Open Access Article
Open Access ArticleAntimicrobial peptide GL13K immobilized onto SLA-treated titanium by silanization: antibacterial effect against methicillin-resistant Staphylococcus aureus (MRSA)†
Yusang Li a,
Ruiying Chend,
Fushi Wangac,
Xinjie Cai
a,
Ruiying Chend,
Fushi Wangac,
Xinjie Cai *ab and
Yining Wang
*ab and
Yining Wang *ab
*ab
aThe State Key Laboratory Breeding Base of Basic Science of Stomatology (Hubei-MOST) and Key Laboratory of Oral Biomedicine Ministry of Education, School and Hospital of Stomatology, Wuhan University, Wuhan, China. E-mail: xinjie.cai@whu.edu.cn; wang.yn@whu.edu.cn
bHospital of Stomatology Wuhan University, Department of Prosthodontics, Wuhan, China
cHospital of Stomatology Wuhan University, Department of Cariology and Endodontics, Wuhan, China
dShanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, College of Stomatology, Shanghai Jiao Tong University, Department of Implant Dentistry, Shanghai, China
First published on 2nd March 2022
Abstract
Infection is the main reason for implant failure, and the incidence of drug-resistant bacterial infection has increased in recent years. Further, methicillin-resistant Staphylococcus aureus (MRSA)-related implant infection has become a serious worldwide threat. New strategies, other than antibiotics, to tackle drug-resistance, are of high clinical significance. Antimicrobial peptides show clear superiority over conventional antibiotics in inhibiting drug-resistant bacteria. In the present study, we combined the antimicrobial peptide, GL13K, with sandblasting and acid-etching (SLA)-treated titanium using a silane coupling agent. Field emission scanning electron microscopy images showed the morphology of the coating. Attenuated total reflectance Fourier transform infrared spectroscopy and X-ray photoelectron spectroscopy results confirmed loading of GL13K, and the hydrophilicity of the SLA-GL13K coating was evaluated by water contact angle analysis. The releasing study of samples showed that the coating has a sustained releasing profile. SLA-GL13K coating exhibited strong contact- and release-killing abilities against MRSA, E. coli, and S. aureus. Meanwhile, Cell Counting Kit 8 analysis and examination of cell morphology demonstrated that the SLA-GL13K coating had good cytocompatibility at antibacterial concentrations. Overall, all these results suggest that SLA-GL13K coating can be successfully fabricated using silanization, and is a promising candidate for controlling MRSA-induced implant-related infection.
Introduction
The superior biological and mechanical properties of titanium for long-term rehabilitation mean that it is among the most popular metallic materials used for biomedical applications.1 Titanium implants have been widely-used in various fields, including oral implantology, hip or knee arthroplasty, and bone fracture fixation;2 however, implant infection has become one of the most common and severe complications of biomaterials. The infection rate of orthopedic surgery is 2–5%,3 rising to approximately 30% after internal fixation of an open fracture.4 Implant-related infections can strongly impede osteogenesis and osseointegration, leading to the need for complex revision procedures, implant failure, or even osteomyelitis.5,6In addition, drug-resistant bacteria have emerged as a severe global healthcare problem.7 Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important and common drug-resistant bacteria in the clinic. Approximately 25–32% of implant infections after fracture fixation are related to MRSA.7 MRSA can synthesize extracellular polymeric substances to form biofilms, which strongly impede penetration of systemically administered antibiotics and inhibition of bacterial growth.8 MRSA can also produce β-lactamase enzymes, which can hydrolyze antibiotic molecules, and penicillin-binding proteins to reduce the binding affinities of antibiotics.9 Relative to normal infections, treatment of patients with MRSA-related infections is more difficult, and the prognosis is significantly worse, due to high recurrence rates and increased sequelae.10,11
Recent studies have focused on local delivery of antibacterial agents, which can maximize in situ antibacterial effects while avoiding potential systemic toxicity and drug resistance.12 Antimicrobial peptides (AMPs) have been introduced into titanium surface modification as effective agents against colonization of microorganisms these years. AMPs are positively-charged bioactive molecules with a few hydrophobic residues.13 As a constituent of the biological innate immune system with broad-spectrum activity, AMPs inhibit bacteria, fungi, and viruses with low levels of host cytotoxicity and bacterial resistance.14 AMPs can induce electrostatic interactions with anionic phospholipids in bacterial cell membranes, resulting in membrane rupture and cell death;15 this unique mechanism decreases the likelihood of bacterial resistance.15 Among various AMPs, the antimicrobial peptide, GL13K, is a 13-amino-acid peptide modified from the human parotid secretory protein (PSP; BPIFA2) with good biocompatibility.13 Similar to other AMPs, GL13K could interact with bacterial membranes in different ways and then leading to its rupture,16,17 and could also inhibit bacteria from forming biofilm.18 Various findings support GL13K as an effective and safe antibacterial agent for titanium surface modification;13,19 however, there has been little research investigating the antibacterial ability of GL13K against drug-resistant bacteria.
Sandblasting and acid-etching (SLA) treatment has been applied for titanium surface modification in recent years. The SLA process creates a hydrophilic titanium surface with micron-scale roughness, which can increase osteoblast differentiation and bone formation.20 SLA-treated implants have higher removal torque values and bone-to-implant contact (BIC) than untreated implants, and the roughened surface can also provide a greater contact area for binding of the antimicrobial peptides.21,22 However, the rough surface with a greater contact area also favors bacterial adhesion and biofilm formation.23 For this reason, various antibacterial agents have been incorporated into SLA-treated titanium surfaces, while no research has combined GL13K and SLA in titanium surface modification to our knowledge. Therefore, we hypothesized that immobilization of GL13K onto SLA-treated titanium surfaces could simultaneously prohibit bacterial growth and favor osseointegration.
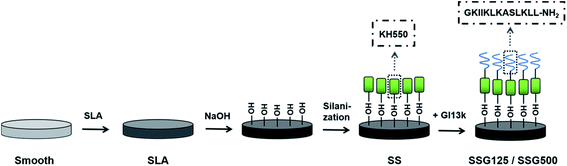
In the present study, we combined GL13K antimicrobial coating with SLA technology for titanium implant surface modification to prevent infection, particularly with drug-resistant bacteria (MRSA), in orthopedic and dentistry applications. Therefore, we covalently immobilized GL13K onto SLA-treated titanium surfaces using the silane coupling agent, 3-aminopropyltriethoxysilane (KH-550). The surface morphology, physicochemical properties, releasing profile, cellular responses, and antimicrobial activities of the coating were evaluated.
Materials and methods
Materials
The antimicrobial peptide, GL13K, was supplied by China Peptides Biotechnology Co., Ltd (Shanghai, China). Commercially pure titanium (grade 2, 99.6% purity) was supplied by Baoji Titanium Industry Co., Ltd (Shanxi, China). The MC3T3-E1 cell line (American Type Culture Collection [ATCC] catalog CRL-2594), Staphylococcus aureus (ATCC catalog 25923), Escherichia coli (ATCC catalog 25922), and MRSA (ATCC catalog 43300) were obtained from the American Type Culture Collection. All other chemical reagents were local products of analytical grade.Sample preparation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) at 60 °C for 30 min. Next, disks were rinsed with deionized water, and ultrasonically cleaned twice with acetone, ethanol, and deionized water for 20 min each procedure.24,25
1) at 60 °C for 30 min. Next, disks were rinsed with deionized water, and ultrasonically cleaned twice with acetone, ethanol, and deionized water for 20 min each procedure.24,25Physicochemical characterization of coatings
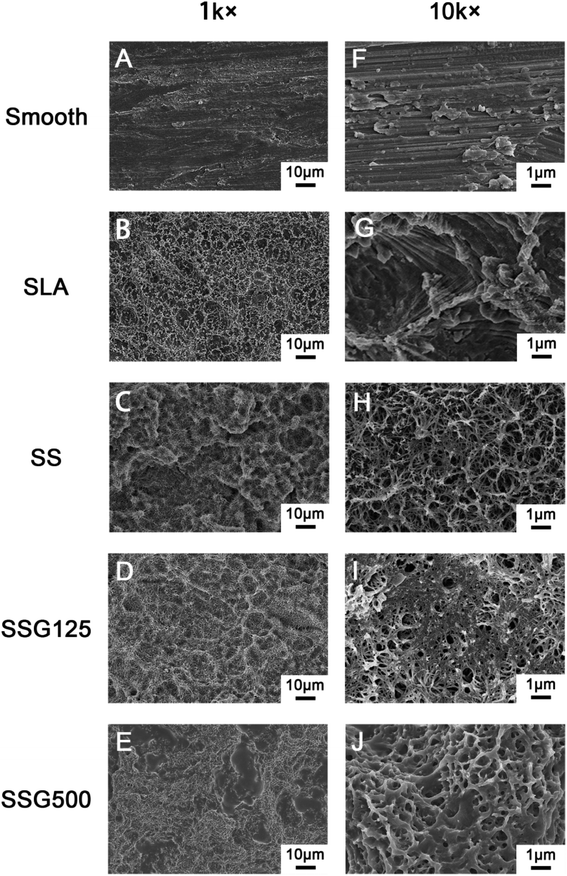
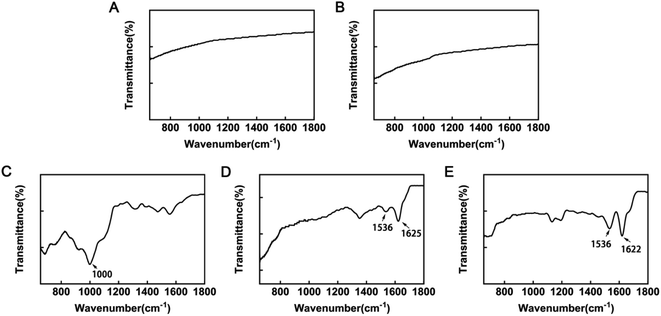
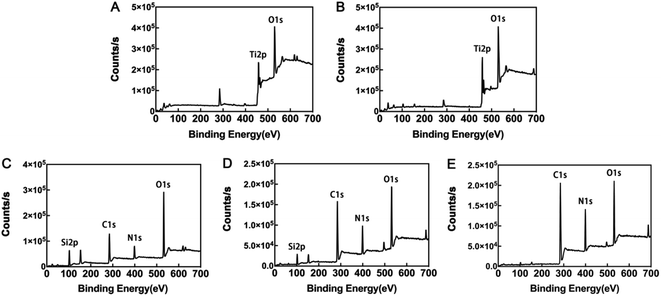
Field emission scanning electron microscopy (Zeiss, Oberkochen, Germany) was used to analyze the surface structure of coatings, which were sputter-coated with gold before microscopic examination. Further, coating surface chemistry was investigated using attenuated total reflectance Fourier transform infrared spectroscopy (ATR-FTIR; Thermo Nicolet 5700, USA). Spectra were obtained at 45 °C at a spectral resolution of 4 cm−1, with 32 scans every 90 s interval, using air as the reference spectrum. The chemical composition of the coatings was analyzed by X-ray photoelectron spectroscopy (XPS; ESCALAB 250Xi, Thermo Fisher Scientific, Britain) with an Al-Kα X-ray source (1486.6 eV). The selected resolution for the general spectra was 100.0 eV of pass energy and 1.0 eV per step, while the resolution for narrow scans was 30.0 eV of pass energy and 0.1 eV per step. Binding energies were based on C 1s at 285.0 eV for calibration.Water contact angles (WCA) of all samples were tested using the drop shape analysis system, CASTs3.0 (SL200KS, KINO Industry Co., Ltd, USA). Milli-Q water (1 μl) was used to conduct tests at room temperature. The contact angle (θ) was defined as the angle between the solid substrate and the liquid phase. The data were tracked and measured for 60 s and the test was repeated five times for each group.
GL13K release study
For the GL13K release study, each sample of SSG125 and SSG500 was immersed into 0.5 ml PBS solution and was incubated at 37 °C. At pre-determined time intervals, 0.5 ml of the solution was removed and the GL13K concentration was measured by Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, US) through absorbance at 205 nm (A205 method). After the removal, 0.5 ml fresh PBS was added into the system. An absorbance/GL13K concentration calibration curve was portrayed using a series of concentrations of GL13K peptide dissolved in PBS.In vitro antibacterial study
To measure the release antibacterial capacity of samples, the OD value of the bacteria suspension after culturing was measured, and serial 10-fold dilutions were performed. Then, viable counts were estimated following the spread plate method. For contact antibacterial capacity, samples cultured with bacteria were removed with sterile forceps, gently washed with PBS three times, and ultrasonicated in 2 ml PBS for 5 min. The obtained suspension was then diluted and re-cultured on solid medium and viable counts were estimated.
In vitro cell analysis
Statistical analysis
Quantitative data are expressed as mean ± standard deviation. Statistical analysis was carried out by one-way analysis of variance (ANOVA) and post hoc testing. The level of significance was set at P < 0.05.Results and discussion
Physicochemical characterization of titanium disk coatings
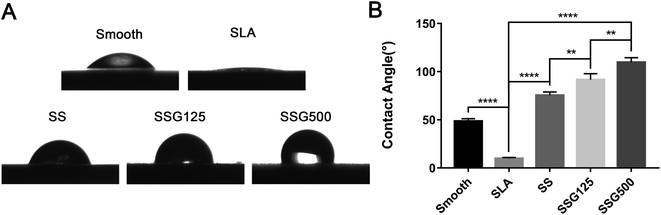
After silanization, the WCA increased to 75.4° ± 1.583°. KH-550 coating is proven to increase the hydrophobicity of materials because it reacts with hydroxyl groups on inorganic surfaces, lowering the surface energy.34 WCA values in the SLAS125 and SLAS500 groups were 91.64° ± 2.843° and 109.7° ± 2.205°, respectively. In general, a solid surface is considered hydrophobic if the WCA is >90°. Like most AMPs, GL13K peptide composition is rich in cationic and hydrophobic amino acids, including Ala, Ile, and Leu.35 From SEM images, the superficial areas of SSG125 and SSG500 were visibly smaller than those in SLA samples; hence, both the amino acid sequence and the organic surface characteristics may contribute to the hydrophobicity of GL13K.
GL13K release study
The release profiles of SSG125 and SSG500 are shown in Fig. 5. Fig. 5A illustrates the standard curve of absorbance of GL13K in PBS at 205 nm (y = 0.01554x − 0.2935, R2 = 0.9972). For SSG125, most of the peptide was released at the first 2 hours of incubation. Then a small amount of peptide (approximately 18%) was constantly released from the second hour to the twelfth hour. After that, the amount of released GL13K became negligible. For SSG500, approximately 68% of the peptide was rapidly released at the first 4 hours, then the speed of releasing gradually slowed down. A sustained release was observed for about 36 hours, followed by a negligible amount of releasing afterward.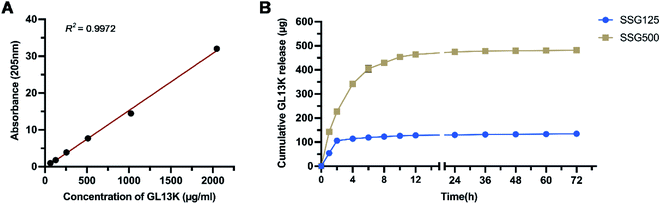 | ||
| Fig. 5 (A) Standard curve of GL13K in PBS (y = 0.01554x − 0.2935, R2 = 0.9972). (B) Cumulative release profile of GL13K from SSG125 and SSG500 groups. | ||
It was speculated that during the sample preparation, a certain amount of GL13K peptide was covalently immobilized onto titanium surface through silane coupling agent, while another amount of peptide was deposited and adsorbed on the surface physically. As a result, there was a rapid release initially, and followed by a sustained release for a longer time. And the sustained release is more obvious in the SSG500 group than the SSG125 group, which might indicate that a higher amount of peptide was covalently bonded to SSG500 samples. The rapid initial release could ensure a functional high concentration locally at an early stage, and then the sustained releasing would help to maintain the biological function of the coating.36
In vitro cellular study
Proliferation of MC3T3-E1 cells in the different experimental groups, determined using the CCK8 assay at 1, 4, and 7 days, are shown in Fig. 6. Cell counts in the SLA group were greater than those of the untreated smooth group at all three time points, with significant differences on days 4 and 7. On day 1, the SSG500 group had a significantly lower cell count than the control group. However, on days 4 and 7, the cell counts in the SSG500 group were significantly higher than those of the control group. To conclude, the incorporation of GL13K did not affect the viability or proliferation of MC3T3-E1 cells.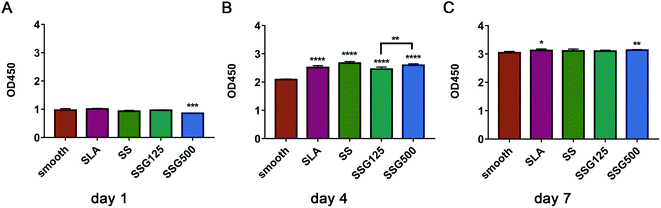 | ||
| Fig. 6 Proliferation of MC3T3-E1 cells culturing with the samples of Smooth, SLA, SS, SSG125, and SSG500 groups. (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001). | ||
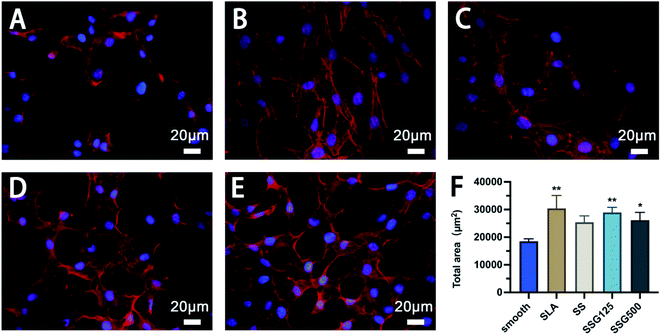
The immunofluorescence images presented in Fig. 7 show the morphology and actin skeleton of cells cultured on different surfaces. Cells grown on the SLA surface showed cellular spreading, with bright and well-pronounced elongated lamellipodia and filopodia protruding from the cell edges (Fig. 7B). By comparison, cells on the smooth titanium surface exhibited irregular morphology with few visible filopodia, and the majority of cells were round-shaped and aggregated together (Fig. 7A). In silanized samples, cells also exhibited clear spreading and interactions, with filopodia extending outward (Fig. 7C). Cells cultured on SSG125 and SSG500 also showed a typical polygonal osteoblastic shape, with numerous membrane channels between adjacent cells (Fig. 7D and E). These tunneling nanotubes can mediate intercellular transfer of organelles, plasma membrane components, and cytoplasmic molecules.37,38 Fig. 7F shows the statistical analysis of total cell area of MC3T3-E1 cells cultured on different samples. The cell area of SLA, SSG125, and SSG500 groups is significantly higher than that of the control group and no significant difference exists between any other two groups. This result is basically in consistent with the result of the CCK8 test. Combining the statistical cell area and proliferation results with the cell morphology in Fig. 7, it is indicated that SSG125 and SSG500 groups have no cytotoxic effect on MC3T3-E1 cells.
The nano-roughness surface of SLA-treated titanium samples has been proven to have desirable effects on osteoblasts.39 In our study, the proliferation and morphology of MC3T3-E1 cells were also significantly better when grown on SLA samples relative to smooth samples, indicating a higher likelihood of stable osseointegration. Further, cells cultured on samples with SSG125 and SSG500 coatings showed favorable proliferation and viability characteristics, with clear cell stretching and interaction. These results indicate that SSG125 and SSG500 have good cytocompatibility. The main factor that has contributed to that is the large amounts of cholesterol and other sterols in animal cell membranes, which could help to inhibit antimicrobial peptide activity and stabilize membranes.40,41 Also, the membranes of animals are not heavily populated with negatively charged molecules such as phosphatidylglycerol, which also reduces the risk of targeting of positively charged AMPs.42
In vitro antibacterial study
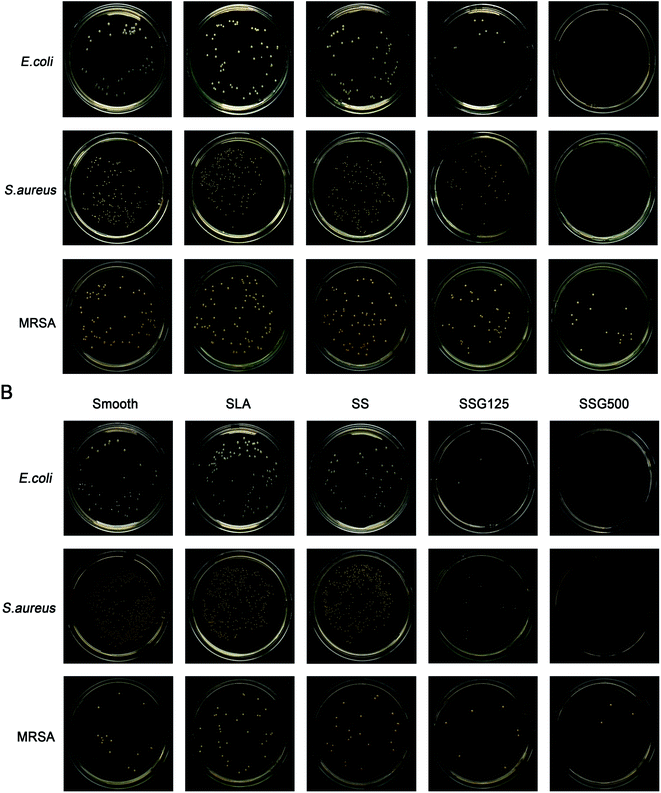
The results of contact and release antibacterial capacity experiments are shown in Fig. 8 and 9. Notably, in results of both the contact and release antibacterial assays, the number of bacteria in the SLA group was higher than that in the smooth group, with significant differences for E. coli and MRSA. By releasing GL13K to bacterial suspensions, bacteria numbers in SSG125 samples reduced by 82.82%, 80.14%, and 43.64% for the E. coli, S. aureus, and MRSA groups, while in the SSG500 samples bacteria numbers were decreased by 100.00%, 100.00%, and 79.09% in the E. coli, S. aureus, and MRSA groups, respectively, compared with SLA samples. Regarding contact-killing capacity, the SSG125 sample significantly reduced the bacteria number by 98.60%, 98.91%, and 73.26% in the E. coli, S. aureus, and MRSA groups, while those in the SSG500 sample were decreased by 100.00%, 100.00%, and 88.37%, respectively, compared with SLA samples. These data demonstrate that GL13K-coated samples exhibit both contact and release antibacterial activity against these three bacteria significantly. Antibacterial and cytotoxicity results of samples with higher concentrations (SSG1000, SSG2000) are shown in Fig. S2 and S3 in the ESI.†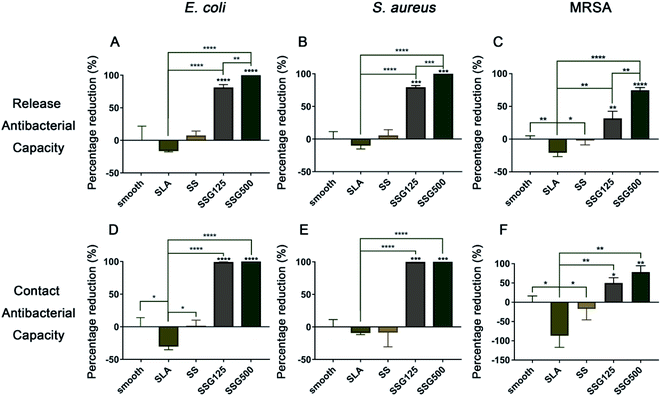 | ||
| Fig. 8 The percentage reduction of E. coli, S. aureus, and MRSA in release and contact antibacterial experiment of different samples. (*p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.0001). | ||
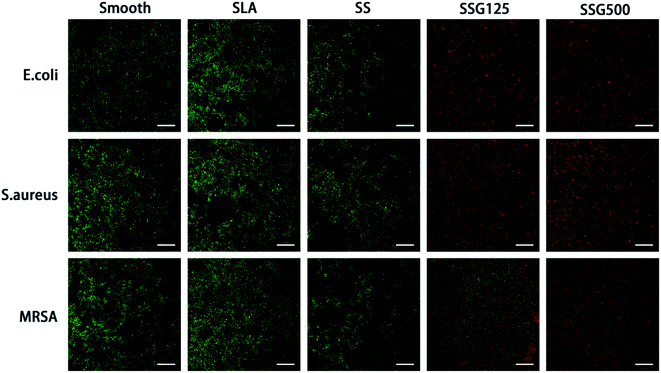
The live and dead staining could help to identify the living and dead bacteria based on the color changing. As shown in Fig. 10, by fluorescence microscopy, live bacteria with intact membranes appeared green, while dead bacteria with damaged membranes were stained red. The fluorescence intensity of bacterial membranes was high in smooth, SLA, and SS samples, indicating that a large number of bacteria had adhered to the surface. Further, imaging revealed almost no living E. coli and S. aureus on the surface of SSG125 and SSG500 samples. However, some living MRSA were present on the surface in SLAS125 samples and green spots were also visible on the surface of SLAS500 samples, consistent with the CFU data.
 | ||
| Fig. 10 Fluorescent images of biofilm formation on sample surfaces obtained by CLSM after live and dead staining. Scale bar = 50 μm. | ||
MRSA is one of the most important drug-resistance bacteria associated with implant infection, while E. coli and S. aureus are also common pathogenic bacteria in implant-related infection. Hence, we chose these three bacteria as representative to examine the antibacterial ability of SLA-GL13K coating against drug-resistant bacteria, Gram-positive bacteria, and Gram-negative bacteria. To our knowledge, this study is the first investigation of the antibacterial properties of GL13K against MRSA.
In addition to promoting the adhesion of osteoblasts, SLA-treated samples can also promote bacterial adhesion;23 however, after coating with GL13K, samples presented significant antibacterial activity and good cytocompatibility. In the high concentration (SSG500) group, E. coli and S. aureus were completely killed and no cytotoxicity to MC3T3-E1 cells was detected. Hence, for common pathogenic bacteria such as E. coli and S. aureus, SLA-GL13K coating showed a significant antibacterial effect. Notably, as for MRSA, GL13K peptides also have significant antibacterial activity, although the effect was weaker compared to the effect against E. coli and S. aureus.
Although AMPs are a diverse group of bioactive molecules, most of them share common properties that related to the antibacterial activity: positive charge, hydrophobicity, and amphiphilicity.43 As a result, the antibacterial mechanism of most AMPs is basically identical and GL13K is not an exception.44 Like most of the AMPs, GL13K is believed to kill bacteria by interacting with the cell membranes.45 As a small molecule, GL13K could easily pass through the cell wall of bacteria as the peptidoglycan mesh is relatively porous.46 As we know, bacterial membranes are negatively charged.23 In contrast, most of the AMPs are cationic peptides,47 so the electrostatic attraction between AMPs and bacterial membranes would ensure the attachment of AMPs, therefore providing the chance for AMPs to interact with the membranes and cause the death of the bacteria.48 Based on that, several models were proposed. For example, barrel-stave model, toroidal-pore model, carpet model and aggregate model are the most studied and widely accepted ones.49,50 Although having minor differences, these models all share the same key point: membrane disruption.
This mechanism is different from that of antibacterial compounds such as β-lactam antibiotics, which usually target a specific site on the cell wall, cell membrane, nucleic acid, or proteins.51 Aiming at it, some bacteria have developed antibiotic resistance by genetic mutation or the acquisition of exogenous resistance genes from other bacterias, which highly decreased the antibacterial efficacy of many antibiotics.52 However, the biochemically simple and efficient mechanism of AMPs like GL13K is predicted to prevent bacteria from developing drug resistance, while no research of the antibacterial properties of GL13K against MRSA was reported before.53
Our study has proved the significant antibacterial effect of GL13K against MRSA, while the effect is relatively weaker compared to that against E. coli and S. aureus. The mutation of MRSA is complex. For example, by blocking murein hydrolysis in cell wall substrates, cell wall turnover and autolysis of MRSA are inhibited, which increases the thickness and density of the cell wall.54 This mutation could partially prevent the AMPs from passing through the cell wall and interacting with the cell membranes, and thus protect MRSA from being disrupted by GL13K. This may explain the reason why the antibacterial effects of GL13K against MRSA were relatively inferior to those against E. coli and S. aureus in our study. Nevertheless, accumulating evidence shows that different AMPs may have synergistic potentiation,55 and some AMPs can also synergize with conventional antibiotics or metal particles, such as ceftriaxone, amoxicillin-clavulanate, linezolid, imipenem, silver nanoparticles (AgNPs), etc.56 The synergy is proved to improve the antibacterial effects, while further reduce the likelihood of resistance.15 So, the combination application of SLA-GL13K coating and some other antibacterial agents might improve its efficacy and could be further studied. Also, as a 13-amino-acid peptide, GL13K has a short sequence. Evidence has revealed that short AMPs have better pharmacological profiles, better metabolic stability, and lower toxicity than long AMPs.54 Therefore, the SLA-GL13K coating might be a promising agent for the control of MRSA-related infection. In addition, for samples as SSG125 and SSG500, GL13K was coated on the titanium surface through silanization and hence gained the property of sustained releasing after a rapid releasing stage. It could ensure an efficient high concentration locally in the early stage and might maintain the biological function through the sustained release.
Conclusions
In summary, the antimicrobial peptide, GL13K, was covalently conjugated onto SLA-treated titanium surfaces using the silane coupling agent, KH-550. The results of SEM, ATR-FTIR, XPS, and WCA confirmed successful immobilization of the peptide, and the release measurement of GL13K indicated a sustained-release property of the samples. SLA-GL13K coating exhibited antibacterial properties against E. coli and S. aureus, as well as significant antibacterial effects against drug-resistant MRSA. Further, the coating had a positive effect on osteoblast proliferation and adhesion in vitro. Based on these findings, and within the limitations of this study, we conclude that SLA-GL13K coating is a promising candidate for early-stage prevention of infections at implant sites; however, further in vivo antibacterial and biocompatible evaluation will be necessary to examine the full potential of SLA-GL13K coatings for biomedical applications.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 81901063) and the Natural Science Foundation of Hubei Province (2017CFB183 and 2018CFB419).Notes and references
- C. N. Elias, J. H. C. Lima, R. Valiev and M. A. Meyers, JOM, 2008, 60, 46–49 CrossRef CAS.
- M. Geetha, A. K. Singh, R. Asokamani and A. K. Gogia, Prog. Mater. Sci., 2009, 54, 397–425 CrossRef CAS.
- K. Mitsutake and K. Niwaya, N. Engl. J. Med., 2004, 351, 193–195 CrossRef PubMed.
- A. Trampuz and W. Zimmerli, Injury, 2006, 37(suppl 2), S59–S66 CrossRef PubMed.
- K. Ma, X. Cai, Y. Zhou, Y. Wang and T. Jiang, Macromol. Biosci., 2017, 17, 1600130 CrossRef PubMed.
- M. B. Zaid, R. J. O'Donnell, B. K. Potter and J. A. Forsberg, J. Am. Acad. Orthop. Surg., 2019, 27, e977–e985 CrossRef PubMed.
- W. J. Metsemakers, N. Emanuel, O. Cohen, M. Reichart, I. Potapova, T. Schmid, D. Segal, M. Riool, P. H. Kwakman, L. de Boer, A. de Breij, P. H. Nibbering, R. G. Richards, S. A. Zaat and T. F. Moriarty, J. Controlled Release, 2015, 209, 47–56 CrossRef CAS PubMed.
- Y. Zhuang, L. Ren, S. Zhang, X. Wei, K. Yang and K. Dai, Acta Biomater., 2021, 119, 472–484 CrossRef CAS PubMed.
- J. T. Murphy, R. Walshe and M. Devocelle, J. Theor. Biol., 2008, 254, 284–293 CrossRef CAS PubMed.
- V. Alt, K. Kirchhof, F. Seim, I. Hrubesch, K. S. Lips, H. Mannel, E. Domann and R. Schnettler, Acta Biomater., 2014, 10, 4518–4524 CrossRef CAS PubMed.
- B. Nie, T. Long, H. Ao, J. Zhou, T. Tang and B. Yue, Antimicrob. Agents Chemother., 2017, 61, e01766 CrossRef CAS PubMed.
- G. T. Boo, D. W. Grijpma, T. F. Moriarty, R. G. Richards and D. Eglin, Biomaterials, 2015, 52, 113–125 CrossRef PubMed.
- M. Abdolhosseini, S. R. Nandula, J. Song, H. Hirt and S. U. Gorr, Peptides, 2012, 35, 231–238 CrossRef CAS PubMed.
- J. M. Ageitos, A. Sánchez-Pérez, P. Calo-Mata and T. G. Villa, Biochem. Pharmacol., 2017, 133, 117–138 CrossRef CAS PubMed.
- R. K. Thapa, D. B. Diep and H. H. Tønnesen, Acta Biomater., 2020, 103, 52–67 CrossRef CAS PubMed.
- V. Balhara, R. Schmidt, S. U. Gorr and C. Dewolf, Biochim. Biophys. Acta, 2013, 1828, 2193–2203 CrossRef CAS PubMed.
- T. Diniz and M. I. Tavares, J. Appl. Polym. Sci., 2003, 90, 2955–2958 CrossRef CAS.
- X. Chen, H. Hirt, Y. Li, S. U. Gorr and C. Aparicio, PLoS One, 2014, 9, e111579 CrossRef PubMed.
- L. Zhou, Z. Lin, J. Ding, W. Huang, J. Chen and D. Wu, Colloids Surf., B, 2017, 160, 581–588 CrossRef CAS PubMed.
- D. Buser, N. Broggini, M. Wieland, R. K. Schenk, A. J. Denzer, D. L. Cochran, B. Hoffmann, A. Lussi and S. G. Steinemann, J. Dent. Res., 2004, 83, 529–533 CrossRef CAS PubMed.
- A. Piattelli, L. Manzon, A. Scarano, M. Paolantonio and M. Piattelli, Int. J. Oral Maxillofac. Implants, 1998, 13, 805–810 CAS.
- D. Buser, T. Nydegger, T. Oxland, D. L. Cochran, R. K. Schenk, H. P. Hirt, D. Snétivy and L. P. Nolte, J. Biomed. Mater. Res., 1999, 45, 75–83 CrossRef CAS PubMed.
- F. Hizal, N. Rungraeng, J. Lee, S. Jun, H. J. Busscher, H. C. van der Mei and C. H. Choi, ACS Appl. Mater. Interfaces, 2017, 9, 12118–12129 CrossRef CAS PubMed.
- Y. Zhou, T. Jiang, M. Qian, X. Zhang, J. Wang, B. Shi, H. Xia, X. Cheng and Y. Wang, Biomaterials, 2008, 29, 461–474 CrossRef PubMed.
- T. Jiang, Z. Zhang, Y. Zhou, Y. Liu, Z. Wang, H. Tong, X. Shen and Y. Wang, Biomacromolecules, 2010, 11, 1254–1260 CrossRef CAS PubMed.
- K. V. Holmberg, M. Abdolhosseini, Y. Li, X. Chen, S. U. Gorr and C. Aparicio, Acta Biomater., 2013, 9, 8224–8231 CrossRef CAS PubMed.
- J. Zhang, J. Liu, C. Wang, F. Chen, X. Wang and K. Lin, Bioact. Mater., 2020, 5, 9–16 CrossRef PubMed.
- R. A. Gittens, T. McLachlan, R. Olivares-Navarrete, Y. Cai, S. Berner, R. Tannenbaum, Z. Schwartz, K. H. Sandhage and B. D. Boyan, Biomaterials, 2011, 32, 3395–3403 CrossRef CAS PubMed.
- J. Li, K. Yu, K. Qian, H. Cao, X. Lu and J. Sun, Nanoscale Res. Lett., 2014, 9, 172 CrossRef PubMed.
- K. Ma, L. Gong, X. Cai, P. Huang, J. Cai, D. Huang and T. Jiang, Int. J. Nanomed., 2017, 12, 3665–3679 CrossRef CAS PubMed.
- C. S. Chang, T. M. Lee, C. H. Chang and J. K. Liu, Clin. Oral Implants Res., 2009, 20, 1178–1184 CrossRef PubMed.
- D. V. Kilpadi and J. E. Lemons, J. Biomed. Mater. Res., 1994, 28, 1419–1425 CrossRef CAS PubMed.
- M. Textor, C. Sittig, V. Frauchiger, S. Tosatti and D. M. Brunette, in Titanium in Medicine: Material Science, Surface Science, Engineering, Biological Responses and Medical Applications, Springer Berlin Heidelberg, Berlin, Heidelberg, 2001, pp. 171–230, DOI:10.1007/978-3-642-56486-4_7.
- J.-C. Liu, M.-J. Xu, T. Lai and B. Li, Ind. Eng. Chem. Res., 2015, 54, 9733–9741 CrossRef CAS.
- Z. Ye, A. C. Kobe, T. Sang and C. Aparicio, Nanoscale, 2020, 12, 20767–20775 RSC.
- F. Wang, P. Huang, D. Huang, Y. Hu, K. Ma, X. Cai and T. Jiang, J. Mater. Chem. B, 2018, 6, 2304–2314 RSC.
- A. Rustom, R. Saffrich, I. Markovic, P. Walther and H. H. Gerdes, Science, 2004, 303, 1007–1010 CrossRef CAS PubMed.
- H. H. Gerdes and R. N. Carvalho, Curr. Opin. Cell Biol., 2008, 20, 470–475 CrossRef CAS PubMed.
- G. Zhao, A. L. Raines, M. Wieland, Z. Schwartz and B. D. Boyan, Biomaterials, 2007, 28, 2821–2829 CrossRef CAS PubMed.
- T. H. Lee, K. N. Hall and M. I. Aguilar, Curr. Top. Med. Chem., 2016, 16, 25–39 CrossRef CAS PubMed.
- L. Zhou, Y. Lai, W. Huang, S. Huang, Z. Xu, J. Chen and D. Wu, Colloids Surf., B, 2015, 128, 552–560 CrossRef CAS PubMed.
- R. Kundu, ChemMedChem, 2020, 15, 1887–1896 CrossRef CAS PubMed.
- P. Bastos, F. Trindade, J. da Costa, R. Ferreira and R. Vitorino, Med. Res. Rev., 2018, 38, 101–146 CrossRef PubMed.
- M. D. Manniello, A. Moretta, R. Salvia, C. Scieuzo, D. Lucchetti, H. Vogel, A. Sgambato and P. Falabella, Cell. Mol. Life Sci., 2021, 78, 4259–4282 CrossRef CAS PubMed.
- J. Lei, L. Sun, S. Huang, C. Zhu, P. Li, J. He, V. Mackey, D. H. Coy and Q. He, Am. J. Transl. Res., 2019, 11, 3919–3931 CAS.
- J. C. Espeche, M. Martinez, P. Maturana, A. Cutro, L. Semorile, P. C. Maffia and A. Hollmann, Arch. Biochem. Biophys., 2020, 693, 108549 CrossRef CAS PubMed.
- Y. Luo and Y. Song, Int. J. Mol. Sci., 2021, 22, 11401 CrossRef CAS PubMed.
- F. Savini, M. R. Loffredo, C. Troiano, S. Bobone, N. Malanovic, T. O. Eichmann, L. Caprio, V. C. Canale, Y. Park, M. L. Mangoni and L. Stella, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183291 CrossRef CAS PubMed.
- Y. Yan, Y. Li, Z. Zhang, X. Wang, Y. Niu, S. Zhang, W. Xu and C. Ren, Colloids Surf., B, 2021, 202, 111682 CrossRef CAS PubMed.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez, A. Ioannidis, M. A. Giulianotti, Y. Apidianakis, S. Bradfute, A. L. Ferguson, A. Cherkasov, M. N. Seleem, C. Pinilla, C. de la Fuente-Nunez, T. Lazaridis, T. Dai, R. A. Houghten, R. E. W. Hancock and G. P. Tegos, Lancet Infect. Dis., 2020, 20, e216–e230 CrossRef CAS PubMed.
- M. Tajbakhsh, A. Karimi, A. Tohidpour, N. Abbasi, F. Fallah and M. M. Akhavan, J. Microbiol., 2018, 56, 128–137 CrossRef CAS PubMed.
- A. Giedraitienė, A. Vitkauskienė, R. Naginienė and A. Pavilonis, Medicina, 2011, 47, 137–146 CrossRef.
- B. Lazzaro, M. Zasloff and J. Rolff, Science, 2020, 368, eaau5480 CrossRef CAS PubMed.
- K. Sieradzki and A. Tomasz, J. Bacteriol., 2003, 185, 7103–7110 CrossRef CAS PubMed.
- A. Moretta, C. Scieuzo, A. M. Petrone, R. Salvia, M. D. Manniello, A. Franco, D. Lucchetti, A. Vassallo, H. Vogel, A. Sgambato and P. Falabella, Front. Cell. Infect. Microbiol., 2021, 11, 668632 CrossRef CAS PubMed.
- X. Liu, Z. Li, X. Wang, Y. Chen, F. Wu, K. Men, T. Xu, Y. Luo and L. Yang, Int. J. Nanomed., 2016, 11, 6781–6794 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04974g |
| This journal is © The Royal Society of Chemistry 2022 |