 Open Access Article
Open Access ArticleBactericidal effect of nanostructures via lytic transglycosylases of Escherichia coli†
Soma Mimura,
Tomohiro Shimizu,
Shoso Shingubara,
Hiroaki Iwaki and
Takeshi Ito
and
Takeshi Ito *
*
Graduate School of Science and Engineering, Kansai University, Yamatecho 3-3-35, Suita, Osaka 564-8680, Japan. E-mail: t.ito@kansai-u.ac.jp
First published on 10th January 2022
Abstract
Nanostructures exhibit a bactericidal effect owing to physical interaction with the bacterial cell envelope. Here, we aimed to identify the mechanism underlying the bactericidal effect of nanostructures based on bacterial autolysis, in contrast to previous reports focusing on structural characteristics. The time profiles of active cell ratios of the Escherichia coli strains (WT, ΔmltA, ΔmltB, Δslt70), incubation time of the wild-type (WT) strains, and autolysis inhibition of WT strains were evaluated with respect to the bactericidal effect of the applied nanostructures. Addition of Mg2+, an autolysis inhibitor, was not found to cause significant cell damage. The incubation phase was significantly associated with envelope damage. The lytic transglycosylase-lacking strain of Slt70 (Δslt70) also showed only minimal envelope damage. Our results indicate that nanostructures may act by triggering bacterial autolysis.
1 Introduction
Natural nanostructures, such as the cicada and dragonfly wing surfaces or gecko spatula, possess bactericidal properties.1–5 These nanostructures exert their bactericidal effects via physical interaction with bacterial cells, leading to cell envelope rupture.6,7 This physical mechanism is fundamentally different from that of chemicals such as antibiotic reagents or nanosized metals. Nanostructures-based bactericidal materials are advantageous since they are non-toxic for animal cells, and their effect is permanent, as long as they maintain their structures. These materials are also attractive for medical applications as their physical mechanism of action prevents the occurrence of antimicrobial resistant (AMR) bacteria.8,9 Although many studies have reported the development of nanostructure-based bactericidal materials and their bactericidal performance,10–14 the mechanism underlying the bactericidal effect of these materials has not been fully clarified due to its complexity.Previous studies have largely focused on structural characteristics such as the pitch, aspect ratio, density,2 and elastic energy of nanostructures, for explaining the bactericidal mechanism.15–17 These studies highlighted three possible bactericidal mechanisms: (i) direct nanostructure penetration into the bacterial cell envelope,1 (ii) stretching and rupture of the envelope in between multiple nanostructures,6,7 (iii) envelope deformation due to interaction between the bacterial cell and the nanostructure.18 In any case, bacteria-nanopattern interaction is certainly the first trigger. Some researchers have tried to analyze the interaction theoretically.6,7,19 In the early stage, the cell envelope was approximated as a thin elastic layer.7 The actual layer thickness and composition were neglected, leading to representation inaccuracies in the actual bacterial cell envelope structure. In a very recent study, Velic et al. analyzed bacteria-nanopattern interaction using 3D finite element modeling (3D-FEM), when the bacterial cell was assumed to be multi-layered.19 They concluded that nondevelopment deformation of the envelope around the pillar tips induced significant in-plane strains, which would trigger the damage to DNA, proteins, or formation of oxidative products. They also pointed out that the membrane rupture could feasibly occur as a result of such significant strains. Several researchers characterized the interaction interface between the nanostructures and bacteria by using the high-resolution microscopy such as helium ion microscopy or transmission electron microscopy (TEM).4,20,21 Bandara et al. found that bacteria adhered to the nanostructure surface using their extracellular polymeric substances (EPS) and the finger-like projections and that initial envelope damage occurs without direct co-tact between the nanostructure and the cell envelope.4 Jenkins et al. reported that TiO2 nanostructure induced deformation and penetration of bacterial envelope of Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus), but did not lead the rupture of cell envelopes of some bacteria. These nanostructures inhibit bacterial cell division and trigger reactive oxygen species (ROS) production, thereby increasing oxidative stress proteins.21 In summary, these studies indicate that bacterial death due to the effect of nanostructures is based on a physical stress-related mechanism.
Based on these findings, we sought to understand the bactericidal mechanism by modifying the structural characteristics of nanostructures and bacterium characteristics. For instance, we prepared Si nanopillar arrays with specific wettability properties and evaluated their bactericidal effect. Our results showed that E. coli preferred to adhere onto the hydrophobic surfaces with the nanostructure bactericidal effect increasing with the water contact angle (WCA), indicating that the hydrophobic nanomaterial surfaces are more effective in killing E. coli.22 Moreover, these results were concerned with motility of the cell:23 E. coli uses hydrophobic flagella for movement, which facilitate cell adhesion onto hydrophobic surfaces. Nevertheless, the complexity of the interaction between the nanostructure and bacterial cell has not allowed a mechanism to be clearly identified.
Here, we focused on bacterial autolysis as a possible bactericidal mechanism of nanostructures. Previous studies have shown that bacterial envelopes lyse upon nanostructure interaction, which led researchers, including us, to conclude that the bactericidal effect was due to the physical penetration. However, previous findings actually may not imply direct physical penetration of bacterial cell envelope by the nanostructures. Indeed, Jenkins et al. reported that some nanostructures did not penetrate the bacterium envelope.21 Thus, we reasoned that the bacterium envelope is lysed by bacteria itself (autolysis), which is considered as a secondary trigger, because of the physical stress due to nanostructures presence.19,24,25 The cell envelope of E. coli is composed of three layers: the cytoplasmic membrane (inner membrane), the peptidoglycan (PG) or murein layer (periplasm), and the outer membrane (OM).26 The combination of PG layer and outer membrane is called the cell wall. The PG layer resides between the cytoplasmic membrane and the outer membrane.27,28 PG is composed of alternating linear glycan strands, β-1,4-linked N-acetyl-β-D-glucosamine (GlcNAc) and N-acetyl-β-D-muramic acid (MurNAc) resides which are interconnected by the cross-linking of their peptide subunits. At least 35 PG hydrases act as autolysis enzymes. Here, we focused on lytic transglycosylases, which are expressed by bacteria to break the strong PG layer during cell division.29–32 Lytic transglycosylases are also known as N-acetylmuramidases, which not only cleave the β-1,4 glycosidic bond between the MurNAc and GlcNAc unit but also concomitantly catalyzes an intramolecular glycosyl transfer reaction whereby a 1,6-anhydro bond is formed between C1 and C6 of N-acetylmuramic acid. We specifically focused on three lytic transglycosylase types, MltA (membrane-bound lytic transglysosylase A),24,25 MltB (membrane-bound lytic transglysosylase B)24–26 and Slt70 (soluble lytic transglysosylases),24,27 which are considered to be major components of the E. coli autolytic system,33 and comparatively evaluated the nanostructure bactericidal performance on strains of E. coli lacking each enzyme with respect to wild-type E. coli. MltA and MltB are membrane proteins located on the outer membrane, and their substrates are soluble component, including muropeptide, peptide, glycan chain and PG. Slt70 is soluble protein located mainly in the periplasm, with 20–30% of the total Slt70 content located in the cytoplasmic membrane. The activity of Slt70 is strictly controlled, and normally this potentially suicidal enzyme does not degrade the cell wall.34 Slt70 acts only in PG. Our results also indicated that bactericidal performance differed depending on the type of enzyme lacking in bacteria. We found that the effect on Δslt70 was lowest, indicating that the location and expression level of lytic enzymes are very important for bacterial death induced due to physical interaction with nanostructures.
2 Experimental
2.1 Materials and chemicals
Propidium iodide (PI), hydrogen peroxide, sulfuric acid, nitric acid, hydrochloric acid, sodium hydrogen carbonate, sodium hydrogen phosphate, sodium dihydrogen phosphate, magnesium chloride, acetone and ammonia water were purchased from Fujifilm-Wako Chemicals (Tokyo, Japan). SYTO 9 was obtained from Thermo Fisher Scientific (Waltham, MA, USA). Hydrofluoric acid was obtained from Daikin Industries, Ltd (Osaka, Japan). 1,1,1,3,3,3-Hexamethyldisilazane (HMDS) was purchased from Shin-Etsu Chemical Co., Ltd (Tokyo, Japan). Polystyrene (PS) beads (diameter: 200 nm) were purchased from Funakoshi Co., Ltd (Tokyo, Japan). A p-type Si wafer (crystal orientation: 100, diameter: 101.6 mm) was purchased from SUMCO Co (Tokyo, Japan).2.2 Preparation of Si nano-pillar array
We fabricated a Si nanopillar array substrate and used it as an artificial nanostructure, which was constructed to mimic the cicada wing surface. We previously reported the manufacturing of Si nanopillar arrays by colloidal lithography and metal-assisted chemical etching (MacEtch),22,35 which allows controlling of nanopillar array dimensions. The gap between the nanopillars was controlled by modifying the diameter of PS beads. Here, the nanopillar pitch was set to 200 nm. Therefore, we chose PS beads with the diameter of 200 nm. The nanopillar diameter was controlled by changing the condition of the oxygen plasma etching of PS beads. The diameter was tuned to 120 nm. The nanopillar depth was controlled by changing the wet etching time on MacEtch. A height of approximately 520 nm was obtained (Fig. 1(a)). The fabricated nanopillars were arranged regularly (Fig. 1(b)). The Si nanopillar array substrate size was 20 mm × 20 mm. Following MachEtch operation, we etched the catalytic layer of Au thin film using nitrohydrochloric acid solution, and PS beads by dipping into the piranha solution (H2O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 = 1
H2SO4 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4). Finally, the nanopillar array wettability was tuned to obtain a hydrophobic structure to allow adherence of E. coli cells.23 Here, the nanopillar array substrate was dipped in HMDS media (HMDS
4). Finally, the nanopillar array wettability was tuned to obtain a hydrophobic structure to allow adherence of E. coli cells.23 Here, the nanopillar array substrate was dipped in HMDS media (HMDS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetone = 19
acetone = 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for 15 min, following the thermal processing at 150 °C for 1 min. The water contact angle (WCA) of the Si nanopillar array increased drastically from 11° to 145° after thermal processing (Fig. 1(c) and (d)), confirming similar hydrophobicity of the obtained material to that of the cicada wing surface.
1) for 15 min, following the thermal processing at 150 °C for 1 min. The water contact angle (WCA) of the Si nanopillar array increased drastically from 11° to 145° after thermal processing (Fig. 1(c) and (d)), confirming similar hydrophobicity of the obtained material to that of the cicada wing surface.
 | ||
| Fig. 1 SEM images of fabricated nanopillar array: (a) cross-sectional image and (b) front view. Photographs of the evaluation of WCA: (c) after dipping in piranha solution, (d) after HMDS coating. | ||
2.3 Preparation of E. coli strains
We used four E. coli strains, three of which were genetically engineered for specific traits. E. coli strain RP437 (courtesy of Dr Y. Sowa, Hosei Univ.) was used as the WT strains. This strain could undergo cell division using autolysis enzymes and swim in the culture media using flagellar rotation. JW2784-KC (ΔmltA), JW2671-KC (ΔmltB), and JW4355-KC (Δslt70), which were obtained from National Institute of Genetics, Microbial Physiology Laboratory, NBRP E. coli lacked lytic transglycosylases.E. coli cells were grown in tryptone broth (TB) (1% bactotryptone, 0.5% NaCl) at 30 °C with shaking at 170 rpm for 3 and 12 hours. In the first three hours of cultivation, E. coli cells were in logarithmic growth phase. This condition is written as the “logarithmic phase”, i.e. active cell division was observed. In the latter condition, cells were in the stationary phase. This condition is written as “stationary phase”, with a constant live E. coli cell number. The cell division rate decreased further and was eventually equal to the rate of cell death due to decrease in nutrient levels. Cultured cells were suspended in motility buffer (10 mM potassium phosphate buffer: PBS, pH 7.0) twice. The cell suspension was diluted with motility buffer to an OD600 = 0.1.
The cells were stained with SYTO 9 and propidium iodide (PI) for the cell envelope damage test. One milliliter of the diluted cell suspension was mixed with 3.3 mM SYTO 9-DMSO solution and 10 mM PI-DMSO solution. After mixing, the cell suspension was allowed to stand for 15 min.
2.4 Observation of attached cells and analysis of active cell ratio
After the cell suspension (10 µl) was dropped onto the Si nanopillar substrates, a coverslip (18 × 18 mm, thickness: 0.15 mm) was placed over it and fixed using double-sided tape (0.1 mm thick). The dropped suspension was pushed out by the coverslip to ensure that the cell suspension covered the entire area of the nanostructured surfaces. After holding for 1 min to allow cell adherence onto the nanostructure surfaces, the sample with coverslip was placed onto the fluorescence microscope stage (Eclipse Ti-E, Nikon, Tokyo, Japan). The microscope was equipped with 20× objective lens (CFI S Plan Fluor ELWD 20X, N.A.0.45, Nikon, Tokyo, Japan) and 2.5× C-mount relay lens (VM2.5X, Nikon).The bactericidal activity of a nanostructure is expected to originate from its physical properties. The cell envelope is stretched by the nanostructured surface, which causes cell breakage. The cell envelope damage after nanostructure surface adhesion was assessed by fluorescence microscopy using commercially available DNA-staining reagents such as SYTO 9 and PI, which provide information about cell envelope damage. Therefore, E. coli cells stained with SYTO 9/PI were illuminated with epi-fluorescence from a mercury lamp. We used the EGFP/FITC filter set (excitation filter: 480/30 nm, dichroic mirror: 505 nm, emission filter: 535/40 nm; 39002 AT, Chroma, MA) to observe SYTO 9 fluorescence and the Texas Red/mCherry filter set (excitation filter: 560/40 nm, dichroic mirror: 600 nm, emission filter: 635/60; 39010 AT, Chroma, MA) to observe PI fluorescence. The fluorescence images were captured every 5 min for 120 min using a CCD camera (DMK23G618; The Imaging Source, Bremen, Germany). Excitation light was irradiated for 1 s for each capturing process to reduce photobleaching. After the bactericidal activity test, we observed the morphology of cells adhering on the nanopillar surface using a scanning electron microscopy (SEM, JSM-7500F, JEOL Ltd).
The time course of the active cell ratio, which was calculated by multiplying the ratio of the number of SYTO 9-stained cells (green-strained cells) to the number of attached cells (green- and red-stained cells) with 100, was analyzed using ImageJ.36,37 Each data shows the average with error bars (N = 3) and was obtained from independent experiments.
3 Results and discussion
3.1 Active cell ratio dependent with growth phase
Gene expression activities of some proteins vary depending on the growth phase. The resulting dependence of active cell ratio on the growth phase is shown in Fig. 2(a) and (b). As the control experiment, the active cell ratio on the flat Si substrate is also shown in Fig. 2(a), when the cells were grown in the logarithmic phase. The active cell ratio on the flat Si substrate did not decrease and remained higher than 96% even after 120 min, indicating clearly that no envelope damage was present on the flat surface. In contrast, the active cell ratio of WT on the nanopillar array decreased gradually with time and was lower than 8% around 90 min, eventually reaching 4.4% after 120 min (Fig. 2(a)), when the cells were in the logarithmic phase. In contrast, when the cells were in the stationary phase, the active cell ratio remained at 93%, even on the nanopillar array. In addition, SEM images of WT cell grown in the logarithmic phase and the stationary phase after the bacterial activity test are shown in Fig. 2(c) and (d), respectively. Fig. 2(c) clearly shows that nanopillars penetrated into the cell and the pillars were deformed around the cell adhesion area. In contrast, Fig. 2(d) shows that the cell adhered on the nanopillar surface and there was no change in its shape. These results may be attributed to bacterial sensitivity to stress, which is often higher in the logarithmic growth phase than in the stationary phase.38 In addition, metabolic activities of actively dividing bacteria, and gene expression levels for lytic transglysosylases in dividing cells are considered high in the logarithmic growth phase as well. Based on these results, subsequent experiments (described in Section 3.2 and 3.3) were carried out with bacterial cells in the logarithmic growth phase.3.2 Active cell ratio of E. coli strains lacking autolysis enzymes
As mentioned in the introduction section, TiO2 nanopillars induced deformation of the bacterial envelope.21 The nanostructures inhibit bacterial cell division, and also trigger ROS production and increased oxidative stress proteins production. Hence, bacterial cell death due to nanostructure is, in fact, caused by reactions to physical stress. We assume that the physical stress induced by nanostructure that leads to cell wall lysis is similar to that occurring during cell division. Thus, we evaluated the active cell ratio according to the location or absence of autolysis-related enzymes using cell envelope damage reagents, PI and SYTO 9. MltA and MltB are located on the outer membrane, whereas Slt70 is mainly located in the periplasm. All of these enzymes can lyse PG. Instances of fluorescent microscopy images and time-course analysis of the active cell ratio are shown in Fig. 3 and 4(a), respectively. Images on Fig. 3 were extracted from the time-lapse imaging as shown in ESI (see Movie S1 to S5†). The data were obtained using Si nanopillar arrays.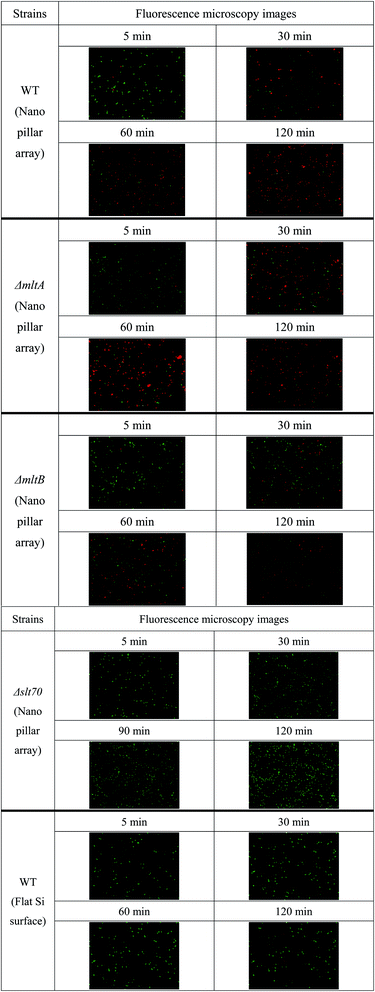 | ||
| Fig. 3 Fluorescent microscopy images of E. coli strains to evaluate the active cell ratio over time. Green and red cells show non-damaged cells and damaged cells, respectively. | ||
As shown in Fig. 4(a), the active cell ratio of WT E. coli decreased gradually and was lower than 8% around 90 min, eventually reaching 4.4% after 120 min. The active cell ratio of ΔmltA strain also decreased gradually, reaching 15% at approximately 80 min. The ΔmltA strain decrease rate was larger than that of WT strain. However, the time-dependent active cell ratios of WT and ΔmltA strains may be regarded similar until 60 minutes due to high data variance. Conversely, this ratio may be regarded to show a clear difference between WT and ΔmltA strains after 60 min due to data variance being smaller. The active cell ratio of ΔmltB strain decreased slowly, reaching 40% at approximately 90 min. In contrast, the time-dependent active cell ratio of Δslt70 strain remained at approximately 84% even at 120 min. This is the highest active cell ratio among the used E. coli strains. The time profile of the active cell ratio of Δslt70 strain was also different from those of the other strains. SEM image of Δslt70 cell grown in the logarithmic phase after the bacterial activity test is shown in Fig. 4(b), which shows that the cell adhered on the nanopillar surface and there was no change in its shape as well as WT cell grown in the stationary phase (Fig. 2(d)). These results indicate that the active cell ratio after adhesion onto the nanostructure varied depending on the location or absence of autolysis-related enzymes, even though the same bacterial species, E. coli, was used in the experiments. In particular, based on the active cell ratio data obtained from Δslt70 strain, Slt70 expression after adhesion may be considered to be the main factor responsible for cell lysis. Unlike MltA and MltB, Slt70 is located mainly in the periplasm. MltA and MltB might not play major catalytic roles in autolysis,33 because they locate on the outer membrane. Distance between these enzymes and PG are too long to degrade PG. The main autolytic enzyme may be Slt70, still this is not yet clarified. Although the activity of Slt70 is known to be strictly controlled and Slt70 does not work normally, stress of contact with the nanostructure might stimulate the activity of the enzyme. To this end, our results indicate that the enzymatic disintegration of the strong PG is responsible for cell death.
Bandara et al. reported that bacteria attach onto the nanostructure surface via the EPS layer, which enables strong adhesion between the bacteria and nanostructure.4 Simultaneously, bacteria experience stress due to unfavorable nanostructured surface. This causes bacteria to attempt to avoid the nanostructure surface, which generates a large amount of shear stress on the bacterium envelope. The authors then assumed that this shear stress would cause separation of inner membrane and outer membrane. However, the tight interaction between the outer and inner membranes renders it difficult to cause damage with mechanical force alone. The authors also pointed out that initial cell envelope damage occurred without direct contact. Velic et al. simulated the deformation of the multilayered bacterial cell, which was well matched to the actual structure, on the bacteria-nanopattern interaction.19 They concluded that nondevelopment deformation of the envelope around the pillar tips induced significant in-plane strains. Considering these findings together with our results here, Slt70 may be considered to be activated when the cell undergoes significant in-plane strains due to bacteria-nanopattern interaction, which subsequently leads to cell wall lysis. Additionally, the starting point of cell wall lysis would not occur near contact point with nanostructure, but at any point of existence of Slt70.
Xie reported that the graphene-based bactericidal material caused autolysis by its strong oxidation.39 The physical stimulation triggers ROS production and increases oxidative stress.21 In our previous study, we showed that envelope lysis occurred within 20 min after cell adhesion on the nanopillar array,22 which is roughly equivalent to the time required for cell division.40 Based on the findings, it can be assumed that physical stimulation with the adhesion on the nanostructure may be considered a cause of cell oxidative stress, which activates the cell wall lysis enzyme. While our results indicated that the PG layer was lysed via enzymatic reaction of lytic transglycosylases as a second trigger of membrane damage, we did not determine whether lysis extends to the OM that has enough stiffness and strength to maintain the cell morphology. However, since the OM is stiffer than the PG layer, and E. coli can switch into a PG layer deficient state, “called L-form”, that grow even though they lack the PG layer,41,42 the process of the cell lysis is expected to include a mechanism of the OM lysis following the PG layer lysis. For example, it is conceivable that some enzymes in the intracellular liquid reach and lyse the OM after the PG layer is lysed. Nevertheless, further research is needed to confirm this hypothesis. As an example, it is necessary to study whether this hypothesis holds for other bacteria.
3.3 Active cell ratio dependent with cultured media
Leduc et al. reported that autolytic system of E. coli was controllable by only changing incubation conditions.43 In fact, 10 mM of Mg2+ efficiently inhibited the autolytic process. To clarify the involvement of autolysis in bacterial death due to nanostructures, the active cell ratio of WT strain on the Si nanopillar array was evaluated upon MgCl2 addition to the motility buffer (10m M PBS, pH = 7.0) and prior to the cell staining with SYTO 9 and PI. Here, concentration of MgCl2 was set to 10 mM. Fig. 5 shows the time-dependent active cell ratio on PBS with 10 mM Mg2+, which indicate no degradation. On the other hands, the active cell ratio with PBS decreased gradually reaching zero at approximately 90 min (Fig. 5 closed circles). These results indicate that cell envelope damage did not occur when the autolysis was inhibited. | ||
| Fig. 5 The time-dependent active cell ratio in PBS (black circles, as same as Fig. 2(a)) and PBS with 10 mM Mg2+ (orange triangles). Here, the growth phase was logarithmic phase for each test. Data show the average of 3 independent experiments with error bars. | ||
From these results, we further conclude that the envelope damage was caused by the physical stimulation of the contact with nanostructures following the generation of oxidative stress and ROS, which trigger the envelope lysis enzyme activation, especially Slt70. In the near future, we plan to conduct a time-series analysis to clarify the mechanisms proposed here.
4 Conclusion
Nanostructures exert bactericidal effect owing to their physical interactions with the bacterial cell envelope. Many researchers have sought to clarify the underlying mechanism based on structural properties of nanostructure; however, only few research groups have focused on bacterial characteristics. In this study, we focused on bacterial autolysis during cell division to explain the mechanism underlying the bactericidal effect of nanostructures. Time profiles of active cell ratios of four E. coli strains (WT, ΔmltA, ΔmltB, Δslt70) showed that Δslt70 strain showed almost no envelope damage. The incubation time of WT was strongly related to the envelope damage. In fact, the most damaged cells were counted in the log-growth phase, which means that cell envelope damage was more likely to occur when the cell division was activated. In addition, cell envelope damage was evaluated with inhibited autolysis in WT strain, addition of Mg2+. The results showed almost no cell damage under this condition. These results also indicate that the bactericidal mechanism due to nanostructures is based on the bacterial autolysis system.Author contributions
SM were responsible for analyzing all data obtained from experiments. TI was the chief investigator and also responsible for the data consideration. HI was responsible for the selection of E. coli and biological experiments. TI and HI were responsible for supervision, funding acquirement, and project administration. TS and SS were responsible for fabrication and observation of the Si nanopillar array. All authors contributed to the writing of the final manuscript and approved the manuscript to be published. All members of study team contributed to the management or administration of this study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Some of E. coli strains (ΔmltA, ΔmltB, and Δslt70) were distributed from NBRP, National Bio Resource Project, Japan. This work was partially supported by JSPS KAKENHI Grant Number 21H01773.References
- E. P. Ivanova, J. Hassan, H. K. Webb, V. K. Truong, G. S. Watson, J. A. Watson, V. A. Baulin, S. Pogodin, J. Y. Wang, M. J. Tobin, C. Löbbe and R. J. Crawford, Small, 2012, 8, 2489–2494 CrossRef CAS PubMed.
- S. M. Keller, O. Habimana, J. Lawler, B. O'Rilly, S. Daniels, E. Casey and A. Cowley, ACS Appl. Mater. Interfaces, 2016, 8, 14966–14974 CrossRef PubMed.
- E. P. Ivanova, J. Hassan, H. K. Webb, G. Gervinskas, S. Juodkazis, V. K. Truong, A. H. F. Wu, R. N. Lamb, V. A. Baulin, G. S. Watson, J. A. Watson, D. E. Mainwaring and R. J. Crawford, Nat. Commun., 2013, 4, 2838 CrossRef PubMed.
- C. D. Bandara, S. Singh, I. O. Afara, A. Wolff, T. Tesfamichael, K. Ostrikov and A. Oloyede, ACS Appl. Mater. Interfaces, 2017, 9, 6746–6760 CrossRef CAS PubMed.
- D. W. Green, K. K. H. Lee, J. A. Watson, H.-Y. Kim, K.-S. Yoon, E.-J. Kim, J.-M. Lee, G. S. Watson and H.-S. Jung, Sci. Rep., 2017, 7, 41023 CrossRef CAS PubMed.
- S. Pogodin, J. hasan, V. A. Baulin, H. K. Webb, V. K. Truong, T. H. P. Nuguyen, V. Boshkovikj, C. J. Gluke, G. S. Watson, J. A. Watson, R. J. Crawford and E. P. Ivanova, Biophys. J., 2013, 104, 835–840 CrossRef CAS PubMed.
- X. Li, Phys. Chem. Chem. Phys., 2016, 18, 1311–1316 RSC.
- L. M. Weiner, A. K. Webb, B. Limbago, M. A. Dudeck, J. Patel, A. J. Kallen, J. R. Edwards and D. M. Sievert, Infect. Control Hosp. Epidemiol., 2016, 37, 1288–1301 CrossRef PubMed.
- L. M. Weiner-Lastinger, S. Abner, J. R. Edwards, A. J. Kallen, M. Karlsson, S. S. Magill, D. Pollock, I. See, M. M. Soe, M. S. Walters and M. A. Dudeck, Infect. Control Hosp. Epidemiol., 2020, 41, 1–18 CrossRef PubMed.
- Y. Xie, X. Qu, J. Li, D. Li, W. Wei, D. Hui, Q. Zhang, F. Meng, H. Yin, X. Xu, Y. Wang, L. Wang and Z. Zhou, Sci. Total Environ., 2020, 738, 139714 CrossRef CAS PubMed.
- S. K. Saini, M. Halder, Y. Singh and R. V. Nair, ACS Biomater. Sci. Eng., 2020, 6, 2778–2786 CrossRef CAS PubMed.
- C. Heo, C. Jeong, H. S. Im, J. U. Kim, J. Woo, J. Y. Lee, B. Park, M. Suh and T. Kim, Nanoscale, 2017, 9, 17743 RSC.
- A. Jaggessar, H. Shahali, A. Mathew and P. K. D. V. Yarlagadda, J. Nanobiotechnol., 2017, 15, 64 CrossRef PubMed.
- A. Hayles, J. Hasan, R. Bright, D. Palms, T. Brown, D. Baker and K. Vasilev, Mater. Today Chem., 2021, 22, 10622 Search PubMed.
- D. P. Linklater, M. De Volder, V. A. Baulin, M. Werner, S. Jessel, M. Golozar, L. Maggini, S. Rubanov, E. hassen, S. Juodkazis and E. P. Ivanova, ACS Nano, 2018, 12, 6657–6667 CrossRef CAS PubMed.
- E. P. Ivanova, D. P. Linklater, M. Werner, V. A. Baulin, X. M. Xu, N. Vranken, S. Rubanov, E. Hassen, J. Wandiyanto, V. K. Truong, A. Elbourne, S. Maclaughlin, S. Juodlazis and R. J. Crawford, Proc. Natl. Acad. Sci. U.S.A., 2020, 117, 12598–126205 CrossRef CAS PubMed.
- A. Vailei, N. Lin, J.-F. Bryche, G. Mckay, M. Canva, P. G. Charette, D. Nguyen, C. Moraes and N. Tufenkji, Nano Lett., 2020, 10, 5720–5727 CrossRef PubMed.
- K. Nowlin, A. Boseman, A. Cowell and D. LaJeunesse, J. R. Soc. Interface, 2015, 12, 20140999 CrossRef PubMed.
- A. Velic, J. Hasan, Z. Li and P. K. D. V. Yarlagadda, Biophys. J., 2021, 120, 217–231 CrossRef CAS PubMed.
- C. D. Bandara, G. Ballerin, M. Leppänen, T. Tesfamichael, K. K. Osrtikov and C. B. Whitcherch, ACS Biomater. Sci. Eng., 2020, 6, 3925–3932 CrossRef CAS PubMed.
- J. Jenkins, J. Mantell, C. Neal, A. Gholinia, P. Verkade, A. H. Nobbs and B. Su, Nat. Commun., 2020, 11, 1626 CrossRef CAS PubMed.
- K. Nakade, K. Jindai, T. Sagawa, H. Kojima, T. Shimizu, S. Shingubra and T. Ito, ACS Appl. Nano Mater., 2018, 1, 5736–5741 CrossRef CAS.
- K. Jindai, K. Nakade, K. Masuda, T. Sagawa, H. Kojima, T. Shimizu, S. Shingubara and T. Ito, RSC Adv., 2020, 10, 5673–5680 RSC.
- K. Lewis, Microbiol. Mol. Biol. Rev., 2000, 64, 503–514 CrossRef CAS PubMed.
- M. A. Kacena, E. E. Smith and P. Todd, Appl. Microbiol. Biotechnol., 1999, 52, 437–439 CrossRef CAS PubMed.
- N. Nanninga, Microbiol. Mol. Biol. Rev., 1998, 62, 110–129 CrossRef CAS PubMed.
- W. Vollmer, D. Blanot and M. A. de Pedro, FEMS Microbiol. Rev., 2008, 32, 149–167 CrossRef CAS PubMed.
- W. Vollmer and U. Bertsche, Biochim. Biophys. Acta, 2008, 1778, 1714–1734 CrossRef CAS PubMed.
- J. van Heijenoort, Microbiol. Mol. Biol. Rev., 2011, 75, 636–663 CrossRef CAS PubMed.
- K. Ehlert, J.-V. Höltje and M. F. Templin, Mol. Microbiol., 1995, 16, 761–768 CrossRef CAS PubMed.
- A. Dijikstra, A. J. F. Hermann and W. Keck, FEBS Lett., 1995, 366, 115–118 CrossRef.
- J.-V. Höltje, D. Mirelman, N. Sharon and U. Schwarz, J. Bacteriol., 1975, 124, 1067–1076 CrossRef PubMed.
- J.-V. Höltje, Arch. Microbiol., 1995, 164, 243–254 CrossRef PubMed.
- A. S. Betzner, L. Ferreira, J.-V. Höltje and W. Keck, FEMS Microbiol. Lett., 1990, 67, 161–164 CrossRef CAS.
- T. Ito, K. Nakade, N. Asai, T. Shimizu and S. Shingubara, ECS Trans., 2017, 75, 1–5 CrossRef CAS.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- M. D. Abramoff, P. J. Magalhães and S. J. Ram, Biophot. Int., 2004, 11, 36–42 Search PubMed.
- T. G. Aldsworth, R. L. Sharman and C. E. R. Dodd, Cell. Mol. Life Sci., 1999, 56, 378–383 CrossRef CAS PubMed.
- C. Xie, P. Zhang, Z. Guo, X. Li, Q. Pang, K. Zheng, X. He, Y. Ma, Z. Zhang and I. Lynch, Sci. Total Environ., 2020, 747, 141546 CrossRef CAS PubMed.
- S. Taheri-Araghi, S. Bradde, J. T. Sauls, N. S. Hill, P. A. Levin, J. Pasulsson, M. Vergassola and S. Jun, Curr. Biol., 2015, 25, 385–391 CrossRef CAS PubMed.
- R. Mercier, Y. Kawai and J. Errington, Elife, 2014, 3, e04629 CrossRef PubMed.
- E. R. Rojas, C. Billings, P. D. Odermatt, G. K. Auer, L. Zhu, A. Miguel, F. Chang, D. B. Weibel, J. A. Theriot and K. C. Huang, Nature, 2018, 559, 617–621 CrossRef CAS PubMed.
- M. Leduc, R. Kasra and J. Heijenoort, J. Bacteriol., 1982, 152, 26–34 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07623j |
| This journal is © The Royal Society of Chemistry 2022 |


