Open Access Article
Open Access ArticleHyperpolarization study on remdesivir with its biological reaction monitoring via signal amplification by reversible exchange†
Hye Jin Jeonga,
Sein Minb,
Sarah Kimb,
Sung Keon Namgoongb and
Keunhong Jeong *a
*a
aDepartment of Physics and Chemistry, Korea Military Academy, Seoul 01805, South Korea. E-mail: doas1mind@kma.ac.kr
bDepartment of Chemistry, Seoul Women's University, Seoul 01797, South Korea
First published on 2nd February 2022
Abstract
Our experiments indicate hyperpolarized proton signals in the entire structure of remdesivir are obtained due to a long-distance polarization transfer by para-hydrogen. SABRE-based biological real-time reaction monitoring, by using a protein enzyme under mild conditions is carried out. It represents the first successful para-hydrogen based hyperpolarization application in biological reaction monitoring.
Nuclear magnetic resonance is a versatile and powerful analytical method for real-time monitoring of significant bio-catalyzed reactions. However, even though NMR has great potential as a reaction monitoring system, providing much structural information, it has not been studied in depth due to low sensitivity. To enhance the sensitivity, the hyperpolarization technique has been suggested which has long been acknowledged as a breakthrough in reaction monitoring by NMR.
There are major hyperpolarization techniques, generating non-Boltzmann population distribution for hyperpolarized signal to noise such as dynamic nuclear polarization,1–5 spin-exchange optical pumping,6–8 para-hydrogen induced polarization (PHIP),9–11 and signal amplification by reversible exchange (SABRE).12–14 In the case of para-hydrogen-based SABRE, the substrate and the para-hydrogen bind to a catalyzing metal complex together, thus allowing polarization to be transferred to the substrate through scalar coupling. To achieve better enhancement, the iridium N-heterocyclic carbene complex15 exhibits the highest polarization transfer efficiency, which delivering an 8100-fold enhancement in 1H NMR signal amplification relative to non-hyperpolarized pyridine. Additionally, enhancements can be increased by employing the bulky electron-donating phosphines of the Crabtree catalyst.16 Most iridium N-heterocyclic carbene catalysts are produced as [Ir(H)2(NHC)(substrate)3]Cl while they are capable of delivering various NMR signal gains such as 1H,17–19 13C,9,20,21 15N,22–24 19F,25,26 31P.27 Recently, a published work has described the extension of the SABRE substrate scope to include a wide range of common drugs such as tuberculosis drugs,28 antifungal,29 and antibiotic agents.30,31 It is mainly N-heterocyclic compounds with low molecular weight. Hyperpolarization experiments using large molecular weight COVID-19 drug candidates have rarely been reported.32 Here, we extend the current scope of biologically relevant SABRE substrates to remdesivir and monitor its enzymatic hydrolysis.
SABRE has been widely considered as a potentially promising reaction monitoring tool for the real-time reaction via hyperpolarization.33,34 After its successful application on the amide-coupling reaction monitoring, it could not be applied to the biological reaction monitoring due to the solvent used for the reaction, mild biological reaction conditions, and mostly, small molecule polarization capacity. Therefore, its direct application on the biological reaction could open up new possibilities in the real-time reaction monitoring via hyperpolarization.
To overcome this COVID-19 pandemic, many researchers have engaged in drug development including drug repurposing. Recently, remdesivir has received emergency use authorization from the FDA, as the first organic medicine to treat COVID-19 around the world. Its prodrug form has been controversial as its intact form is not detectable even after two hours post-injection and as its efficiency for all clinical treatment should also be catalyzed by several key biological reactions including esterase hydrolysis reaction.35–37 Therefore, further research on its molecular level of understanding is still required for enhanced drug development. Along with that, repurposing nucleoside analogue drugs have been considered as the attractive future drug candidates to overcome this pandemic and beyond. They target the RNA polymerase and prevent viral RNA synthesis in a broad spectrum of RNA viruses, including human coronaviruses.38,39 Thus, to develop the most appropriate repurposing nucleoside analogue drug candidates for COVID-19, in-depth timely research on its pharmacokinetics and pharmacodynamics at the molecular level is also essential to overcome this pandemic and its consequent recurrences.
In our previous study, we reported the hyperpolarization on the several anti-viral drug candidates of COVID-19.32 However, these drug candidates have been reported rather ineffective in treating COVID-19.40 Furthermore, even though those unprecedented hyperpolarization on the large drug molecules are new, their polarizations are only done in methanol solvent, which is not applicable in a biological form.32 Our research results indicate successful hyperpolarization of remdesivir via SABRE under mild conditions and hyperpolarization performed in DMSO, a more non-toxic solvent for in vitro application. To provide a more clinical perspective of using this technique, its biological reaction by enzyme was successfully monitored by signal enhancement via SABRE. Moreover, to widen its future applications, we added one more Ir-catalyst by matching external magnetic field condition for efficient polarization transfer to remdesivir. Our findings will expand newly applicable research areas, not only in biological reaction monitoring via NMR, but also in other biomedicine research, in order to cope with dreadful diseases in the future.
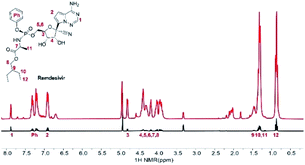
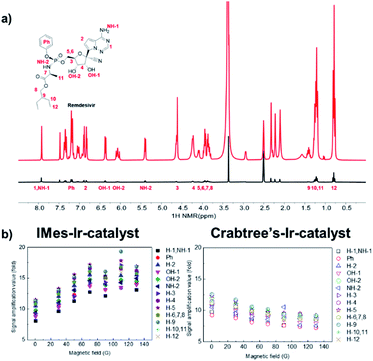
Remdesivir structure has several potential key polarization sources for SABRE: nitrile, amine, and triazine. However, its complex structure and large molecular weight have been considered as the limiting factors for hyperpolarization. Fig. 1 depicts the normal signal and its hyperpolarized signal after SABRE using IMes-Ir-catalyst, which shows the different extent of hyperpolarization of remdesivir. Among the amplified proton signals, proton 6 represents the highest hyperpolarization attached to the 5′-carbon of the nucleoside. However, its polarization indicates no major difference compared to those protons in the whole structure.
Therefore, we can anticipate its polarization transfer is mostly from the SABRE-Relay41–43 or SPINOE,44–46 which are discussed more on conclusion. To optimize hyperpolarization, the external magnetic field is changed, and its polarization is maximized at around 130 G.
However, no major difference was noted in the extent of hyperpolarization in different magnetic fields, which could have been due to Ir-catalyst's fast exchange. Furthermore, its polarization transfer could be from other factors such as SPINOE, other than from the Zeeman effect and J-coupling matching condition. Referring to a recent study on the SABRE hyperpolarization difference between Ir-catalysts,47 we tested the SABRE with Crabtree's-Ir-catalyst, which indicates the different polarization number. Interestingly, its polarization confirmed that the number of hyperpolarization using the Crabtree's-Ir-catalyst was slightly higher than IMes-Ir-catalyst. The hyperpolarization patterns in the different magnetic field also present no major changes from the different Ir-catalyst (Fig. 2 and S1† for structures of Ir-catalysts). Remdesivir is considered to be one of the most important treatments amid this pandemic due to its usage in blocking viral RNA production, leading to an additional study on hyperpolarization in which a more biological solvent has been conducted. Interestingly, SABRE in the CD3SOCD3 with IMes-Ir-catalyst shows the highest polarization efficiency with approximately 17-fold enhanced signal (Fig. 3). DMSO has various biological impacts, such as the ability to increase the skin penetration of chemicals. It can pass through biological membranes, including human skin, probably by changing lipid packing structure and producing breaks in the bilayer.48,49 This result observed in the current research opens a new possibility for applying the in vitro experiment with biological tests since DMSO has been widely used in the in vitro drug test.50,51 Its polarization result of remdesivir among the Ir-catalyst led to higher IMes-Ir-catalyst than Crabtree's-Ir-catalyst, different from the CD3OD. This indicates the polarization transfer mechanism with chelating structure is not solely dependent on the solvent or catalyst. Furthermore, the polarization trend in its structure is the highest in the proton of 9, followed by the proton of 5, which bonded with 5′-carbon of the nucleoside. This different trend in each Ir-catalyst indicates that the polarization transfer efficiency varies depending on solvents in different external magnetic fields and different solvent systems.
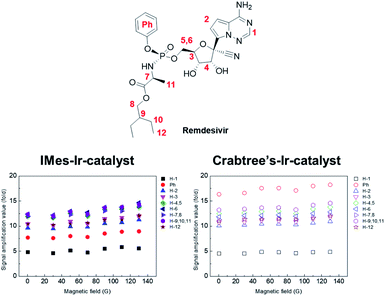 | ||
| Fig. 2 Signal amplification value (SE) of individual protons from hyperpolarized remdesivir using IMes-Ir-catalyst and Crabree's-Ir-catalyst. | ||
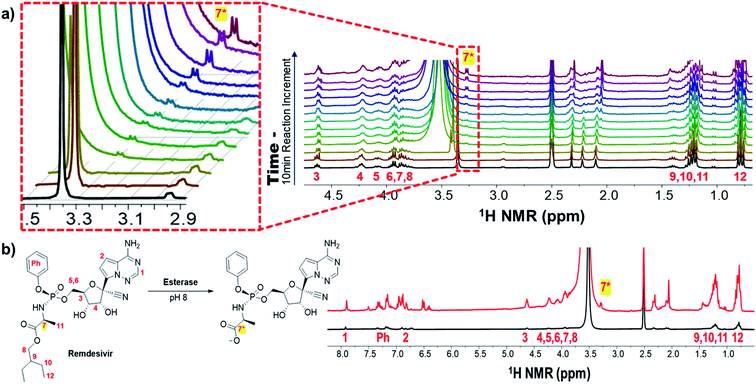
Remdesivir activates analogue, inhibits RNA-dependent RNA polymerase, and prevents viral RNA synthesis as a phosphoramidate prodrug.52 The activation pathway of remdesivir has been proposed to have four steps: (1) cell entrance; (2) enzymatic elimination of masking group with 2-ethylbutyl ester and phenoxy; (3) phosphorylation; and (4) incorporation into COVID-19 RNA.53 The masking group of remdesivir performs increasing hydrophobicity to facilitate cellular entry.54 Also its inventor found that the proton of 7 was shifted to 3 to 4 ppm when the masking group of remdesivir was removed.55,56 Monitoring hydrolysis of a 2-ethylbutyl ester by esterase confirmed the proton of 7* in 3.2 ppm via hyperpolarization (Fig. 4a). The splitting pattern of 7* is controversial because of its doublet instead of quartet. It attributes to several factors. Because of the 5-membered ring intermediate from remdesivir, the splitting pattern of 7* could be affected. The initial metabolism of remdesivir produces an intermediate of cyclopentane containing 7* is made.57 Therefore, stereochemical relations in the equilibrium among protons in 5-membered rings cannot be determined by simply measuring coupling constants, except in cases where the substitution pattern of the specific ring system has been carefully investigated. It could be covered by a water peak close to 7*. Another possibility is that hyperpolarized proton 8 of the 2-ethyl butyl group that is released by enzymatic hydrolysis can be observed. However, the cleavage of the phenoxy group is hard to prove via spectra due to little variation in the signal of aromatic region (Fig. S2†). Its normal cleaved remdesivir signal after the same reaction time and condition was not shown in a scan and its maximum polarization was calculated by ∼22 enhancement after comparing with the multiple scan average (Fig. 4b and S3†). To the best of our knowledge, this is the first study that conducts hyperpolarization reaction monitoring via SABRE in biological condition is conducted for the first time. Furthermore, its NMR-based reaction monitoring on a biologically important prodrug suggests that significantly wide applications in biomedical research. Its application can be significantly widened in the biomedical researches.
Conclusions
SABRE-based hyperpolarization technique has been mainly focused on small organic molecules due to its polarization transfer limitation. Based on the long-distance polarization transfer on the drug molecule, this study expands the possibility of SABRE-based hyperpolarization on the wide range of large drug and biological molecules. By investigating the polarization transfer optimization with external magnetic fields and different Ir-catalyst structures, more detailed SABRE-based polarization transfer studies on remdesivir could be performed.Polarization transfer mechanism needs to be investigated further in the future, which might be explained by SABRE-Relay41–43 or/and SPINOE44–46 for intermolecular dipolar interaction-mediated spin polarization and long-range interaction between hydride and protons in the molecule. The most interesting and important fact that we observed here is that the polarization transfer on remdesivir's entire molecular structure takes less than a minute in the DMSO-d6 solvent compared to methanol-d4. Moreover, this SABRE-based biological real-time reaction monitoring by using protein enzyme via polarization technique under mild conditions (room temperature and 1 atm), among other well-known hyperpolarization techniques, represents the very first successful hyperpolarization application to identify the hidden signal in the biological reaction monitoring-NMR field. Therefore, this finding creates new opportunities, as an analytical tool, for future biomedical applications. The efficient long-range hyperpolarization transfer to the whole structure of remdesivir investigated in this research may facilitate more practical applications in the future, which could be used as NMR- and MRI-based analyses for COVID-19 treatment after removing the toxic Ir-catalyst in the solvent.58 Furthermore, its polarization can be enhanced by higher concentration of para-hydrogen and other nuclei such as 15N and 13C, polarized via SABRE, which will also drastically widen its potential usage with longer relaxation time, avoiding the signal overlap with solvents. Therefore, this real-time hyperpolarization research on the biological molecules with reaction monitoring can also serve as a foundation for future applications in pharmacokinetics and biomedicine chemistry.
Author contributions
H. J. J, S. M, and S. K. conducted experiments and analyzed experimental data. N. K. S and K. J. designed the research. H. J. J and K. J. drafted the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1C1C1007888). This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (No. 2020R1I1A1A01072828).Notes and references
- T. Maly, G. T. Debelouchina, V. S. Bajaj, K. N. Hu, C. G. Joo, M. L. Mak-Jurkauskas, J. R. Sirigiri, P. C. A. Wel, J. Herzfeld, R. J. Temkin and R. G. Griffin, J. Chem. Phys., 2008, 128, 052211–052231 CrossRef PubMed.
- A. J. Rossini, A. Zagdoun, M. Lelli, A. Lesage, C. Copéret and L. Emsley, Acc. Chem. Res., 2013, 46, 1942–1951 CrossRef CAS PubMed.
- Q. Z. Ni, E. Daviso, T. V. Can, E. Markhasin, S. K. Jawla, T. M. Swager, R. J. Temkin, J. Herzfeld and R. G. Griffin, Acc. Chem. Res., 2013, 46, 1933–1941 CrossRef CAS PubMed.
- A. Abragam and M. Goldman, Rep. Prog. Phys., 1978, 41, 395–467 CrossRef CAS.
- J. H. Ardenkjaer-Larsen, B. Fridlund, A. Gram, G. Hansson, L. Hansson, M. H. Lerche, R. Servin, M. Thaning and K. Golman, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 10158–10163 CrossRef CAS PubMed.
- E. Babcock, S. Kadlecek, B. Driehuys, L. W. Anderson, F. W. Hersman and T. G. Walker, Phys. Rev. Lett., 2003, 91, 123003–123006 CrossRef PubMed.
- S. Appelt, A. B. A. Beranga, C. J. Erickson, M. V. Romalis, A. R. Young and W. Happer, Phys. Rev. A, 1998, 58, 1412–1439 CrossRef CAS.
- T. G. Walker and W. Happer, Rev. Mod. Phys., 1997, 69, 629–642 CrossRef CAS.
- F. Reineri, T. Boi and S. Aime, Nat. Commun., 2015, 6, 1–6 Search PubMed.
- K. V. Kovtunov, I. E. Beck, V. I. Bukhtiyarov and I. V. Koptyung, Angew. Chem., 2008, 120, 1514–1517 CrossRef.
- S. B. Duckett and R. E. Mewis, Acc. Chem. Res., 2012, 45, 1247–1257 CrossRef CAS PubMed.
- R. W. Adams, J. A. Aguilar, K. D. Atkinson, M. J. Cowley, P. I. P. Elliott, S. B. Duckett, G. G. R. Green, I. G. Khazal, J. López-Serrano and D. C. Williamson, Science, 2009, 323, 1708–1711 CrossRef CAS PubMed.
- W. Iali, P. J. Rayner, A. Alshehri, A. J. Holmes, A. J. Ruddlesden and S. B. Duckett, Chem. Sci., 2018, 9, 3677–3684 RSC.
- L. S. Lloyd, R. W. Adams, M. Bernstein, S. Coombes, S. B. Duckett, G. G. R. Green, R. J. Lewis, R. E. Mewis and C. J. Sleigh, J. Am. Chem. Soc., 2012, 134, 12904–12907 CrossRef CAS PubMed.
- M. J. Cowley, R. W. Adams, K. D. Atkinson, M. C. R. Cockett, S. B. Duckett, G. G. R. Green, J. A. B. Lohman, R. Kerssebaum, D. Kilgour and R. E. Mewis, J. Am. Chem. Soc., 2011, 133, 6134–6137 CrossRef CAS PubMed.
- J. F. P. Colell, A. W. J. Logan, Z. Zhou, J. R. Lindale, R. Laasner, R. V. Shchepin, E. Y. Chekmenev, V. Blum, W. S. Warren, S. J. Malcolmson and T. Theis, Chem. Commun., 2020, 56, 9336–9339 RSC.
- E. B. Ducker, L. T. Kuhn, K. Munnemann and C. Griesinger, J. Magn. Reson., 2012, 214, 159–165 CrossRef PubMed.
- P. J. Rayner, M. J. Burns, A. M. Olaru, P. Norcott, M. Fekete, G. G. R. Green, L. A. R. Highton, R. E. Mewis and S. B. Duckett, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E3188–E3194 CrossRef CAS PubMed.
- P. J. Rayner, P. Norcott, K. M. Appleby, W. Iali, R. O. John, S. J. Hart, A. C. Whitwood and S. B. Duckett, Nat. Commun., 2018, 9, 4251–4262 CrossRef PubMed.
- W. Iali, S. S. Roy, B. J. Tickner, F. Ahwal, A. J. Kennerley and S. B. Duckett, Angew. Chem., 2019, 131, 10377–10381 CrossRef.
- Z. Zhou, J. Yu, J. F. P. Colell, R. Laasner, A. Logan, D. A. Barskiy, R. V. Shchepin, E. Y. Chekmenev, V. Blum, W. S. Warren and T. Theis, J. Phys. Chem. Lett., 2017, 8, 3008–3014 CrossRef CAS PubMed.
- K. V. Kovtunov, L. M. Kovtunova, M. E. Gemeinhardt, A. V. Bukhtiyarov, J. Gesiorski, V. I. Bukhtiyarov, I. V. Koptyug and B. M. Goodson, Angew. Chem., Int. Ed., 2017, 56, 10433–10437 CrossRef CAS PubMed.
- W. Iali, G. A. I. Moustafa, L. Dagys and S. S. Roy, Magn. Reson. Chem., 2021, 59(12), 1199–1207 CrossRef CAS PubMed.
- R. V. Shchepin, D. A. Barskiy, A. M. Coffey, T. Theis, F. Shi, W. S. Warren, B. M. Goodson and E. Y. Chekmenev, ACS Sens., 2016, 1, 640–644 CrossRef CAS PubMed.
- M. Plaumann, U. Bommerich, T. Trantzschel, D. Lego, S. Dilenberger, G. Sauer, J. Bargon, G. Buntkowsky and J. Bernarding, Chemistry, 2013, 19, 6334–6339 CrossRef CAS PubMed.
- A. M. Olaru, T. B. R. Robertson, J. S. Lewis, A. Antony, W. Iali, R. E. Mewis and S. B. Duckett, ChemistryOpen, 2017, 7, 97–105 CrossRef PubMed.
- V. V. Zhivonitko, I. V. Skovpin and I. V. Koptyug, Chem. Commun., 2015, 51, 2506–2509 RSC.
- H. Zeng, J. Xu, J. Gillen, M. T. McMahon, D. Artemov, J. M. Tyburn, J. A. B. Lohman, R. E. Mewis, K. D. Atkinson, G. G. R. Green, S. B. Duckett and P. C. M. Ziji, J. Magn. Reson., 2013, 237, 73–78 CrossRef CAS PubMed.
- K. MacCulloch, P. Tomhon, A. Browning, E. Akeroyd, S. Lehmkuhl, E. Y. Chekmenev and T. Theis, Magn. Reson. Chem., 2021, 59(12), 1225–1235 CrossRef CAS PubMed.
- O. G. Salnikov, N. V. Chukanov, A. Svyatova, I. A. Trofimov, M. S. H. Kabir, J. G. Gelovani, K. V. Kovtunov, I. V. Koptyug and E. Y. Chekmenev, Angew. Chem., Int. Ed., 2021, 60, 2406–2413 CrossRef CAS PubMed.
- D. A. Barskiy, R. V. Shchepin, A. M. Coffey, T. Theis, W. S. Warren, B. M. Goodson and E. Y. Chekmenev, J. Am. Chem. Soc., 2016, 138, 8080–8083 CrossRef CAS PubMed.
- H. J. Jeong, S. Min, H. Chae, S. Kim, G. Lee, S. K. Namgoong and K. Jeong, Sci. Rep., 2020, 10, 1–13 CrossRef PubMed.
- H. Chae, S. Min, H. J. Jeong, S. K. Namgoong, S. Oh, K. Kim and K. Jeong, Anal. Chem., 2020, 92, 10902–10907 CrossRef CAS PubMed.
- K. Jeong, S. Min, H. Chae and S. K. Namgoong, Magn. Reson. Chem., 2019, 57, 44–48 CrossRef CAS PubMed.
- S. Jomah, S. M. B. Asdaq and M. J. Al-Yamani, J. Infect. Public Health, 2020, 13, 1187–1195 CrossRef PubMed.
- V. C. Yan and F. L. Muller, ACS Med. Chem. Lett., 2020, 11, 1361–1366 CrossRef CAS PubMed.
- Y. Zhu, Z. Teng, L. Yang, S. Xu, J. Liu, Y. Teng, Q. Hao, D. Zhao, X. Li, S. Lu and Y. Zeng, medRxiv, 2020 DOI:10.1101/2020.06.22.20136531.
- G. Li and E. D. Clercq, Nat. Rev. Drug Discovery, 2020, 19, 149–150 CrossRef CAS PubMed.
- E. D. Clercq, Chem.–Asian J., 2019, 14, 3962–3968 CrossRef PubMed.
- M. S. Khuroo, Int. J. Antimicrob. Agents, 2020, 56, 106101–106112 CrossRef CAS PubMed.
- S. S Roy, K. M. Appleby, E. J. Fear and S. B. Duckett, J. Phys. Chem. Lett., 2018, 9, 1112–1117 CrossRef PubMed.
- P. M. Richardson, R. O. John, A. J. Parrott, P. J. Rayner, W. Iali, A. Nordon, M. E. Halse and S. B. Duckett, Phys. Chem. Chem. Phys., 2018, 20, 26362–26371 RSC.
- P. J. Rayner, B. J. Tickner, W. Iali, M. Fekete, A. D. Robinson and S. B. Duckett, Chem. Sci., 2019, 10, 7709–7717 RSC.
- Y. Song, Concepts Magn. Reson., 2021, 12, 6–20 CrossRef.
- I. Marco-Rius, S. E. Bohndiek, M. I. Kettunen, T. J. Larkin, M. Basharat, C. Seeley and K. M. Brindle, Contrast Media Mol. Imaging, 2014, 9, 182–186 CrossRef CAS PubMed.
- G. Navon, Y.-Q. Song, T. Rõõm, S. Appelt, R. E. Taylor and A. Pines, Science, 1996, 271, 1848–1851 CrossRef CAS.
- J. F. P. Colell, A. W. J. Logan, Z. Zhou, J. R. Lindale, R. Laasner, R. V. Shchepin, E. Y. Chekmenev, V. Blum, W. S. Warren, S. J. Malcolmson and T. Theis, Chem. Commun., 2020, 56, 9336–9339 RSC.
- T. J. Anchordoguy, J. F. Carpenter and L. M. Crowe, Biochim. Biophys. Acta, 1992, 1104, 117–122 CrossRef CAS.
- B. W. Barry, J. Controlled Release, 1987, 6, 85–97 CrossRef CAS.
- N. C. Santos, J. Figueira-Coelho, J. Martins-Silva and C. Saldanha, Biochem. Pharmacol., 2003, 65, 1035–1041 CrossRef CAS PubMed.
- M. Colucci, F. Maione, M. C. Bonito, A. Piscopo, A. D. Giannuario and S. Pieretti, Pharmacol. Res., 2008, 57, 419–425 CrossRef CAS PubMed.
- T. K. Warren, R. Jordan, M. K. Lo, A. S. RAy, R. L. Mackman, V. Soloveva, D. Siegel, M. Perron, R. Bannister, H. C. Hui, N. Larson, R. Strickley, J. Wells, K. S. Stuthman, S. A. V. Tongeren, N. L. Garza, G. Donnelly, A. C. Shurtleff, C. J. Retterer, D. Gharaibeh, R. Zamani, T. Keeny, B. P. Eaton, E. Grimes, L. S. Welch, L. Gomba, C. L. Wilhelmsen, D. K. Nichols, J. E. Nuss, E. R. Nagle, J. R. Kugelman, G. Palacios, E. Doerffler, S. Neville, E. Carra, M. O. Clarke, L. Zhang, W. Lew, B. Ross, Q. Wang, K. Chun, L. Wolfe, D. Babusis, Y. Park, K. M. Stray, I. Trancheva, J. Y. Feng, O. Barauskas, Y. Xu, P. Wong, M. R. Braun, M. Flint, L. K. McMullan, S. S. Chen, R. Fearns, S. Swaminathan, D. L. Mayers, C. F. Spiropoulou, W. A. Lee, S. T. Nichol, T. Cihlar and S. Bavari, Nature, 2016, 531, 381–385 CrossRef CAS PubMed.
- Z. Wang and L. Yang, New J. Chem., 2020, 44, 12417–12429 RSC.
- S. J. Hecker and M. D. Erion, J. Med. Chem., 2008, 51, 2328–2345 CrossRef CAS PubMed.
- B. K. Chun, M. O. Clarke, E. Doerffler, H. C. Hui, R. Jordan, R. L. Mackman, J. P. Parrish and A. S. Ray, US Pat., 9,724,360 B2, 2017.
- D. Bao, W. Chang and D. Nagarathnam, AU Pat., 2009329917 B2, 2013.
- R. T. Eastman, J. S. Roth, K. R. Brimacombe, A. Simeonov, M. Shen, S. Patnaik and M. D. Hall, ACS Cent. Sci., 2020, 6, 672–683 CrossRef CAS PubMed.
- A. Manoharan, P. J. Rayner, W. Iali, M. J. Burns, V. H. Perry and S. B. Duckett, ChemMedChem, 2018, 13, 352–359 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2ra00062h |
| This journal is © The Royal Society of Chemistry 2022 |