Open Access Article
Open Access ArticleTransforming waste into value: pomelo-peel-based nitrogen-doped carbon dots for the highly selective detection of tetracycline
Haiyan Qi *a,
Demin Huanga,
Jing Jing*b,
Maoxia Rana,
Tao Jinga,
Ming Zhaoc,
Chenqi Zhanga,
Xiaona Suna,
Rokayya Samid and
Nada Benajibae
*a,
Demin Huanga,
Jing Jing*b,
Maoxia Rana,
Tao Jinga,
Ming Zhaoc,
Chenqi Zhanga,
Xiaona Suna,
Rokayya Samid and
Nada Benajibae
aCollege of Chemistry and Chemical Engineering, Qiqihar University, Qiqihar, Heilongjiang Province 161006, China. E-mail: qhy120@sina.com
bSchool of Medicine and Health, Harbin Institute of Technology, No. 92, West Dazhi Street, Harbin, 150000, P. R.China
cHeilongjiang Industrial Hemp Processing Technology Innovation Center, Qiqihar University, Qiqihar, Heilongjiang Province 161006, China
dDepartment of Food Science and Nutrition, College of Sciences, Taif University, P.O. 11099, Taif-21944, Saudi Arabia
eDepartment of Basic Health Sciences, Deanship of Preparatory Year, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh 11671, Saudi Arabia
First published on 8th March 2022
Abstract
Tetracycline (TC) is widely used as a veterinary drug, and its residue in livestock products could enter the human body and cause damage. In this study, we developed an eco-friendly approach that utilized pomelo peel as a carbon source to synthesize new water-soluble N-doped carbon dots (P-NCDs) with blue fluorescence, obtaining a high quantum yield of up to 76.47% and achieving the goal of turning waste into value. Our prepared P-NCDs can selectively recognized TC, and their fluorescence was quenched based on the IFE. P-NCDs could measure the TC concentration in the linear range of 0–100 μmol L−1 with a detection limit (LOD, S/N = 3) as low as 0.045 μmol L−1. Furthermore, we have successfully applied our P-NCDs to the detection of TC in milk samples with convincing results within 90 s. Overall, our newly synthesized fluorescent sensor, P-NCDs, demonstrated huge potential to become an alternative way to detect TC in a simple, efficient, sensitive way without using any special instruments.
1 Introduction
Tetracycline (TC), as a broad-spectrum antibiotic, has strong disinfection and sterilization efficacy at low cost. Currently, it is widely used in animal husbandry to treat a wide range of disease-causing bacteria, including chlamydia, mycoplasma, and rickettsia. Although TC use in livestock can help treat, control, and prevent bacterial diseases in animals, antibiotic residue will remain in the animal, resulting in the existence of trace amounts of antibiotics in edible livestock products.1 The long-term consumption of dairy products containing TC residue may lead to liver and kidney damage, allergies and toxic reactions, and yellow teeth, seriously affecting human health.2 In fact, it is difficult for TC to degrade in a short period of time, and it easily accumulates in the human body. Recently, this problem relating to residue has attracted attention worldwide. Therefore, it is very important to monitor TC concentrations in animals and pharmaceutical products at all times. Presently, several methods are used to detect TC residue, including HPLC,3 SERS,4 and enzyme-linked immunosorbent assays.5 However, these methods often require laborious sample preparation procedures, they can be complicated and expensive, and they need professional operators, creating huge obstacles for practical detection. In contrast, the use of fluorescence analysis could avoid the above shortcomings, and this approach has simple operation procedures, rapid analysis times, strong stability, and high selectivity and specificity. However, traditional fluorescent materials, including perovskite-type nanocrystals, organic fluorescent dyes, and metal quantum dots, generally have strong toxicity. Therefore, it is urgent and necessary to develop novel fluorescent sensors with low toxicity for the identification and detection of TC, while retaining high sensitivity and selectivity.6In recent years, carbon dots (CDs) have been widely studied. They have unique properties, including excellent optical properties, high stability, easy synthesis, low biotoxicity, and good biocompatibility.7,8 Therefore, they have been applied in a variety of fields, including in sensing and detecting,9,10 drug-loaded therapeutics,11,12 bioimaging,13 medical diagnosis,14 and catalysis reactions.15 At present, various methods have been exploited for CD synthesis. Among these methods, hydrothermal methods are the most commonly used because of the ease of preparation, high efficiency, and environmental friendliness.16,17 Many organic compounds, such as amino acids, citric acid, phenylenediamine, etc., can be used as carbon sources to synthesize CDs. Compared to these materials, biowaste has attracted large amounts of attention recently because of its abundance, nontoxicity, ease of obtainment, low cost, and environmentally friendly properties.18–20 Therefore, numerous types of biowaste, including orange peel,21 leaves,22 banana peel,23–25 soybean roots,26 and flowers,27 have been utilized as carbon sources for CD preparation. These materials have been considered as rising stars in the field of fluorescent materials. However, it remains a big challenge to synthesize CDs with a high fluorescence quantum yield using biowaste.
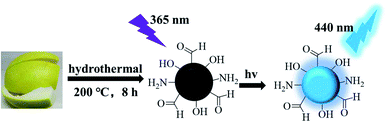
Here, we used pomelo peel as the carbon source to synthesize water-soluble nitrogen carbon dots (P-NCDs) with blue fluorescence, obtaining a high quantum yield of up to 76.47%, via a hydrothermal method (Fig. 1).28–30 Our functional P-NCDs could specifically identify TC, and their fluorescence was quenched due to the inner filter effect (IFE). Furthermore, we have successfully applied our P-NCDs to detect TC in milk samples.
2 Experimental
2.1 Chemicals and instruments
Grapefruit skins were obtained from local supermarkets. Chloramphenicol (Cl), levofloxacin (Lev), kanamycin (Kan), benzylpenicillin potassium (Bep), streptomycin (Str), amoxicillin (Aml), erythromycin (Ery), tetracycline (TC), and ascorbic acid (AA) were purchased from Aladdin Reagent Co., Ltd (Shanghai, China). Tryptophan (Trp), tyrosine (Tyr), thiourea (THU), histidine (His), cysteine (Cys), arginine (Arg), serine (Ser), glycine (Gly), aspartic (Asp), glutamic acid (Glu), threonine (Thr), sulfanilamide (Sul), and phenylalanine (Phe) were obtained from Tianjin Institute of Fine Chemical Industry Co., Ltd. (Tianjin, China). HCl and NaOH were acquired from Beijing Chemical Plant (Beijing, China). Quinine sulfate was purchased from Beijing Enoch Technology Co., Ltd. (Beijing, China). The above reagents are analytically pure, and ultrapure water was used for experiments.A JEM-2100F transmission electron microscope (JEOL, Japan) was used to obtain HRTEM images of P-NCDs.
XRD spectra of samples were measured using a D8-FOCUS German X-ray powder diffractometer (BRUKER-AXS, Germany).
An ESCALAB 250Xi XPS spectrometer (Thermo, USA) was used to analyze the component elements and surface functional groups of the P-NCDs.
An AS380 (Thermo, USA) FTIR spectrometer was used to obtain FT-IR spectra of the P-NCDs. KBr particles were used as the background, the spectral resolution is 4 cm−1, and the scanning range is 4000–500 cm−1.
The UV-vis spectra of the P-NCDs were acquired with a TU-1901 UV-vis spectrophotometer (Purkinje, China). We used xenon lamps that automatically provide excitation wavelengths as a light source for testing, with a scan range from 200–600 nm, a spectral bandwidth of 2 mm, a variable slit width of 2.0 nm, a scan interval of 1 nm, and a medium scanning speed.
The fluorescence spectra were obtained using a Shimadzu RF4301-PC fluorescence spectrometer (Shimadzu, Japan).
Fluorescence lifetimes were recorded using a FLUOROMAX-4 high sensitivity fluorescence spectrometer (HORIBA, USA).
All optical tests were conducted at room temperature. The experimental conditions were set as follows: the excitation wavelength λmax was 325 nm; the wavelength scanning range was 300–600 nm; the incident slit width was 5 nm; the shot slit width was 5 nm; and the scan speed was medium.
2.2 Synthesis of P-NCDs
Fresh grapefruits were bought from a local supermarket near Qiqihar University in China. The pomelo peel of the grapefruits was washed with deionized water, cut into small pieces, and then stored in a dry and ventilated place. Next, pomelo peel was ground into fine particles. 1.00 g of pomelo peel fine particles was weighed and left in a beaker, with the addition of 15 mL of ultrapure water. The mixture was transferred into a 30 mL Teflon-lined stainless-steel autoclave and heated at 180 °C for 8 h. The brown solution obtained upon centrifugation at 5000 rpm for 15 min was dialyzed for 24 h using a dialysis bag (MWCO = 1000 Da). Then, the dialyzed solution was filtered through a 0.22 μm filter membrane. Lastly, solid P-NCDs were obtained after drying the solution via vacuum freezing.2.3 Fluorescence quantum yields
Fluorescence quantum yields were measured using quinine sulfate in sulfuric acid (Ys = 0.54) as a fluorescence standard. The absorbance of the sample in solution at the excitation wavelength was matched with the standard.31 The quantum yield was calculated using the below equation:
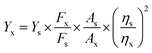 | (1) |
2.4 Detection of tetracycline (TC)
5 mg of P-NCDs was dissolved in 100 mL of deionized water to obtain stock solution with a concentration of 50 μg mL−1. Then, TC stock solution was prepared with a concentration of 0.01 mol L−1. 2 mL of P-NCD stock solution was added into a quartz fluorescence cell, following by the addition of 1 μL of various TC concentrations. After the mixtures were incubated for 90 s at room temperature, fluorescence emission spectra were recorded at an excitation wavelength of 345 nm.2.5 Real sample assays
The method for TC detection in milk was as follows. Briefly, extract solution was prepared using 0.1 mol L−1 Na2EDTA–McIlvaine buffer (pH = 4.0 ± 0.05) and methanol (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Then, 5 g of milk sample was added to 20 mL of extract solution, and the mixture were swirled using a vortex oscillator for 1 min, sonicated at room temperature for 10 min, and left overnight at 4 °C. The next day, the supernatant was collected after centrifugation at 5000 rpm for 15 min. The residue was then extracted twice, using 15 mL and 10 mL, separately. After extraction, the supernatants were combined and transferred to a 50 mL volumetric flask. The samples were then stored at 4 °C for further analysis. To detect TC in milk samples, we performed similar steps to those outlined above and collected the analysis data (n = 3).
1). Then, 5 g of milk sample was added to 20 mL of extract solution, and the mixture were swirled using a vortex oscillator for 1 min, sonicated at room temperature for 10 min, and left overnight at 4 °C. The next day, the supernatant was collected after centrifugation at 5000 rpm for 15 min. The residue was then extracted twice, using 15 mL and 10 mL, separately. After extraction, the supernatants were combined and transferred to a 50 mL volumetric flask. The samples were then stored at 4 °C for further analysis. To detect TC in milk samples, we performed similar steps to those outlined above and collected the analysis data (n = 3).
3 Results and discussion
3.1 Structural characterization of the P-NCDs
The HRTEM image of P-NCDs indicated that the CDs were approximately spherical (Fig. 2A), with a particle size distribution from 1.8 to 5.2 nm and an average particle size of 3.29 nm (Fig. 2B). The XRD spectrum of P-NCDs showed only one obvious diffraction peak, near 2θ = 21.24° (d = 0.419 nm), suggesting an amorphous structure32,33 (Fig. 2C). Furthermore, the FTIR spectrum of P-NCDs displayed several strong and weak absorption peaks. For example, the strong absorption peak at around 3430 cm−1 corresponds to O–H and N–H stretching vibrations;34 the absorption peaks at 2920 and 2980 cm−1 were associated with saturated C–H stretching vibrations; and the absorption peak at 1400 cm−1 was caused by saturated C–N bending vibrations. For weak absorption peaks, the weak absorption peak at 1450 cm−1 was related to C–H bending vibrations; the absorption peaks at 1710 cm−1 and 1050 cm−1 were caused by C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations and C–O flexural vibrations,35,36 respectively; and the absorption peak at 1630 cm−1 was related to O–H and N–H stretching vibrations (Fig. 2D). Lastly, we evaluated additional functional groups based on XPS spectra results.
O stretching vibrations and C–O flexural vibrations,35,36 respectively; and the absorption peak at 1630 cm−1 was related to O–H and N–H stretching vibrations (Fig. 2D). Lastly, we evaluated additional functional groups based on XPS spectra results.
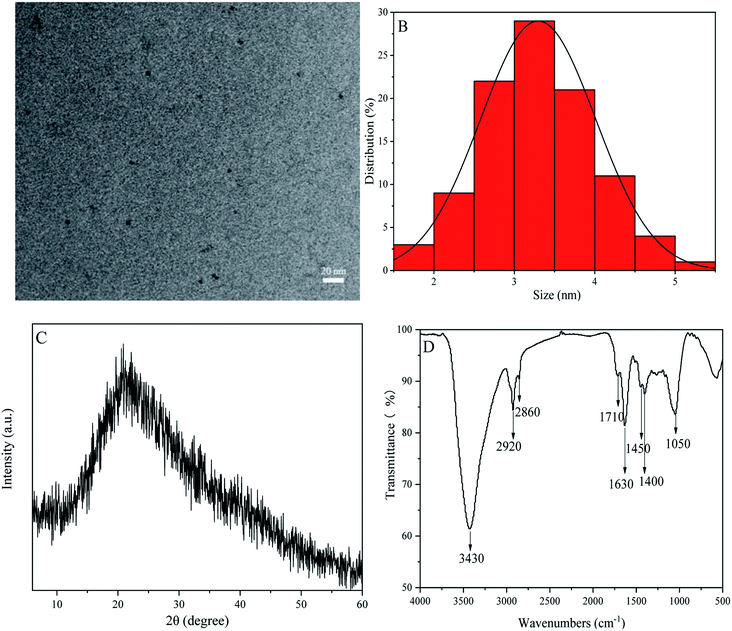 | ||
| Fig. 2 P-NCD structural characterization: (A) a high-resolution TEM (HRTEM) image; (B) the particle size distribution; (C) the XRD spectrum; and (D) the Fourier-transform infrared (FTIR) spectrum. | ||
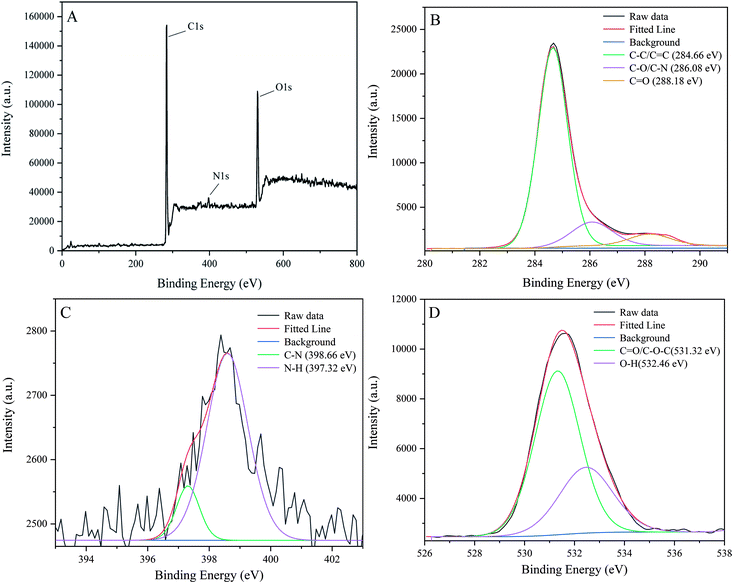
To reveal more functional groups and the element composition at the surface of P-NCDs, we performed XPS analysis. As shown in Fig. 3A, the full survey spectrum indicated that P-NCDs displayed peaks from C 1s, N 1s, and O 1s at 284.66 eV, 399.53 eV, and 531.32 eV, respectively. In addition, the atomic ratios of C, N, and O were 78.60%, 3.25%, and 18.15%, respectively. The C 1s characteristic peak could be deconvoluted into three peaks at 284.66 eV, 286.08 eV, and 288.18 eV, which related to C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–O/C–N, and C
C, C–O/C–N, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, respectively (Fig. 3B). The deconvoluted N 1s peaks were at 398.66 and 397.32 eV, which were associated with C–N and N–H bonds, respectively (Fig. 3C). Moreover, the deconvoluted O 1s peaks are at 531.32 eV and 532.46 eV, which corresponded to C
O, respectively (Fig. 3B). The deconvoluted N 1s peaks were at 398.66 and 397.32 eV, which were associated with C–N and N–H bonds, respectively (Fig. 3C). Moreover, the deconvoluted O 1s peaks are at 531.32 eV and 532.46 eV, which corresponded to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/C–O and O–H bonds, respectively (Fig. 3D).37–39 Combining these results with the infrared analysis results, we concluded that the surface of P-NCDs contained C
O/C–O and O–H bonds, respectively (Fig. 3D).37–39 Combining these results with the infrared analysis results, we concluded that the surface of P-NCDs contained C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, –OH, NH2, and other structures, indicating the easy modification and high water solubility.
O, –OH, NH2, and other structures, indicating the easy modification and high water solubility.
3.2 Optical properties of P-NCDs
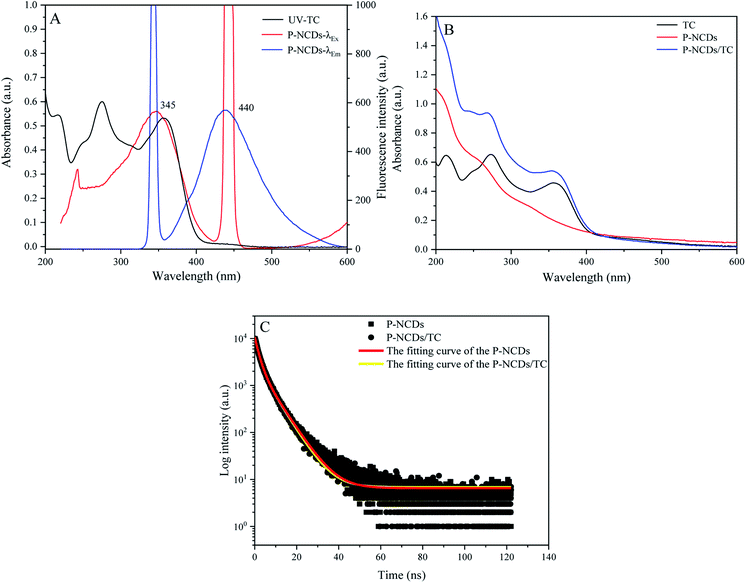
When we measured the fluorescence quantum yield of P-NCDs via the reference method, it was as high as 76.47%. The UV-absorption spectrum of P-NCDs showed strong absorption peaks at 278 nm and 335 nm, which corresponded to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O conjugation and R absorption bands, respectively. This indicated that the structure of the CDs involved an n-electron chromophore group (carbonyl group) and a conjugated structure40–42 (Fig. 4A). An aqueous solution of P-NCDs is colorless under sunlight but emits blue fluorescence under a UV lamp (inset, Fig. 4A). The maximum excitation and emission wavelengths of P-NCDs were 345 nm and 440 nm, respectively (Fig. 4B).
O conjugation and R absorption bands, respectively. This indicated that the structure of the CDs involved an n-electron chromophore group (carbonyl group) and a conjugated structure40–42 (Fig. 4A). An aqueous solution of P-NCDs is colorless under sunlight but emits blue fluorescence under a UV lamp (inset, Fig. 4A). The maximum excitation and emission wavelengths of P-NCDs were 345 nm and 440 nm, respectively (Fig. 4B).
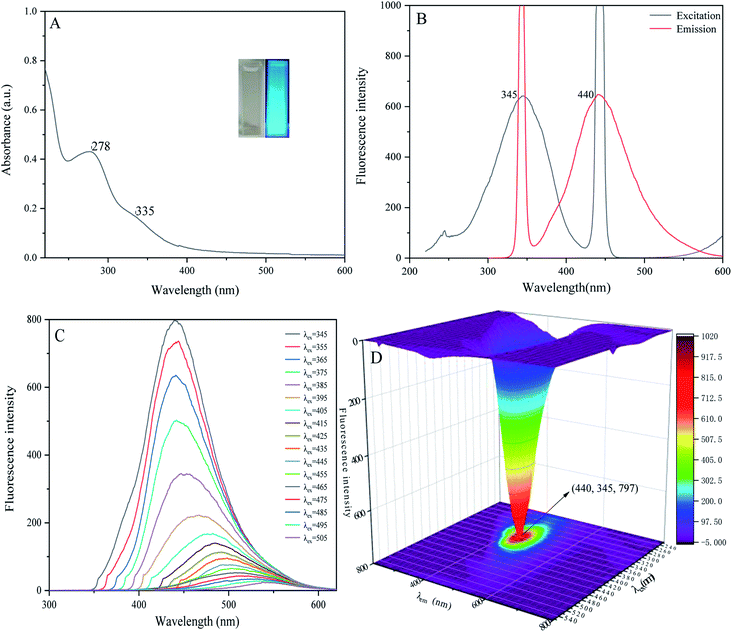 | ||
| Fig. 4 Optical properties of P-NCDs: (A) UV-absorption spectrum; (B) fluorescence spectra; (C) fluorescence emission spectra; and (D) the excitation–emission 3D matrix. | ||
When the excitation wavelength of P-NCDs was increased from 345 nm to 505 nm, the emission peak red-shifted from 440 nm to 550 nm, and the fluorescence intensity was reduced from a relative value of around 800 to as low as 22, suggesting strong excitation wavelength dependence (Fig. 4C).43–45 To further elucidate the changes in the emission spectra, we performed a 3D fluorescence matrix scan of P-NCDs (Fig. 4D). As shown in Fig. 4D, P-NCDs show a broad emission spectrum ranging from 220 to 700 nm, while the excitation wavelengths range from 345 to 555 nm in 10 nm increments. One emission center at 440 nm was detected.
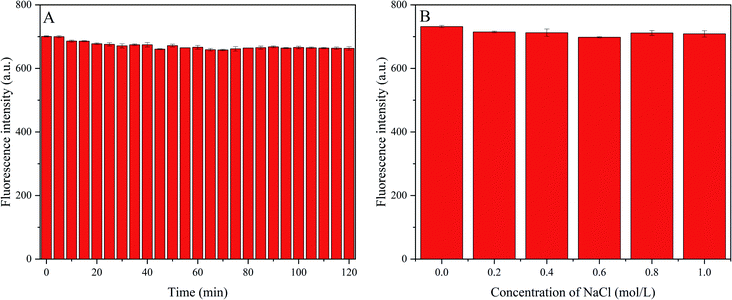
Next, we investigated the fluorescence stability of P-NCDs under different conditions, including various ionic strengths and long-term light exposure. Under ultraviolet light (365 nm) irradiation, the change in the fluorescence intensity was recorded for 120 min (Fig. 5A). We observed a resistance to light drift, with no obvious change in the fluorescence intensity, indicating benefits for analysis and testing. To explore the influence of ionic strength on the fluorescence intensity, we produced 50 μg mL−1 P-NCD solution via adding 200 μL of 450 μg mL−1 P-NCD solution to 2 mL of NaCl (pH = 7) at various concentrations, and we then recorded the change in fluorescence intensity (Fig. 5B). Even when the ionic strength was as high as 1.0 mol L−1, the fluorescence intensity did not change significantly, suggesting a high tolerance to salt. These results suggested the stability and excellent optical performance of P-NCDs, even under extreme environmental conditions. Our findings indicated that P-NCDs could have significant potential for sensing applications in a physiological environment.
3.3 Mechanism of tetracycline (TC) sensing
In the presence of tetracycline, the fluorescence intensity of P-NCDs was decreased by about 94.15% at 440 nm (Fig. 6A). The UV absorption spectrum of TC showed a broad band overlapping with the excitation band of P-NCDs near 345 nm (Fig. 6A). Furthermore, the absorption peak intensity was strengthened in the presence of TC, with no change in the location of the absorption peaks, indicating that no new substances were formed from TC and P-NCDs (Fig. 6B). To better comprehend the mechanism of tetracycline detection, we examined the fluorescence lifetime of P-NCDs with TC. Without TC, the average fluorescence lifetime of P-NCDs was 8.01 ns. In the presence of TC, this was 7.96 ns, suggesting no obvious change in the average fluorescence lifetime (Fig. 6C). Taken together, our results suggest that the fluorescence quenching of P-NCDs was possibly caused by an inner filter effect (IFE) in the presence of TC.46–493.4 Optimization of the detection conditions
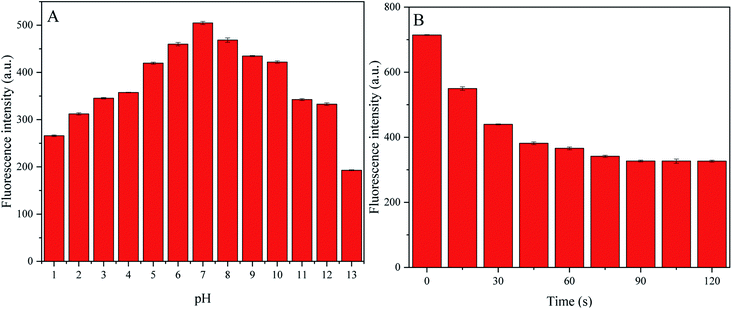
To conduct quantitative analysis of TC, we first evaluated the detection conditions, including pH and response time. Considering that pH is a key factor in actual TC detection, we decided to optimize the pH value to achieve better sensitivity during TC detection. The fluorescence intensity gradually improved when the pH value was increased from 1 to 7, then the intensity dropped when the pH value was increased from 8 to 13, suggesting stable fluorescence intensity even under strongly acidic and alkaline conditions (Fig. 7A). We observed that P-NCDs had wide-ranging pH stability, with the maximum fluorescence intensity at pH 7. In addition, we also examined the response time of P-NCDs toward TC. The fluorescence intensity decreased within 75 s and remained nearly constant up to 120 s (Fig. 7B). Thus, we selected a pH of 7 and a response time of 90 s for subsequent experiments.3.5 Detection of TC
Under the optimum conditions, we explored the use of P-NCDs to detect various concentrations of TC. When the TC concentration increased from 0 to 100 μM, the fluorescence intensity of P-NCDs gradually decreased (Fig. 8A). Specifically, when the TC concentration was in the range of 0–100 μM, a good linear relationship was obtained, with the linear fitting equation (F0 − F)/F0 = 0.0088[TC] − 0.01297 (R2 = 0.9993), with a TC detection limit (S/N = 3) of 0.045 μmol L−1 (Fig. 8B). This indicates that P-NCDs could sensitively detect TC. Moreover, the use of P-NCDs as a fluorescent sensor for detecting TC was also compared with previous works, as shown in Table 1. Our P-NCD sensor system presented a wider linear range and lower detection limit toward TC. Furthermore, we obtained results within 90 s, which was more rapid than other methods. Based on the above results, P-NCDs in this work have huge advantages for the detection and analysis of TC.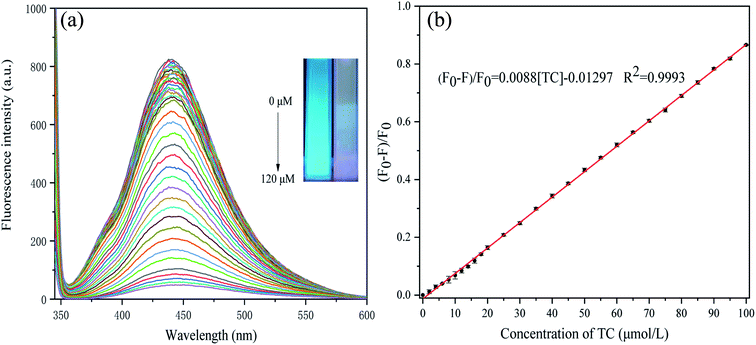 | ||
| Fig. 8 Response of P-NCDs to TC: (A) fluorescence intensity in the presence of various concentrations of TC; and (B) fitting curve of (F0 − F)/F0 vs. TC concentration. | ||
| Sensor | Preparation method | Detection mechanism | Linear range (μmol L−1) | LOD (μmol L−1) | Ref. |
|---|---|---|---|---|---|
| C-dots | Solvothermal | Inner filter effect | 0.5–40 | 0.26–0.48 | 50 |
| CDs | Hydrothermal method | On–off–on | — | 0.22 | 51 |
| F-CQDs | Highly efficient RT synthesis | Inner filter effect | 0.1–25 | 0.085 | 52 |
| N-CDs | Hydrothermal method | Inner filter effect | 0–80 | 0.06 | 53 |
| N/S/P-CDs | Ultrasonic-assisted synthesis | Inner filter effect | 0.1–20 | 0.0444 | 54 |
| CDs | Microwave synthesis | Inner filter effect | 0.1–100 | 0.033 | 55 |
| N,S-CDs | Typical hydrothermal | Inner filter effect | 1.24–165 | 0.0324 | 56 |
| CDs | Amine-aldehyde condensation | Inner filter effect | 10.0–400.0 | 6.00 | 57 |
| P-NCDs | Hydrothermal method | Inner filter effect | 0–100 | 0.045 | This work |
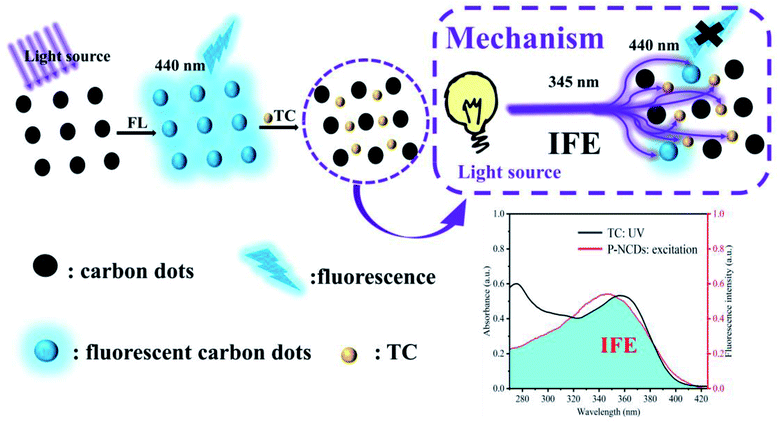
To evaluate the specificity of P-NCDs for TC detection, we evaluated the fluorescence intensity of P-NCDs in the presence of other substances, including various antibiotics (Chl, Flo, Kan, Bep, Str, Amo, and Ery) and organic small molecules (Trp, Tyr, THU, AA, His, Cys, Arg, Ser, Gly, Asp, Glc, Thr, Sul, and Phe). As expected, we observed no obvious effects in the presence of other substances (Fig. 9). The UV absorption spectrum of TC overlaps with the excitation spectrum of P-NCDs at 345 nm, which will lead to the inner filter effect (IFE) phenomenon. When TC was added to P-NCD solution, the excitation light energy of the light source could be absorbed by TC. This reduced the effective actual effect of the excitation light on P-NCDs and the fluorescence of P-NCDs was quenched in the presence of TC (Fig. 10). Therefore, our P-NCDs had the specific capacity to recognize TC. Combined with the good fluorescence stability of P-NCDs in extreme environments, they could be used for the detection of TC in actual samples.
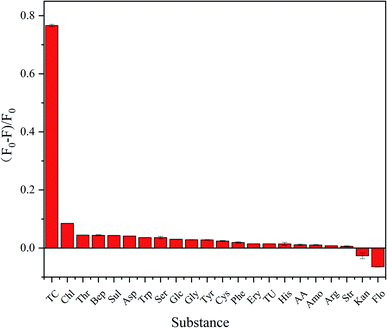 | ||
| Fig. 9 The specificity of using P-NCDs to detect TC over various antibiotics and organic small molecules (concentration: 100 μmol L−1). | ||
3.6 Detection of TC in milk samples
To evaluate the reliability and practical application of P-NCDs, we decided to examine the TC content in milk with our P-NCDs. As described, we obtained milk samples from a local supermarket. Considering the maximum residue limit of tetracycline in milk is as low as 0.232 μmol L−1, our TC detection method could be successfully used to detect the concentration of TC in milk, since our detection limit for TC was 0.045 μmol L−1 (Fig. 8B). A preconditioned sample was tested and TC was not detected. Further, we mixed five different concentrations of TC (40, 45, 50, 55, and 60 μmol L−1) with milk samples and evaluated the performance of our P-NCDs. Unsurprising, the recovery rate was 99.89–101.54% (RSD: <2.1%), showing that our P-NCD sensor had high practicability and accuracy (Table 2).| No. | Added (μmol L−1) | Found (μmol L−1) | Recovery (%) | RSD (%, n = 3) |
|---|---|---|---|---|
| 1 | 40 | 40.31 | 100.8 | 2.3 |
| 2 | 45 | 46.20 | 102.7 | 4.2 |
| 3 | 50 | 50.43 | 100.9 | 4.6 |
| 4 | 55 | 54.36 | 98.8 | 3.8 |
| 5 | 60 | 60.31 | 100.5 | 2.3 |
4 Conclusions
In summary, we proposed an eco-friendly and waste-into-value approach to prepare new water-soluble N-doped carbon dots (P-NCDs) with blue fluorescence using pomelo peel as the carbon source, obtaining a high quantum yield of up to 76.47%. Compared with existing methods, our P-NCDs did not require additional surface modification, and we could successfully detect TC in milk samples with convincing results based on the IFE. In addition, our results were obtained within 90 s, suggesting a more rapid process than other detection methods. Above all, our newly proposed P-NCD fluorescent sensor demonstrated potential to be used for examining TC in livestock products without any special instruments.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work is funded by the National Natural Science Foundation of China (52172092); the Fundamental Research Funds in Heilongjiang Provincial Universities (145109507); the Program for Science and Technology of Qiqihar (GYGG-201903). Taif University Researchers Supporting Project Number (TURSP-2020/140), Taif University, Taif, Saudi Arabia. Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2022R43), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.References
- R. Parker and R. Patel, Analyst, 1994, 119, 2577–2579 RSC.
- S. T. Yang, L. Cao, P. G. J. Luo, F. S. Lu, X. Wang, H. F. Wang, M. J. Meziani, Y. F. Liu, G. Qi and Y. P. Sun, J. Am. Chem. Soc., 2009, 131, 11308–11309 CrossRef CAS PubMed.
- V. G. Silveira, M. S. Oliveira, C. A. A. Almeida, R. Hoff and C. A. Mallmann, Food Anal. Methods, 2018, 11, 2181–2194 CrossRef.
- S. Dhakal, K. L. Chao, Q. Huang, M. Kim, W. Schmidt, J. W. Qin and C. L. Broadhurst, Sensors, 2018, 18, 424–432 CrossRef PubMed.
- L. Naik, R. Sharma, B. Mann, K. Lata, Y. S. Rajput and B. S. Nath, Food Chem., 2017, 219, 85–92 CrossRef CAS PubMed.
- T. C. Wareing, P. Gentile and A. N. Phan, ACS Nano, 2017, 219, 85–92 Search PubMed.
- H. T. Wang, J. R. Bi, B. W. Zhu and M. Q. Tan, Curr. Med. Chem., 2018, 25, 2894–2909 CrossRef CAS PubMed.
- K. Wang, Z. C. Gao, G. Gao, Y. Wo, Y. X. Wang, G. X. Shen and D. X. Cui, Nanoscale Res. Lett., 2013, 8, 122 CrossRef PubMed.
- A. Boruah, M. Saikia, T. Das, R. L. Goswamee and B. K. Saikia, J. Photochem. Photobiol., B, 2020, 209, 111940 CrossRef CAS PubMed.
- Y. D. Xie, D. D. Cheng, X. L. Liu and A. X. Han, Sensors, 2019, 19, 3169 CrossRef CAS PubMed.
- D. Li, Y. Fan, M. W. Shen, I. Banyai and X. Y. Shi, J. Mater. Chem. B, 2019, 7, 277–285 RSC.
- S. L. D'souza, S. S. Chettiar, J. R. Koduru and S. K. Kailasa, Optik, 2018, 158, 893–900 CrossRef.
- X. D. Wang, P. Yan, P. Kerns, S. Suib, L. M. Loew and J. Zhao, J. Electrochem. Soc., 2020, 167, 147515 CrossRef CAS.
- C. X. Lu and L. P. Li, New Carbon Mater., 2018, 33, 12–18 CAS.
- H. Z. Sun, G. D. Y ang and B. Yang, Synthesis, Chem. J. Chin. Univ., 2021, 42, 349–365 Search PubMed.
- Y. S. Zhao, S. S. Jing, X. W. Peng, Z. H. Chen, Y. J. Hu, H. Zhuo, R. C. Sun and L. X. Zhong, Cellulose, 2020, 27, 415–428 CrossRef CAS.
- S. S. Jing, Y. S. Zhao, R. C. Sun, L. X. Zhong and X. W. Peng, ACS Sustainable Chem. Eng., 2019, 7, 7833–7843 CrossRef CAS.
- X. Y. Zhang, M. Y. Jiang, N. Niu, Z. J. Chen, S. J. Li, S. X. Liu and J. Li, ChemSusChem, 2018, 11, 11–24 CrossRef PubMed.
- P. Bhartiya, A. Singh, H. Kumar, T. Jain, B. K. Singh and P. K. Dutta, J. Indian Chem. Soc., 2016, 93, 759–766 CAS.
- S. F. Ou, Y. Y. Zheng, S. J. Lee, S. T. Chen, C. H. Wu, C. T. Hsieh, R. S. Juang, P. Z. Peng and Y. H. Hsueh, Crystals, 2021, 11, 789 CrossRef CAS.
- M. Wang, R. Shi, M. J. Gao, K. L. Zhang, L. L. Deng, Q. F. Fu, L. J. Wang and D. Gao, Food Chem., 2020, 318, 126506 CrossRef CAS PubMed.
- M. Zaib, A. Akhtar, F. Maqsood and T. Shahzadi, Arabian J. Sci. Eng., 2021, 46, 437–446 CrossRef CAS.
- T. N. Nguyen, P. A. Le and V. B. T. Phung, Biomass Convers. Biorefin., 2020 Search PubMed.
- H. Kumar, K. Bhardwaj, R. Sharma, E. Nepovimova, K. Kuca, D. S. Dhanjal, R. Verma, P. Bhardwaj, S. Sharma and D. Kumar, Molecules, 2020, 25, 2812 CrossRef CAS PubMed.
- X. Rong, M. Xie, L. S. Kong, V. Natarajan, L. Ma and J. H. Zhan, Chem. Eng. J., 2019, 372, 294–303 CrossRef CAS.
- N. N. Guo, M. Li, Y. Wang, X. K. Sun, F. Wang and R. Yang, ACS Appl. Mater. Inter., 2016, 8, 33626–33634 CrossRef CAS PubMed.
- K. Huang, Q. He, R. S. Sun, L. L. Fang, H. Y. Song, L. Li, Z. Li, Y. Tian, H. X. Cui and J. B. Zhang, Chem. Phys. Lett., 2019, 731, 136586 CrossRef CAS.
- X. Z. Huo, Y. X. He, S. Q. Ma, Y. Y. Jia, J. M. Yu, Y. Li and Q. Cheng, J. Nanomater., 2020, 2020, 8601307 Search PubMed.
- C. Quan, R. R. Su and N. B. Gao, Int. J. Energy Res., 2020, 44, 4335–4351 CrossRef CAS.
- H. Yang, B. X. Zhou, Y. Zhang, H. M. Liu, Y. W. Liu, Y. He and S. B. Xia, Waste Biomass Valorization, 2021, 12, 2109–2117 CrossRef CAS.
- Y. Zhang, J. J. Xing, S. J. Sun, D. Wu and L. Y. Zeng, Chin. J. Lumin., 2020, 41, 1249–1254 Search PubMed.
- N. Sudhan, K. Subramani, M. Karnan, N. Ilayaraja and M. athish, Energy Fuels, 2017, 31, 977–985 CrossRef CAS.
- R. Venkadeshkumar, M. Shaheer and B. Karthikeyan, Sol. Energy, 2018, 169, 120–127 CrossRef.
- X. L. Yuan, S. J. Liu, G. R. Feng, Y. Y. Liu, Y. D. Li, H. F. Lu and B. Liang, Korean J. Chem. Eng., 2016, 33, 2134–2141 CrossRef CAS.
- J. J. Zhou, Z. H. Sheng, H. Y. Han, M. Q. Zou and C. X. Li, Mater. Lett., 2012, 66, 222–224 CrossRef CAS.
- Y. L. Liu, Q. X. Zhou, Y. Y. Yuan and Y. L. Wu, Carbon, 2017, 115, 550–560 CrossRef CAS.
- H. J. Qi, M. Teng, M. Liu, S. X. Liu, J. Li, H. P. Yu, C. B. Teng, Z. H. Huang, H. Liu, Q. Shao, A. Umar, T. Ding, Q. Gao and Z. H. Guo, J. Colloid Interface Sci., 2019, 539, 332–341 CrossRef CAS PubMed.
- Y. J. Feng, D. Zhong, H. Miao and X. M. Yang, Talanta, 2015, 140, 128–133 CrossRef CAS PubMed.
- Q. M. Yang, J. L. Duan, W. Yang, X. M. Li, J. H. Mo, P. Z. Yang and Q. W. Tang, Appl. Surf. Sci., 2018, 434, 1079–1085 CrossRef CAS.
- S. Thambiraj and D. R. Shankaran, Appl. Surf. Sci., 2016, 390, 435–443 CrossRef.
- L. Jiang, H. H. Ding, M. S. Xu, X. L. Hu, S. L. Li, M. Z. Zhang, Q. Zhang, Q. Y. Wang, S. Y. Lu, Y. P. Tian and H. Bi, Small, 2020, 16, 2000680 CrossRef CAS PubMed.
- Y. Pu, J. N. Leng, D. Wang, J. X. Wang, N. R. Foster and J. F. Chen, Chin. J. Chem. Eng., 2018, 25, 2206–2218 CrossRef.
- R. Mohan, J. Drbohlavova and J. Hubalek, Chem. Phys. Lett., 2018, 692, 196–201 CrossRef CAS.
- L. J. Ren, P. Zhang, R. B. Qi, J. Yin, S. Liu, J. T. Zhang, Q. H. Chen and L. Y. Jiang, Spectrosc. Spectral Anal., 2017, 37, 3354–3359 CAS.
- N. X. Li, F. Lei, D. D. Xu, Y. Li, J. L. Liu and Y. Shi, Opt. Mater., 2021, 111, 110618 CrossRef CAS.
- X. M. Tang, L. S. Wang, H. Ye, H. Q. Zhao and L. S. Zhao, J. Photochem. Photobiol., A, 2022, 424, 113653 CrossRef CAS.
- L. Wang, Y. D. Bi, J. Hou, H. Y. Li, Y. Xu, B. Wang, H. Ding and L. Ding, Talanta, 2016, 160, 268–275 CrossRef CAS PubMed.
- Y. F. Hu, J. J. Zhao, X. F. Li and S. L. Zhao, New J. Chem., 2019, 43, 11510–11516 RSC.
- J. Shen, T. Zhang, Y. Cai, X. Y. Chen, S. M. Shang and J. Li, New J. Chem., 2017, 41, 11125–11137 RSC.
- L. H. Wu, R. Q. Long, T. Li, C. Tang, X. Tong, Y. Guo, S. Y. Shi, H. Y. Xiang and C. Y. Tong, Anal. Bioanal. Chem., 2020, 412, 7481–7489 CrossRef CAS PubMed.
- J. A. O. Granados, P. Thangarasu, N. Singh and J. M. Vazquez-Ramos, Food Chem., 2019, 278, 523–532 CrossRef CAS PubMed.
- Y. Q. Huang, X. S. Huang, H. S. Lin, Z. K. Liu and Y. Zong, Opt. Mater., 2021, 114, 110967 CrossRef CAS.
- Z. M. Zhao, Y. Z. Guo, T. Zhang, J. L. Ma, H. M. Li, J. H. Zhou, Z. W. Wang and R. C. Sun, Int. J. Biol. Macromol., 2020, 164, 4289–4298 CrossRef CAS PubMed.
- Z. D. Gao, M. Y. Liu, K. Xu, M. L. Tang, X. W. Lin, S. W. Hu and X. Q. Ren, Anal. Methods, 2020, 12, 2551–2554 RSC.
- Y. J. Feng, D. Zhong, H. Miao and X. M. Yang, Talanta, 2015, 140, 128–133 CrossRef CAS PubMed.
- S. J. Cheng, J. Q. Zhang, Y. M. Liu, Y. T. Wang, Y. T. Xiao and Y. Zhang, Microchim. Acta, 2021, 188, 325 CrossRef CAS PubMed.
- Y. Yuan, J. H. Liu, R. S. Li, Y. F. Li, C. Z. Huang and S. J. Zhen, Anal. Chim. Acta, 2019, 1063, 144–151 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |