Open Access Article
Open Access ArticlePeroxidase catalytic activity of carbon nanoparticles for glutathione detection†
Lijuan Chena,
Xiang Lib,
Zezhi Lic,
Kejian Liub and
Jianping Xie *b
*b
aCollege of Food and Bioengineering, Zhengzhou University of Light Industry, Zhengzhou, 450001, China
bZhengzhou Tobacco Research Institute of CNTC, Zhengzhou, 450001, China. E-mail: ztridicp@126.com; Tel: +86371-67672113
cBeijing Technology and Business University, Beijing, 100048, China
First published on 22nd December 2021
Abstract
Peroxidases are present widely in microorganisms and plants, and catalyze many reactions. However, the activity of natural peroxidases is susceptible to external conditions. We prepared carbon nanoparticles (CNPs) using an environmentally friendly and simple method. These CNPs were demonstrated to possess intrinsic peroxidase-like activity. CNPs could catalyze the reaction of a peroxidase substrate, 3,3,5,5-tetramethylbenzidine (TMB), in the presence of H2O2 to produce a blue solution at 652 nm. CNPs exhibited higher peroxidase activity than that of other carbon-based nanomaterials. Moreover, CNPs retained their high peroxidase activity after being reused several times. Glutathione (GSH) can change the blue color of oxidized TMB into a colorless hue at 652 nm. Based on this fact, qualitative and quantitative approaches were employed to detect GSH using a colorimetric method. This method showed a broad detection range (2.5–50 μM) with a limit of detection of 0.26 μM. This method was shown to be accurate for GSH detection in a cell culture medium compared with that using a commercial assay kit. Our findings could facilitate application of CNPs in biomedical areas.
1 Introduction
Enzymes are produced by living cells. An enzyme is highly specific and has high catalytic efficiency for its substrate. Peroxidases are a large group of enzymes which have a role in various biological processes. Peroxidases catalyze the oxidation of substrates with hydrogen peroxide as the electron acceptor.1 A peroxidase exists mainly in the peroxisome of the carrier with iron porphyrin as the auxiliary group. A peroxidase can catalyze hydrogen peroxide (H2O2), the oxidation of phenols and amines, and the oxidation products of hydrocarbons. However, the storage conditions for commercial peroxidases are rigorous (−20 °C) and the cost of production is extremely high.Recently, great efforts have been made to synthesize nanomaterial-based mimetic enzymes (“nanozymes”) due to their superiority over natural enzymes, such as excellent stability, cost-effectiveness, and environmentally friendly production.2 Nanozymes, with their large specific surface area, exhibit excellent peroxidase catalytic activity, such as for metal oxides,3–5 carbon nanomaterials,6–8 metal–organic frameworks9 and composite materials (e.g., Co3O4 complex,10 CeO2-wrapped Ag2S microspheres11). These peroxidase-like entities can oxidize the substrate 3,3,5,5-tetramethylbenzidine (TMB) in the presence of H2O2 with a color change from colorless to blue.
Recently, graphite-based conductive nanomaterials or N-doped carbon nanomaterials (e.g., carbon nanotubes,12 graphene oxide (GO),7 carbon nanodots8) have been applied extensively in electrochemical energy-storage or catalysis areas,13–15 and been found to possess intrinsic peroxidase mimetic activity. Preparation of carbon nanomaterials is cost-effective and environmentally friendly. Carbon-based nanomaterials are low cost and easy to obtain. These features broaden their application in bioassays and medical-diagnostic areas, such as detection of dopamine,11 glucose,8,16 L-lactic acid,17 and glutathione (GSH).18,19 Carbon nanoparticles (CNPs) have been exploited due to their good biocompatibility in biological and biomedical areas in recent years. They have been used as probes for biomolecule detection in cells,20 dual-palladium and mercury sensors for live-cell imaging,21 and oral vaccine adjuvants.22 CNPs with sp2-bonded carbon atoms have been prepared via electrochemical oxidation of graphite in deionized water.23 In addition, CNPs have good solubility in water due to abundant of carboxyl and hydroxy functional groups on their surface. CNPs possess abundant surface sites with many functional groups and excellent electron-transport properties, which enable them donate or accept electrons. The outstanding electron-transfer performance of CNPs may enable them to display excellent peroxidase-mimicking activity and can be used to detect H2O2-related bioactive molecules. Few studies have focused on whether CNPs possess peroxidase-like catalytic activity.
GSH is a vital tripeptide thiol antioxidant. It is present widely in cells and involved in many metabolic processes.18 It plays an important part in the biochemical defense system in the human body by removing free radicals, protecting protein structure and enzyme activity, and regulating immune function.24 Various methods have been developed for GSH detection: chromatography,25 mass spectrometry,26 absorbance spectroscopy,27 capillary electrophoresis,28 and immunoiluminescence.29 Among these methods, absorbance spectroscopy (also called a “colorimetric” method) has received increasing attention due to its low cost as well as its quick and easy operation. The sulfhydryl group in GSH, cysteine (Cys), and homocysteine (Hcy) can cause fading of the color of a reaction solution (mimic enzyme + TMB + H2O2 system).30,31 The change in color intensity varies with the amount of thiol agents. Based on this reaction mechanism, a colorimetric method for detection of GSH was established.
In the present study, CNPs were found to possess peroxidase-like activity. They displayed considerable catalytic activity to TMB compared with that of horseradish peroxidase (HRP) according to kinetic studies. Based on the peroxidase-like activity of CNPs, a convenient and rapid method for colorimetric detection of the cellular GSH level was proposed (Scheme 1). The chemical reaction could be monitored by the naked eye as well as recorded by an ultraviolet-visible (UV-vis) spectrometer. These results indicate that CNPs can be used widely in biomedical areas.
2 Experimental
2.1 Materials
GSH and these types of amino acid were purchased from Acros Organics (The Hague, Belgium). TMB was obtained from TCI (Shanghai) Development (Shanghai, China). H2O2 was from Aladdin (Shanghai, China). GO and multiwalled carbon nanotubes (MWCNTs) were purchased from Timesnano (Chengdu, China). CNPs were prepared according to the method described by Chen and colleagues.23 The solution of CNPs was 3 mg mL−1. A commercial GSH assay kit was purchased from Solarbio Science & Technology (Beijing, China).2.2 Measurement of catalytic activity
In this assay, the reaction parameters were identical to those of a previous report30 with some modification. Briefly, 100 μL of TMB solution (15 mM in dimethyl sulfoxide) and 20 μL of H2O2 (30%) were added to phosphate-buffered saline (PBS), followed by addition of a 20 μL aliquot of CNPs suspension (3 mg mL−1) and the total volume made up to 2 mL. After 5 min, the absorbance at 652 nm was measured by a UV-vis spectrophotometer (Cary 300; Agilent Technologies, Santa Clara, CA, USA). For measurement of the optimum pH value, the pH value was set as 4.0, 5.0, 6.0, or 7.0. Kinetic measurements were recorded in time-course mode by monitoring the absorbance change at 652 nm with different concentrations of CNPs. The catalytic activity of carbon-based nanomaterials was determined with a fixed concentration (30.0 μg mL−1), including CNPs(E) (i.e., CNPs solution mixed with ethanol at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (v/v) followed by removal of ethanol from the solution), GO, and MWCNTs on a UV-vis spectrophotometer without a time delay. The reusability of CNPs was evaluated by catalyzing TMB and centrifugation to collect the blue reaction solution after 5 min for four times. The Michaelis–Menten constant was calculated according to the Lineweaver–Burk plot:
2 (v/v) followed by removal of ethanol from the solution), GO, and MWCNTs on a UV-vis spectrophotometer without a time delay. The reusability of CNPs was evaluated by catalyzing TMB and centrifugation to collect the blue reaction solution after 5 min for four times. The Michaelis–Menten constant was calculated according to the Lineweaver–Burk plot:  , where v is the initial velocity, Km is the Michaelis constant, Vmax is the maximal reaction velocity, and C is the substrate concentration.16
, where v is the initial velocity, Km is the Michaelis constant, Vmax is the maximal reaction velocity, and C is the substrate concentration.16
2.3 Detection of GSH
Briefly, 20 μL of GSH at different concentrations was added to solution 0, 2.5, 5, 10, 15, 20, 25, 50, 100, 150, 200 and 400 μmol L−1. The other parameters were set the same as for measurement of catalytic activity, and the total volume was made up to 2 mL (pH = 4.0). After 5 min, the absorbance change at 652 nm was measured by UV-vis spectrophotometry. HepG2 cells were treated with various concentrations of nicotine for 12 h to evaluate GSH production in the cell culture medium.3 Results and discussion
3.1 Optimization of experimental conditions
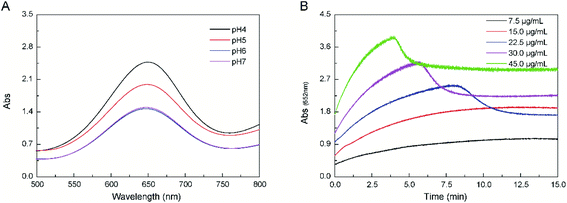
CNPs were prepared via an electrochemical oxidization method according to our previous study.23 The physicochemical properties of CNPs were characterized by different methods. CNPs had an average diameter of 30 nm and carried a potential of −25 mv. H2O2 could be reduced to generate OH˙ by CNPs, along with oxidation of TMB into oxTMB with a blue color, which could be observed by recording absorbance changes at 652 nm (Fig. 1A). This result indicated that CNPs possessed peroxidase catalytic activity. Like peroxidases, the catalytic activity of CNPs was dependent upon pH, and the optimal pH was 4.0. This pH value is consistent with the value for natural HRP.31 The absorbance changes of the catalytic reaction of CNPs were time-dependent during the first 10 min, and then reached an equilibrium (≤15 μg mL−1) (Fig. 1B). The reaction rate increased with an increasing CNPs concentration. Then, the reaction rate decreased to a certain extent after 5 min (probably due to adsorption by CNPs) at CNPs ≥22.5 μg mL−1. The peroxidase-like activity of CNPs stems from its own intrinsic property. Peroxidases are sensitive to temperature and liable to inactivation. Compared with peroxidases and other nanoparticles, CNPs are low-cost, easily accessible, and stable.32,33Structural analyses of CNPs revealed that they possessed a nanocrystalline core of graphitic sp2 carbon atoms, functionalized with peripheral hydroxy and carboxylic/carbonyl moieties.34 Functionalization of graphene surfaces with carboxylic groups is vital for the reduction of H2O2 according to density functional theory. Hence, a similar effect may occur on CNPs due to functionalized carboxylic/carbonyl moieties. Hence, we deduced that the catalytic mechanism likely originated from an increase in the electron density and mobility on the surface of CNPs because of electron transfer from lone-pair electrons in the amino groups of TMB to CNPs, as well as few layers of graphene and GO.16,35
3.2 Comparison and reusability of CNPs
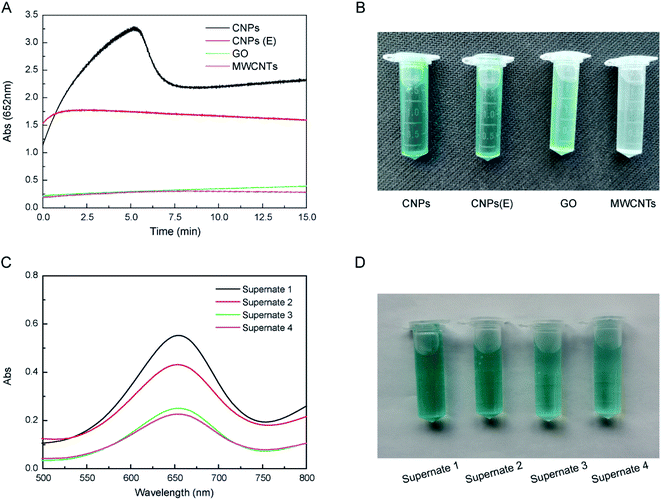
CNPs exhibited higher catalytic activity than that of other carbon-based nanomaterials at the same concentration, including CNPs(E), GO and MWCNTs (Fig. 2A). The catalytic activity of CNPs(E) was decreased compared with that of CNPs. Moreover, the catalytic activity of CNPs was increased almost tenfold compared with that of GO and MWCNTs. These results may be due to the unique structure and dispersity of carbon-based nanomaterials, and the latter improved interactions with substrates to some extent.35 CNPs of average diameter 30 nm had excellent dispersity in aqueous solution compared with that of the three other materials in the order CNPs > CNPs(E) > GO > MWCNTs. In addition, the functional groups on the surface accelerated the electron transfer from CNPs to H2O2, compared with that using GO and MWCNTs. CNPs appeared to aggregate and precipitate after 30 min (Fig. 2B). CNPs were reusable during the catalytic reaction. Even though CNPs were recycled four times, they continued to exhibit high catalytic activity (Fig. 2C and D).3.3 Steady-state kinetics of CNPs
The catalytic-reaction mechanism of CNPs was analyzed by measuring steady-state kinetic parameters.36 Double-reciprocal plots of initial velocity were obtained by fixing one substrate concentration, and the concentration of the second substrate varied. Vmax and Km were obtained according to the Lineweaver–Burk plot30 (Fig. 3). As shown in Table 1, the Km of CNPs with TMB as the substrate was 0.198 mM, similar to that of HRP (Km = 0.434 mM) at pH = 4.0, which indicated that CNPs had higher affinity to the TMB substrate than the HRP substrate. The Km of CNPs (1.112 ± 0.062) with H2O2 was less than half of that for HRP. A metal catalyst was not doped in CNPs, so the catalytic effect was attributed to the electronic structure of CNPs, due to provision of electrons originally from the graphene structure (for which the fast electron transfer was confirmed by electrochemical characterization).35| Catalyst | Substrate | Km (mM) | Vmax (10−8M s−1) |
|---|---|---|---|
| CNPs | TMB | 0.198 ± 0.052 | 4.478 ± 0.858 |
| CNPs | H2O2 | 1.112 ± 0.062 | 1.872 ± 0.040 |
| GO-COOH16 | TMB | 0.024 ± 0.001 | 3.450 ± 0.310 |
| GO-COOH16 | H2O2 | 3.990 ± 0.670 | 3.850 ± 0.220 |
| Fe3O4 nanoparticles36 | TMB | 0.098 | 3.440 |
| Fe3O4 nanoparticles36 | H2O2 | 154.000 | 9.780 |
| HRP36 | TMB | 0.434 | 1.000 |
| HRP36 | H2O2 | 3.700 | 0.871 |
3.4 GSH detection
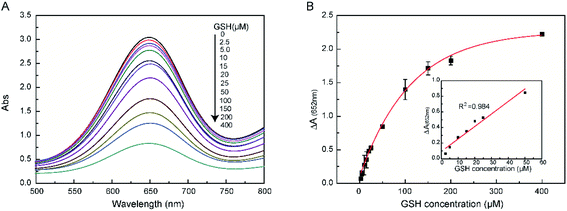
We wished to determine the reducing ability of GSH against the catalytic action of TMB. Hence, the colorimetric change at an absorbance at 652 nm was documented for GSH detection. The changes in absorption at 652 nm were observed in the catalytic reaction using different concentrations of GSH (Fig. 4A). The maximum absorption decreased with increasing concentrations of GSH, which indicated that oxTMB could be reduced to TMB by GSH. The GSH concentration could be determined by CNPs at a wide concentration range (2.5–50 μM) with a limit of detection (LoD) of 0.26 μ M (Fig. 4B, inset). This LoD range indicated that CNPs could be applied widely in biological fields for GSH detection. Other types of nanozyme-detection systems are summarized in Table 2. Comparison of our CNP-based system for GSH detection with that in previous reports (Table 2) revealed our system to have a relatively lower LoD and wider linear range.| Materials | Catalytic property | LOD (μM) | Linearity range (μM) | Ref. |
|---|---|---|---|---|
| Co3O4-MMT | Peroxidase-like activity | 0.088 | 0.1–20.0 | 27 |
| TiO2/MoS2 hybrid nanofibers | Peroxidase-like activity | 0.10 | 0.05–1.00 | 30 |
| H2TCPP-Co9S8 nanocomposites | Peroxidase-like activity | 0.194 | 0.3–10.0 | 38 |
| CuS-PDA-Au composite | Peroxidase-like activity | 0.42 | 0.5–100 | 3 |
| Co,N-HPC | Oxidase-like activity | 0.036 | 0.05–30 | 39 |
| CNPs | Peroxidase-like activity | 0.26 | 2.5–50 | This work |
In the GSH-detection system, CNPs catalyze H2O2 decomposition to generate ˙OH radicals. Then, ˙OH radicals can oxidize the colorless TMB into blue ox-TMB, and ox-TMB can be reduced by GSH to form TMB again (Scheme 1). Besides, the abundant active oxygen species (COOH, OH) on the surface of CNPs layers act as catalytic sites, just like enzymes. Therefore, the structural stability for CNPs had a negligible effect after GSH detection.
3.5 Selectivity of our colorimetric method for GSH detection
To evaluate the selectivity of our colorimetric method for GSH detection, the effects of amino acids (Gln, Asp, Met, Phe, Val, Ser, Cys) were investigated under identical conditions. The inhibitory effect of GSH was much stronger than that of the other substances tested (Fig. 5). The reducing ability of Cys was stronger than that of other amino acids but weaker than that of GSH. Cys could also decrease the absorbance at 652 nm upon addition to the CNPs reaction system, which suggested that the thiol agent was responsible for the catalytic reduction of oxidized TMB into a colorless solution.37 In biological samples, the concentration of Cys (micromolar level) is far lower than the concentration of GSH (millimolar level). Therefore, these results indicated that the influence of amino acids for GSH detection was negligible. GSH detection using a colorimetric approach had excellent selectivity when using biological samples.3.6 Detection of GSH in a real sample
We wished to verify the applicability of our method for GSH detection in real samples. Hence, the GSH concentration in HepG2 cell culture medium was measured after treatment with different concentrations of nicotine. Our colorimetric method for GSH detection was applied to real samples. HepG2 cells were treated with various concentrations of nicotine for 12 h to evaluate GSH production in the cell culture medium. The culture medium was collected for the GSH assay. The GSH concentration varied upon nicotine treatment compared with that in the control group (Fig. 6). A commercial GSH assay kit was employed as a control, and extracellular GSH levels were evaluated by our method: an obvious difference between the two methods was not observed. These results indicated that our colorimetric method for GSH detection was reliable and accurate. Compared with the commercial GSH assay kit, our method was time-saving, low-cost, and easy to use. In addition, the reagents in our method did not need to be stored under difficult conditions.4 Conclusions
Our results demonstrated that CNPs possess peroxidase activity and that their catalytic activity is strongly affected by pH. A pH of 4 is the optimum value for the catalytic reaction. The peroxidase catalytic activity of CNPs was 12-times higher than that of GO and MWCNTs at an identical concentration (30.0 μg mL−1). CNPs could be recycled four times yet continue to retain their catalytic activity. CNPs had the same catalytic activity as that of TMB and HRP. Moreover, a convenient method for colorimetric detection of GSH was developed, which could be applied to real samples. These CNPs displayed promising applications for colorimetric sensing in biological fields.Conflicts of interest
There are no conflicts of interest to declare.References
- X. Zhu, Y. Xue, S. Han, W. Chen, M. Fu and Y. Gao, et al., Appl. Clay Sci., 2020, 195 Search PubMed.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42(14), 6060–6093 RSC.
- Y. Wang, Y. Liu, F. Ding, X. Zhu, L. Yang and P. Zou, et al., Anal. Bioanal. Chem., 2018, 410(20), 4805–4813 CrossRef CAS PubMed.
- J. Feng, P. Huang, S. Shi, K. Y. Deng and F. Y. Wu, Anal. Chim. Acta, 2017, 967, 64–69 CrossRef CAS PubMed.
- T. Rohani Bastami and Z. Dabirifar, RSC Adv., 2020, 10(59), 35949–35956 RSC.
- R. Li, M. Zhen, M. Guan, D. Chen, G. Zhang and J. Ge, et al., Biosen. Bioelectron., 2013, 47, 502–507 CrossRef CAS PubMed.
- F. Qu, T. Li and M. Yang, Biosens. Bioelectron., 2011, 26(9), 3927–3931 CrossRef CAS PubMed.
- W. Shi, Q. Wang, Y. Long, Z. Cheng, S. Chen and H. Zheng, et al., Biomol. Concepts, 2011, 47(23), 6695–6697 CAS.
- H. Yu, H. Wu, X. Tian, Y. Zhou, C. Ren and Z. Wang, RSC Adv., 2021, 11(43), 26963–26973 RSC.
- J. Yin, H. Cao and Y. Lu, J. Mater. Chem., 2012, 22(2), 527–534 RSC.
- J. Lian, P. Liu, X. Li, B. Bian, X. Zhang and Z. Liu, et al., Colloids Surf., A, 2019, 565, 1–7 CrossRef CAS.
- R. Cui, Z. Han and J. J. Zhu, Chemistry, 2011, 17(34), 9377–9384 CrossRef CAS PubMed.
- X. Wang, A. Dong, Z. Zhu, L. Chai, J. Ding and L. Zhong, et al., Small, 2020, 16(43), e2004614 CrossRef PubMed.
- X. Wang, A. Dong, Y. Hu, J. Qian and S. Huang, Biomol. Concepts, 2020, 56(74), 10809–10823 CAS.
- X. Wang, Z. Zhu, L. Chai, J. Ding, L. Zhong and A. Dong, et al., J. Power Sources, 2019, 440 CAS.
- Y. Song, K. Qu, C. Zhao, J. Ren and X. Qu, Adv. Mater., 2010, 22(19), 2206–2210 CrossRef CAS PubMed.
- D. Zhou, K. Zeng and M. Yang, Mikrochim. Acta, 2019, 186(2), 121 CrossRef PubMed.
- J. Liu, L. Meng, Z. Fei, P. J. Dyson, X. Jing and X. Liu, Biosens. Bioelectron., 2017, 90, 69–74 CrossRef CAS PubMed.
- Y. Wang, X. Liu, M. Wang, X. Wang, W. Ma and J. Li, Sens. Actuators, B, 2021, 329 Search PubMed.
- M. Lan, J. Zhang, Y.-S. Chui, H. Wang, Q. Yang and X. Zhu, et al., J. Mater. Chem. B., 2015, 3(1), 127–134 RSC.
- V. Sharma, A. K. Saini and S. M. Mobin, J. Mater. Chem. B., 2016, 4(14), 2466–2476 RSC.
- T. Wang, M. Zou, H. Jiang, Z. Ji, P. Gao and G. Cheng, Eur. J. Pharm. Sci., 2011, 44(5), 653–659 CrossRef CAS PubMed.
- L. Chen, H. Wang, X. Li, C. Nie, T. Liang and F. Xie, et al., RSC Adv., 2018, 8(61), 35246–35256 RSC.
- R. R. Perry, JoA. Mazetta, M. Levin and S. C. Barranco, Cancer, 1993, 72(3), 783–787 CrossRef CAS PubMed.
- D. Giustarini, I. Dalle-Donne, R. Colombo, A. Milzani and R. Rossi, Free Radical Biol. Med., 2003, 35(11), 1365–1372 CrossRef CAS PubMed.
- Y. Huang and H. Chang, Anal. Chem., 2007, 79, 4852–4859 CrossRef CAS PubMed.
- Y. Gao, K. Wu, H. Li, W. Chen, M. Fu and K. Yue, et al., Sens. Actuators, B, 2018, 273, 1635–1639 CrossRef CAS.
- Q. Yang, C. Krautmacher, D. Schilling, M. R. Pittelkow and S. Naylor, Biomed. Chromatogr., 2002, 16(3), 224–228 CrossRef CAS PubMed.
- T. Kamidate and H. Watanabe, Talanta, 1996, 43, 1733–1738 CrossRef CAS PubMed.
- W. Zhu, M. Chi, M. Gao, C. Wang and X. Lu, J. Colloid Interface Sci., 2018, 528, 410–418 CrossRef CAS PubMed.
- S. Chen, M. Chi, Y. Zhu, M. Gao, C. Wang and X. Lu, Appl. Surf. Sci., 2018, 440, 237–244 CrossRef CAS.
- Y. Ma, M. Zhao, B. Cai, W. Wang, Z. Ye and J. Huang, Biosens. Bioelectron., 2014, 59, 384–388 CrossRef CAS PubMed.
- A. Li, X. Mu, T. Li, H. Wen, W. Li and Y. Li, et al., Nanoscale, 2018, 10(25), 11948–11954 RSC.
- L. Chen, J. Hao, L. Xu, X. Meng, X. Li and C. Nie, et al., Int. J. Biol. Macromol., 2019, 138, 29–36 CrossRef CAS PubMed.
- Z. Wang, X. Lv and J. Weng, Carbon, 2013, 62, 51–60 CrossRef CAS.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang and N. Gu, et al., Nat. Nanotechnol., 2007, 2(9), 577–583 CrossRef CAS PubMed.
- W. Yang, J. Hao, Z. Zhang and B. Zhang, New J. Chem., 2015, 39(11), 8802–8806 RSC.
- Y. Gao, C. Jin, X. Li, K. Wu, L. Gao and X. Lyu, et al., Colloids Surf., A, 2019, 568, 248–258 CrossRef CAS.
- S. Li, L. Wang, X. Zhang, H. Chai and Y. Huang, Sens. Actuators, B, 2018, 264, 312–319 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07601a |
| This journal is © The Royal Society of Chemistry 2022 |