davenport, H. Teshima, D. Bruce, C. Detter, R. Tapia, S. Han, M. Land and L. R. Lynd, Complete genome sequence of Clostridium clariflavum DSM 19732, Stand. Genomic Sci., 2012, 6(1), 104–115, DOI:10.4056/sigs.2535732.
- S. S. Ahmed, M. Akhter, M. Sajjad, R. Gul and S. Khurshid, Soluble production, characterization, and structural aesthetics of an industrially important thermostable β-glucosidase from Clostridium thermocellum in Escherichia coli, BioMed Res. Int., 2019, 2019 DOI:10.1155/2019/9308593.
- P. de Oliveira Rodrigues, L. V. A. Gurgel, D. Pasquini, F. Badotti, A. Góes-Neto and M. A. Baffi, Lignocellulose-degrading enzymes production by solid-state fermentation through fungal consortium among Ascomycetes and Basidiomycetes, Renewable Energy, 2020, 145, 2683–2693, DOI:10.1016/j.renene.2019.08.041.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72(1–2), 248–254, DOI:10.1016/0003-2697(76)90527-3.
- U. K. Laemmli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature, 1970, 227, 680–685, DOI:10.1038/227680a0.
- A. Sluiter, R. Ruiz, C. Scarlata, J. Sluiter and D. Templeton, Determination of extractives in biomass. Laboratory analytical procedure (LAP), 2005, pp. 1–9 Search PubMed.
- A. Zafar, M. N. Aftab, Z. ud Din, S. Aftab, I. Iqbal, A. Shahid, A. Tahir and I. ul Haq, Cloning, expression, and purification of xylanase gene from Bacillus licheniformis for use in saccharification of plant biomass, Appl. Biochem. Biotechnol., 2016, 178(2), 294–311, DOI:10.1007/s12010-015-1872-z.
- D. Dodd and I. K. Cann, Enzymatic deconstruction of xylan for biofuel production, GCB Bioenergy, 2009, 1, 2–17, DOI:10.1111/j.1757-1707.2009.01004.x.
- Q. Li, T. Wu, Z. Qi, L. Zhao, J. Pei and F. Tang, Characterization of a novel thermostable and xylose-tolerant GH 39 β-xylosidase from Dictyoglomus thermophilum, BMC Biotechnol., 2018, 18(1), 1–11, DOI:10.1186/s12896-018-0440-3.
- N. Mhetras, V. Mapare and D. Gokhale, Production of Xylanase and β-Xylosidase Enzymes by Pseudozyma hubeiensis in Solid State Fermentation, Adv. Microbiol., 2019, 9, 467–478, DOI:10.4236/aim.2019.95028.
- B. D. M. Souto, A. Bitencourt, P. Hamman, A. Bastos, E. Noronha and B. Quirino, Characterization of a novel GH3 β-xylosidase from a caatinga goat rumen metagenomic library. In Embrapa Agroenergia-Resumo em anais de congresso (ALICE). In: Congresso Brasileiro De Microbiologia, 30, 2019, 2019, Maceió, Anais, Maceió, SBM Search PubMed.
- A. Zafar, M. N. Aftab, A. Asif, A. Karadag, L. Peng, L. H. U. Celebioglu, M. S. Afzal, A. Hamid and I. Iqbal, Efficient biomass saccharification using a novel cellobiohydrolase from Clostridium clariflavum for utilization in biofuel industry, RSC Adv., 2021, 11(16), 9246–9261, 10.1039/D1RA00545F.
- M. Wahjudi, C. Catherina, N. M. Wangunhardjo, E. Suryadjaja and X. Daniel, Cloning and Over-expression of xynB Gene of Bacillus subtilis subsp. spizizenii W23 into Escherichia coli Origami Host Cells, KnE Life Sciences, 2017, DOI:10.18502/kls.v3i5.986.
- M. N. Aftab, A. Zafar and A. R. Awan, Expression of thermostable β-xylosidase in Escherichia coli for use in saccharification of plant biomass, Bioengineered, 2017, 8(5), 665–669, DOI:10.1080/21655979.2016.1267884.
- S. Silaban, M. Simorangkir, S. Gaffar, I. P. Maksum and T. Subroto, Temperature effect on expression of recombinant human prethrombin-2 in Escherichia coli BL21 (DE3) ArcticExpress, Jurnal Pendidikan Kimia, 2019, 11(3), 122–128, DOI:10.24114/jpkim.v11i3.15779.
- R. Bellaouchi, H. Abouloifa, Y. Rokni, A. Hasnaoui, N. Ghabbour, A. Hakkou, A. Bechchari and A. Asehraou, Characterization and optimization of extracellular enzymes production by Aspergillus niger strains isolated from date by-products, J. Gen. Eng. Biotechnol., 2021, 19, 1–8, DOI:10.1186/s43141-021-00145-y.
- W. E. Bentley, R. H. Davis and D. S. Kompala, Dynamics of induced CAT expression in E. coli, Biotechnol. Bioeng., 1991, 38, 749–760, DOI:10.1002/bit.260380709.
- W. E. Bentley, N. Mirjalili, D. C. Andersen, R. H. Davis and D. S. Kompala, Plasmid-encoded protein: the principal factor in the “metabolic burden” associated with recombinant bacteria, Biotechnol. Bioeng., 1990, 35, 668–681, DOI:10.1002/bit.260350704.
- B. R. Glick, Metabolic load and heterologous gene expression, Biotechnol. Adv., 1995, 13, 247–261, DOI:10.1016/0734-9750(95)00004-A.
- A. Fazaeli, A. Golestani, M. Lakzaei, S. S. Rasi Varaei and M. Aminian, Expression optimization, purification, and functional characterization of cholesterol oxidase from Chromobacterium sp. DS1, PloS One, 2019, 14, e0212217, DOI:10.1371/journal.pone.0212217.
- M. Mühlmann, E. Forsten, S. Noack and J. Büchs, Optimizing recombinant protein expression via automated induction profiling in microtiter plates at different temperatures, Microb. Cell Fact., 2017, 16, 1–12, DOI:10.1186/s12934-017-0832-4.
- F.-L. Du, H.-L. Yu, J.-H. Xu and C.-X. Li, Enhanced limonene production by optimizing the expression of limonene biosynthesis and MEP pathway genes in E. coli, Bioresour. Bioproc., 2014, 1, 1–10, DOI:10.1186/s40643-014-0010-z.
- M. Gutiérrez-González, C. Farías, S. Tello, D. Pérez-Etcheverry, A. Romero, R. Zúñiga, C. H. Ribeiro, C. Lorenzo-Ferreiro and M. C. Molina, Optimization of culture conditions for the expression of three different insoluble proteins in Escherichia coli, Sci. Rep., 2019, 9, 1–11, DOI:10.1038/s41598-019-53200-7.
- C. Moradian, M. R. Fazeli and D. Abedi, Over expression of the Interferon β-1b by optimizing induction conditions using response surface methodology, J. Biol. Today's World, 2013, 2(4), 217–226 Search PubMed.
- Y. Wang, Z. Wang, Y. Duo, X. Wang, J. Chen and J. Chen, Gene cloning, expression, and reducing property enhancement of nitrous oxide reductase from Alcaligenes denitrificans strain TB, Environ. Pollut., 2018, 239, 43–52, DOI:10.1016/j.envpol.2018.04.005.
- M. Maruthamuthu and J. D. van Elsas, Molecular cloning, expression, and characterization of four novel thermo-alkaliphilic enzymes retrieved from a metagenomic library, Biotechnol. Biofuels, 2017, 10(1), 1–17, DOI:10.1186/s13068-017-0808-y.
- A. Hamid and M. N. Aftab, Cloning, purification, and characterization of recombinant thermostable β-xylanase Tnap_0700 from Thermotoga naphthophila, Appl. Biochem. Biotechnol., 2019, 189(4), 1274–1290, DOI:10.1007/s12010-019-03068-0.
- G. L. Rosano and E. A. Ceccarelli, Recombinant protein expression in Escherichia coli: advances and challenges, Front. Microbiol., 2014, 5, 172, DOI:10.3389/fmicb.2014.00172.
- T. R. Whitehead and M. A. Cotta, Identification of a broad-specificity xylosidase/arabinosidase important for xylooligosaccharide fermentation by the ruminal anaerobe Selenomonas ruminantium GA192, Curr. Microbiol., 2001, 43(4), 293–298, DOI:10.1007/s002840010304.
- N. A. Valdez-Cruz, L. Caspeta, N. O. Pérez, O. T. Ramírez and M. A. Trujillo-Roldán, Production of recombinant proteins in E. coli by the heat inducible expression system based on the phage lambda pL and/or pR promoters, Microb. Cell Fact., 2010, 9, 1–16, DOI:10.1186/1475-2859-9-18.
- J. Choi and S. Lee, Secretory and extracellular production of recombinant proteins using Escherichia coli, Appl. Microbiol. Biotechnol., 2004, 64, 625–635, DOI:10.1007/s00253-004-1559-9.
- T. Xu, X. Huang, Z. Li, C. S. Ki Lin and S. Li, Enhanced purification efficiency and thermal tolerance of Thermoanaerobacterium aotearoense β-xylosidase through aggregation triggered by short peptides, J. Agri. Food Chem., 2018, 66(16), 4182–4188, DOI:10.1021/acs.jafc.8b00551.
- S. Sumarsih, Expression of b-xylosidase encoding gene in PHIS/Bacillus megaterium MS system, Ind. J. Trop. Infect. Dis., 2011, 2(1), 25–29, DOI:10.20473/ijtid.v2i1.185.
- P. Turner, G. Mamo and E. N. Karlsson, Potential and utilization of thermophiles and thermostable enzymes in biorefining, Microb. Cell Fact., 2007, 6, 1–23, DOI:10.1186/1475-2859-6-9.
- S. Thapa, H. Li, J. OHair, S. Bhatti, F.-C. Chen, K. A. Nasr, T. Johnson and S. Zhou, Biochemical characteristics of microbial enzymes and their significance from industrial perspectives, Mol. Biotechnol., 2019, 61, 579–601, DOI:10.1007/s12033-019-00187-1.
- F. Rigoldi, S. Donini, A. Redaelli, E. Parisini and A. Gautieri, Engineering of thermostable enzymes for industrial applications, APL Bioeng., 2018, 2, 011501, DOI:10.1063/1.4997367.
- S. Zhang, J. Xie, L. Zhao, J. Pei, E. Su, W. Xiao and Z. Wang, Cloning, overexpression and characterization of a thermostable β-xylosidase from Thermotoga petrophila and cooperated transformation of ginsenoside extract to ginsenoside 20 (S)-Rg3 with a β-glucosidase, Bioorg. Chem., 2019, 85, 159–167, DOI:10.1016/j.bioorg.2018.12.026.
- W. Don and D. Karunathilaka, Biochemical Characterization of the Highly Thermostable β-Xylosidase from Caldicellulosiruptor saccharolyticus, Doctoral dissertation, University of Dayton, 2019, pp. 1–76. http://rave.ohiolink.edu/etdc/view?acc_num=dayton1565064459365423 Search PubMed.
- A. Tomazini, P. Higasi, L. R. Manzine, M. Stott, R. Sparling, D. B. Levin and I. Polikarpov, A novel thermostable GH5 β-xylosidase from Thermogemmatispora sp. T81, New Biotechnol., 2019, 53, 57–64, DOI:10.1016/j.nbt.2019.07.002.
- M. P. Martins, R. Z. Ventorim, R. R. Coura, G. P. Maitan-Alfenas, R. F. Alfenas and V. M. Guimarães, The β-xylosidase from Ceratocystis fimbriata RM35 improves the saccharification of sugarcane bagasse, Biocatal. Agric. Biotechnol., 2018, 13, 291–298, DOI:10.1016/j.bcab.2018.01.009.
- D. R. Kim, H. K. Lim, K. I. Lee and I. T. Hwang, Identification of a New 1, 4-beta-D-xylosidase Pae1263 from the whole genome sequence of Paenibacillus terrae HPL-003, Biotechnol. Bioprocess Eng., 2018, 23(2), 1–23, DOI:10.1007/s12257-017-0246-5.
- S. D. Kane and C. E. French, Characterisation of novel biomass degradation enzymes from the genome of Cellulomonas fimi, Enzyme Microb. Technol., 2018, 113, 9–17, DOI:10.1016/j.enzmictec.2018.02.004.
- D. W. K. Chin, S. Lim, Y. L. Pang, C. H. Lim, S. H. Shuit, K. M. Lee and C. T. Chong, Effects of Organic Solvents on the Organosolv Pretreatment of Degraded Empty Fruit Bunch for Fractionation and Lignin Removal, Sustainability, 2021, 13, 6757, DOI:10.3390/su13126757.
- L. Shuai and J. Luterbacher, Organic solvent effects in biomass conversion reactions, ChemSusChem, 2016, 9, 133–155, DOI:10.1002/cssc.201501148.
- K. Rattanaporn, P. Tantayotai, T. Phusantisampan, P. Pornwongthong and M. Sriariyanun, Organic acid pretreatment of oil palm trunk: effect on enzymatic saccharification and ethanol production, Bioprocess Biosyst. Eng., 2018, 41, 467–477, DOI:10.1007/s00449-017-1881-0.
- A. Oliva-Taravilla, C. Carrasco, L. J. Jönsson and C. Martín, Effects of biosurfactants on enzymatic saccharification and fermentation of pretreated softwood, Molecules, 2020, 25, 3559, DOI:10.3390/molecules25163559.
- D. L. Nelson and M. M. Cox, Princípios de Bioquímica de Lehninger-7, Artmed Editora, 2018 Search PubMed.
- K. C. Shin, M. J. Seo and D. K. Oh, Characterization of β-xylosidase from Thermoanaerobacterium thermosaccharolyticum and its application to the production of ginsenosides Rg 1 and Rh 1 from notoginsenosides R 1 and R 2, Biotechnol. Lett., 2014, 36(11), 2275–2281, DOI:10.1007/s10529-014-1604-4.
- N. Kirikyali, J. Wood and I. F. Connerton, Characterisation of a recombinant β-xylosidase (xylA) from Aspergillus oryzae expressed in Pichia pastoris, AMB Express, 2014, 4(1), 1–7, DOI:10.1186/s13568-014-0068-1.
- Q. J. Yan, L. Wang, Z. Q. Jiang, S. Q. Yang, H. F. Zhu and L. T. Li, A xylose-tolerant β-xylosidase from Paecilomyces thermophila: characterization and its co-action with the endogenous xylanase, Bioresour. Technol., 2008, 99(13), 5402–5410, DOI:10.1016/j.biortech.2007.11.033.
- C. Vieille and G. J. Zeikus, Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability, Microbiol. Mol. Biol. Rev., 2001, 65(1), 1–43, DOI:10.1128/MMBR.65.1.1-43.2001.
- A. Zhou, Y. Hu, J. Li, W. Wang, M. Zhang and S. Guan, Characterization of a recombinant β-xylosidase of GH43 family from Bacteroides ovatus strain ATCC 8483, Biocatal. Biotransform., 2020, 38(1), 46–52, DOI:10.1080/10242422.2019.1631813.
- L. Marcolongo, F. La Cara, G. Del Monaco, S. M. Paixão, L. Alves, I. P. Marques and E. Ionata, A novel β-xylosidase from Anoxybacillus sp. 3M towards an improved agro-industrial residues saccharification, Int. J. Biol. Macromol., 2019, 122, 1224–1234, DOI:10.1016/j.ijbiomac.2018.09.075.
- I. U. Haq, M. Riaz, A. N. Asad-Ur-Rehman, H. Mukhtar and S. Y. E. D. Qurrat-Ul-Ain, Enzymatic hydrolysis of Saccharum officinarum lignocellulosic biomass by genetically modified hyperthermophilic cellulases, Pak. J. Bot., 2020, 52(1), 311–315, DOI:10.30848/PJB2020-1(26.
- C. Dong, Y. Wang, H. Wang, C. S. K. Lin, H. Y. Hsu and S. Y. Leu, New generation urban biorefinery toward complete utilization of waste derived lignocellulosic biomass for biofuels and value-added products, Energy Procedia, 2019, 158, 918–925, DOI:10.1016/j.egypro.2019.01.231.
- M. Dong, S. Wang, F. Xu, J. Wang, N. Yang, Q. Li, J. Chen and W. Li, Pretreatment of sweet sorghum straw and its enzymatic digestion: insight into the structural changes and visualization of hydrolysis process, Biotechnol. Biofuels, 2019, 12(1), 1–11, DOI:10.1186/s13068-019-1613-6.
- P. R. Waghmare, R. V. Khandare, B. H. Jeon and S. P. Govindwar, Enzymatic hydrolysis of biologically pretreated sorghum husk for bioethanol production, Biofuel Res. J., 2018, 5(3), 846–853, DOI:10.18331/BRJ2018.5.3.4.
- C. Rodriguez, A. Alaswad, K. Y. Benyounis and A. G. Olabi, Pretreatment techniques used in biogas production from grass, Renewable Sustainable Energy Rev., 2017, 68, 1193–1204, DOI:10.1016/j.rser.2016.02.022.
- R. Huang, R. Su, W. Qi and Z. He, Understanding the key factors for enzymatic conversion of pretreated lignocellulose by partial least square analysis, Biotechnol. Prog., 2010, 26, 384–392, DOI:10.1002/btpr.324.
- Z. Wang, J. Xu and J. J. Cheng, Modeling biochemical conversion of lignocellulosic materials for sugar production: a review, BioRes, 2011, 6, 5282–5306 Search PubMed.
- I. Ben Taher, P. Fickers, S. Chniti and M. Hassouna, Optimization of enzymatic hydrolysis and fermentation conditions for improved bioethanol production from potato peel residues, Biotechnol. Prog., 2017, 33, 397–406, DOI:10.1002/btpr.2427.
- D. Kumar and G. S. Murthy, Stochastic molecular model of enzymatic hydrolysis of cellulose for ethanol production, Biotechnol. Biofuels, 2013, 6, 1–20, DOI:10.1186/1754-6834-6-63.
- J. P. O'Dwyer, L. Zhu, C. B. Granda, V. S. Chang and M. T. Holtzapple, Neural network prediction of biomass digestibility based on structural features, Biotechnol. Prog., 2008, 24, 283–292, DOI:10.1021/bp070193v.
- T. Vancov and S. Mcintosh, in Cellulase: Types and Action, Mechanism and Uses, Nova Science Publishers, 2011 Search PubMed.
- S. Lu, Q. Wang, Z. Liang, W. Wang, C. Liang, Z. Wang, Z. Yuan, P. Lan and W. Qi, Saccharification of sugarcane bagasse by magnetic carbon-based solid acid pretreatment and enzymatic hydrolysis, Ind. Crops Prod., 2021, 160, 113159, DOI:10.1016/j.indcrop.2020.113159.
- H. Zhang and S. Wu, Efficient sugar release by acetic acid ethanol-based organosolv pretreatment and enzymatic saccharification, J. Agri. Food Chem., 2014, 62(48), 11681–11687, DOI:10.1021/jf503386b.
- A. L. Zimbardi, C. Sehn, L. P. Meleiro, F. H. Souza, D. C. Masui, M. S. Nozawa, L. H. Gulmaraeas, J. A. Jorge and R. P. Furriel, Optimization of β-glucosidase, β-xylosidase and xylanase production by Colletotrichum graminicola under solid-state fermentation and application in raw sugarcane trash saccharification, Int. J. Mol. Sci., 2013, 14(2), 2875–2902, DOI:10.3390/ijms14022875.
- T. Matsuzawa, S. Kaneko and K. Yaoi, Screening, identification, and characterization of a GH43 family β-xylosidase/α-arabinofuranosidase from a compost microbial metagenome, Appl. Microbiol. Biotechnol., 2015, 99(21), 8943–8954, DOI:10.1007/s00253-015-6647-5.
- J. M. Corrêa, E. L. dos Santos, M. R. Simões, M. K. Kadowaki, R. F. Gandra and R. D. C. G. Simão, Optimization of C. crescentus β-xylosidases and expression of xynB1–5 genes in response to agro-industrial waste, Waste Biomass Valorization, 2020, 11(11), 6169–6178, DOI:10.1007/s12649-019-00881-w.
Footnote |
| † These authors have equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |