Open Access Article
Open Access ArticleFungal–fungal co-culture: a primer for generating chemical diversity†
Sonja L.
Knowles
 ,
Huzefa A.
Raja
,
Huzefa A.
Raja
 ,
Christopher D.
Roberts
,
Christopher D.
Roberts
 and
Nicholas H.
Oberlies
and
Nicholas H.
Oberlies
 *
*
Department of Chemistry and Biochemistry, University of North Carolina at Greensboro, Greensboro, NC, USA. E-mail: nicholas_oberlies@uncg.edu
First published on 9th February 2022
Abstract
Covering: 2002 to 2020
In their natural environment, fungi must compete for resources. It has been hypothesized that this competition likely induces the biosynthesis of secondary metabolites for defence. In a quest to discover new chemical diversity from fungal cultures, a growing trend has been to recapitulate this competitive environment in the laboratory, essentially growing fungi in co-culture. This review covers fungal–fungal co-culture studies beginning with the first literature report in 2002. Since then, there has been a growing number of new secondary metabolites reported as a result of fungal co-culture studies. Specifically, this review discusses and provides insights into (1) rationale for pairing fungal strains, (2) ways to grow fungi for co-culture, (3) different approaches to screening fungal co-cultures for chemical diversity, (4) determining the secondary metabolite-producing strain, and (5) final thoughts regarding the fungal–fungal co-culture approach. Our goal is to provide a set of practical strategies for fungal co-culture studies to generate unique chemical diversity that the natural products research community can utilize.
1 Introduction and background
Can you imagine a world where Alexander Fleming did not observe the effects of Penicillium rubens1,2 on bacteria (Staphylococcus aureus)? Fleming was working to understand the biological properties of these bacteria. He went on vacation, and when he returned, he saw a mould growing on the same plate as his bacteria. He observed that the bacteria that were near the mould had died, but the bacteria farther away were not affected.3 This may have been one of the most important co-culture experiments of the 20th century, eventually leading to the isolation of penicillin by Florey and colleagues.4 How many people would have died from bacterial infections if penicillin had not been discovered? One way to gauge this is to examine the differences in survival rates during WWI vs. WWII, where penicillin was available to the Allies during the latter. Before the discovery of antibiotics, tens of thousands of service men died during WWI, not from the damage caused by their combat wounds, but from the resulting incurable infections.5 In contrast, it has been estimated that penicillin saved the lives of about a 1/6th of all Allied forces during WWII.6 While Fleming is attributed to the discovery of penicillin (and shared the Nobel Prize in Physiology or Medicine in 1945 with Florey and Chain),7 another impact he had was demonstrating the value of co-culturing organisms for discovering new chemical diversity.1.1 Fungal secondary metabolites are an untapped resource for new drug leads, but do we need new ways to ‘turn on’ their biosynthesis?
Fungi are a hyper diverse kingdom of life, with millions of species estimated worldwide.8,9 Of those, probably less than 10% (approximately 148![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species10) have been described taxonomically,11,12 and thus, perhaps 90% of fungi are unknown. Out of all the described and undescribed fungi, roughly only 7% have been studied for the chemistry of secondary metabolites.13 Thus, fungi have, and continue to be, a ripe source for the discovery of new chemical diversity.
000 species10) have been described taxonomically,11,12 and thus, perhaps 90% of fungi are unknown. Out of all the described and undescribed fungi, roughly only 7% have been studied for the chemistry of secondary metabolites.13 Thus, fungi have, and continue to be, a ripe source for the discovery of new chemical diversity.
The secondary metabolites of fungi can be subdivided into various categories, such as antibiotics, mycotoxins, and pigments, and while these designations apply to their uses either for or against humans, such small molecules are likely biosynthesized to allow fungi to survive in their environments.14,15 For example, fungi can be pathogenic to animals (including humans) and plants, and fungal secondary metabolites are implicated in disease causing interactions.16,17 However, other fungal secondary metabolites can be utilized by humans for beneficial medical uses,18 including: penicillin (the first antibiotic), cyclosporine (used for organ transplants), and statins (to reduce cholesterol).19,20
While many of these facts are well known, especially to those that study fungi, a growing question is: are there more fungal metabolites to discover? Another way to think about this question is: if we study fungi in a manner that has been replicated since the 1960s (i.e., growing monocultures of fungi in the lab and isolating major metabolites), will we discover new chemical diversity or will we simply uncover slight variations on well-known metabolites? Alternatively, if fungi are grown in a manner that approximates what they are forced to do in Nature, will we uncover unprecedented chemical diversity that those fungi are using to help fight for their survival?
1.2 Interaction driven secondary metabolite discovery: mimicking the competitive environment of nature in the lab
In Nature, fungi grow in somewhat hostile environments, often competing with other microorganism, such as those found in plants, the rhizosphere, soil, water, and human fluids and tissues.21,22 They can overcome competition in various ways, such as, rapid growth, sporulation, stress recovery, and the use and negation of inhibitors.23,24 An emerging technique, which is often termed ‘co-culturing’, imparts stress on microorganisms by forcing them to compete for resources, and this may serve to stimulate the biosynthesis of unique secondary metabolites,25 whose role could be to provide a competitive advantage to these organisms.26–28 It has been shown that co-culturing can activate novel biosynthetic pathways in a way quite different than simply supplementing media with signaling molecules, indicating that the physical interactions between fungi are important for activation of new metabolites.29,30 In one recent example, the co-culturing of two fungal species activated the biosynthesis of a novel cytotoxic metabolite, wheldone (104), but when the producer strain was grown as a monoculture, this unique metabolite was not observed.31Under standard laboratory conditions, fungi have been shown to produce only a fraction of their potential secondary metabolites.32,33 Thus, co-culturing fungi in a controlled environment can be a strategic way to turn on the biosynthesis of unique secondary metabolites through interspecies crosstalk (and this has been termed as ‘interspecific interactions’).32,34 Co-culturing confines two fungal species to an area with both limited nutrients and territory, which will cause them to interact in two key ways. First, they may biosynthesize metabolites that could inhibit the growth of the other fungus (and of course, vice versa). In addition, the rate of growth of a fungus can be modulated, perhaps sped up, so that it uses the resources faster than the competing fungus, essentially invoking another kind of stress.35 In contrast, without the stimulation needed to activate the silenced biosynthetic gene clusters, the true wealth of secondary metabolites may be under realized. The significance of co-culturing is that it more closely mimics the natural environment of fungi, forcing them to ‘fight’ for their survival, which may uncover new secondary metabolites that are not produced under standard monoculture conditions.
1.3 Genomics and biosynthetic gene clusters
The scientific premise that fungi yield promising compounds and vast chemical diversity is evidenced in their genetics too.36 As sequencing technology platforms have become less expensive and newer technologies emerge, our capacity to access genomes has been revolutionized, providing insights into the biosynthetic potential of fungi.37,38 This has led to the identification of between 30 to 40 biosynthetic gene clusters per Aspergillus spp. with the vast majority of the encoded metabolites being unknown. These types of genes are referred to as being either: cryptic (the secondary metabolite was not identified), silent (there is little to no expression of that biosynthetic gene cluster), or orphan (the biosynthetic gene cluster has not been linked to a secondary metabolite).39,40Genomic tools for studying fungal biosynthesis are growing,20 and there are >8000 genome assemblies for fungi available via NCBI,41 which has led to the identification of genes putatively responsible for secondary metabolite production.42,43 Annotations of those genomes have shown that genes involved in secondary metabolite production are clustered in the chromosome.44,45 This indicates that individual biosynthetic pathways for secondary metabolites can be annotated and identified,46 although doing so is not trivial.47 Sequences for approximately 30 biosynthetic gene cluster per fungus have been detected,48 and a recent review noted 97 DNA loci that are likely associated with secondary metabolite biosynthesis in Pestalotiopsis fici.49 An authoritative review on biosynthetic gene clusters in fungi approximates even higher numbers for prolific genera of fungi.20 However, not all biosynthetic gene clusters are connected to the production of secondary metabolites.32,38,42,50–53 The lack of genotype and chemotype connections suggests that there are numerous silent, cryptic, and/or orphan biosynthetic gene clusters. Discovering such biosynthetic genes can help gain insights into heretofore-undiscovered pathways as well as new secondary metabolites. This, in turn, can guide us toward the frontiers of drug discovery as we can begin to uncover and activate these gene clusters to obtain structures with novel scaffolds for new drug leads. Since gene clusters in genomes are evolutionary tools for chemical innovation,37 linking the phenotype (i.e. secondary metabolites) to the genotype would increase the knowledge of chemical diversity and how secondary metabolites are being biosynthesized in Nature.36,55–57
The power of co-culturing fungi, specifically forcing them to interact in a defined environment, is that it can provide a pragmatic way to ‘turn on’ biosynthetic gene clusters that may be otherwise silent when growing fungi as monocultures. There is growing evidence in the literature that co-culturing results in new chemical diversity (Fig. 1). However, the rationale for choosing strains for the “fungal fight club” can be varied, and for those interested in new compound discovery, details on how to scale observations in a Petri dish to a level that results in meaningful amounts of structurally diverse pure compounds, are both areas where more clarity is warranted.
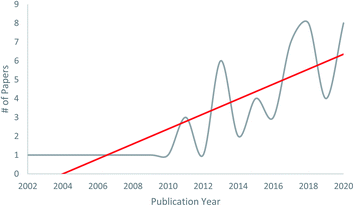 | ||
| Fig. 1 Manuscripts on fungal–fungal co-cultures published between 2002 to 2020. The first six to eight years after the publication of the seminal work of Degenkolb et al.54 resulted in about one manuscript per year on this topic, but after 2010, the trend has increased. While there are still relatively few papers on this topic to date, we believe this will expand greatly in the ensuing years, especially as varied rationales for pairing competitive co-cultures evolve, and as techniques for scaling the production of molecules of interest become more refined. | ||
2 Rationale for pairing fungal strains for co-culturing
There are likely two main goals when pairing fungi for co-culturing: to either activate silent biosynthetic gene clusters to discover new secondary metabolites and/or to characterize how fungi chemically respond to competition.58 While those endpoints may seem different, the protocols and tools for carrying out such experiments are quite similar. In either case, one must decide how to pair the fungi, ranging from focusing on fungi from the same environment all the way to completely random combinations, and shades of grey in-between.One logical method is to pair fungi that have similar ecological niches. The rationale is that since they live and reproduce in the same ecological habitat in Nature, they will likely be competing for similar nutrient resources, which will lead to interactions that may activate silent biosynthetic gene clusters that encode for metabolites to overcome, or at least survive, those interactions. Soil dwelling fungi are also good candidates, as many slow growing and fast growing fungi co-occur in the soil.59 But in general, there are many different ecological niches for fungi,22 and fungal cultures from any of these ecological groups can likely serve as candidates for co-culture experiments.
2.1 Pairings from endophytic environments
Fungal endophytes are plant-associated fungi that occur asymptomatically within the photosynthetic tissues of plants.60 Since multiple species (e.g., often 16 ± 3) can coexist within small segments (i.e., 2 × 2 cm) of tissues as a plant matures (Fig. 2),60–63 these fungi provide a logical source of fungi for co-culturing experiments for harvesting novel chemistry.64,65 When endophytes from similar environments were paired, the activated secondary metabolites encompassed a broad range of structural diversity, such as: polyketides (1–17),66–69 anthrones (18–20),70 citrinins (21–22),71 azaphilones (23–32),72,73 meroterpenoids (33–37),74 terpenes (28–37),72,74 and alkaloids (38–41) (Fig. 3).75–77 These metabolites also demonstrated a wide range of biological activities, including: antifungal,66,74 antibacterial,70,75,76 anti-proliferative,74 and antiviral.71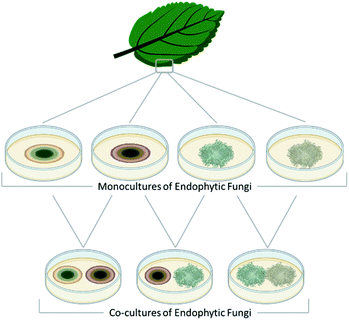 | ||
| Fig. 2 Fungi can be isolated from many different ecological niches. This figure details how one group of endosymbiotic fungi (i.e., endophytes) are used for co-culture. A plant substrate is identified, and then aseptically, the endophytic fungi are grown under standard laboratory conditions after surface sterilization of leaf segments.78 These isolated fungi are then combined in co-culture, the logic being that since they exist in the same ecological niche and occur in close physical distance, they have evolved ways to interact with each other, including the biosynthesis of small molecules. Several research teams have used variations of this approach and reported the generation of new chemical diversity.66–77,79–84 | ||
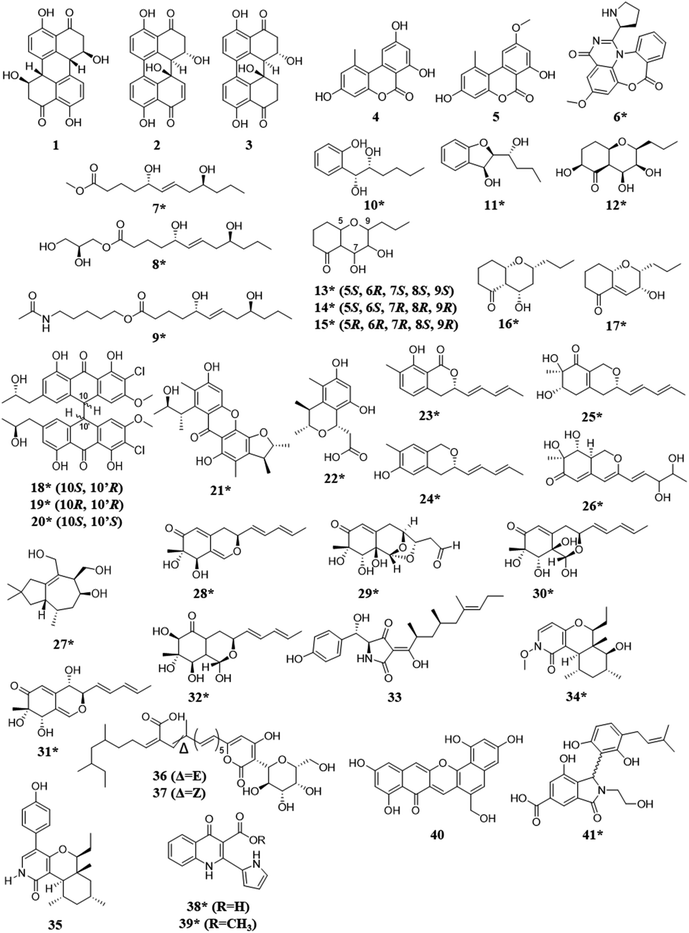 | ||
| Fig. 3 Structures of compounds isolated from the co-culturing of endophytic fungi.66–77 The new secondary metabolites are indicated by an asterisk (*). See Table S1 for more details (ESI†). | ||
2.2 Pairings from marine environments
When fungi isolated from marine environments were co-cultured, it resulted in the biosynthesis of approximately 13 new metabolites (Fig. 4). A variety of structural classes were produced from these ecological pairings, including: alkaloids (42–57),91–93 peptides (58–60),25,94 xanthones (61),95 coumarins (62),96 and polyketides (63).97 In competition, the activation of the biosynthesis of such compounds was likely for inhibitory purposes, as noted biological activities included cytotoxicity,93 inhibition of colony formation,92 and antifungal activity.25,91,94–97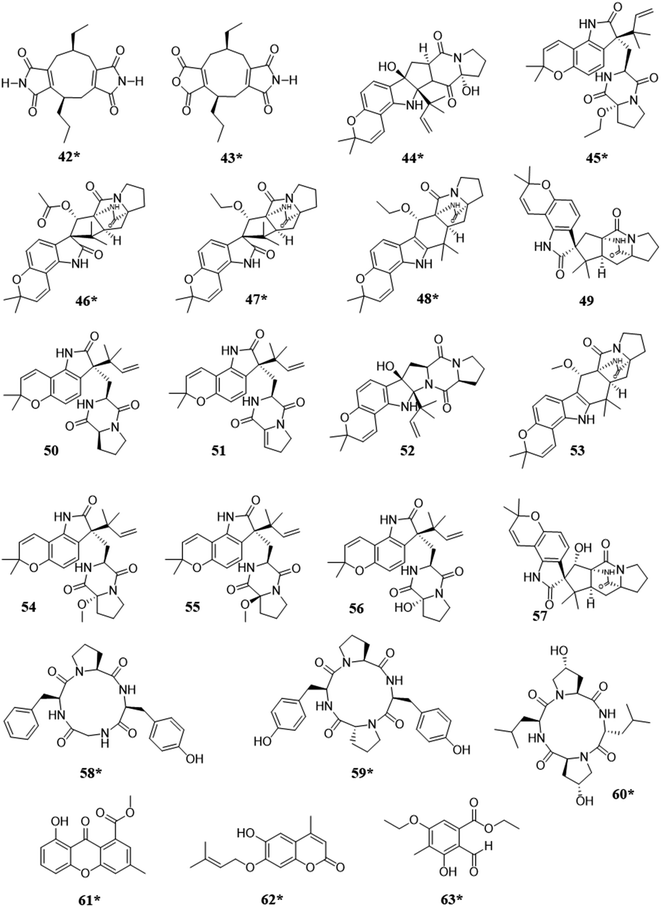 | ||
| Fig. 4 Structures of compounds isolated from the co-culturing of fungi from marine habitats.25,91–97 The new secondary metabolites are indicated by an asterisk (*). See Table S1 for more details (ESI†). | ||
2.3 Pairings from unique environments
Fungi have also been isolated from humans, vertebrate and invertebrate animals, and extreme environments. For example, when fungi isolated from invertebrate animals were co-cultured the biosynthesis of a variety of structural classes, such as, alkaloids (64), meroterpenoids (65–66), and polyketides (70–78), were observed, including a rare class of 2-alkenyl-tetrahydropyrans (78)104 and compounds with unique metal alloy associations (79 and 80) (Fig. 5).105 The co-culture of two extremophile fungi, both of which were isolated from a former copper mine pit, demonstrated the biosynthesis of sulfur containing macrolides (70–77) with potent antimicrobial activities.28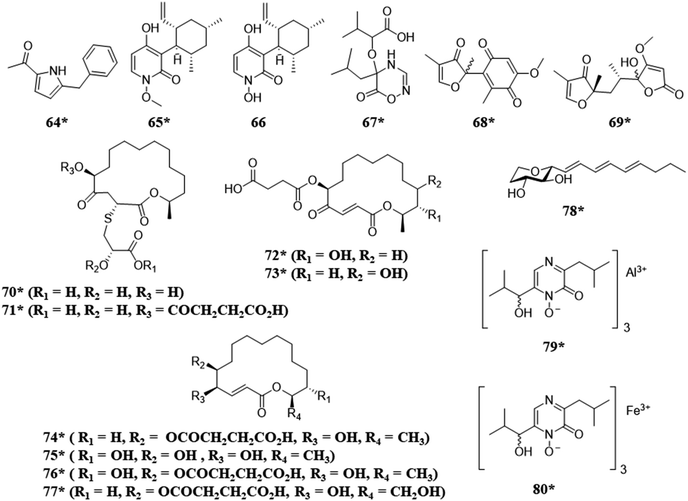 | ||
| Fig. 5 Structures of compounds isolated from the co-culturing of fungi from unique environments.28,104,105 The new secondary metabolites are indicated by an asterisk (*). See Table S1 for more details (ESI†). | ||
2.4 Pairings with and/or between pathogens
Fungi chemically communicate within the environment for a variety of reasons, for example, to gain a competitive advantage over other microbes in the fight for resources. Co-culturing can help shed light on chemical communication. When phytopathogens that compete for the same plant were co-cultured, the activation or upregulation (i.e., higher metabolite abundance) of toxic secondary metabolites with antifungal and cytotoxic capabilities have been observed.79–82 Interestingly, these interactions have been observed with both volatile and non-volatile compounds (discussed below) (Fig. 6).79 These co-culture experiments also indicate that biosynthetic gene clusters are activated by an endophyte trying to protect itself from a phytopathogen of the host.80,81 It also sheds light on the protective act of biotransformation, the process of transforming a toxic metabolite to a nontoxic form.111 When an endophyte was co-cultured with a corresponding phytopathogen, the phytopathogen activated the production of beauvericin, a potent mycotoxin. However, the authors of that study purport that the endophyte was able to overcome this competition by the biotransformation of beauvericin, as its concentration decreased as the endophyte grew.80,81 An interesting idea that emerged from such studies is: could a better understanding of the chemical communication between these fungi (i.e., phytopathogens and endophytes) lead to the development of biocontrol agents?83,112–114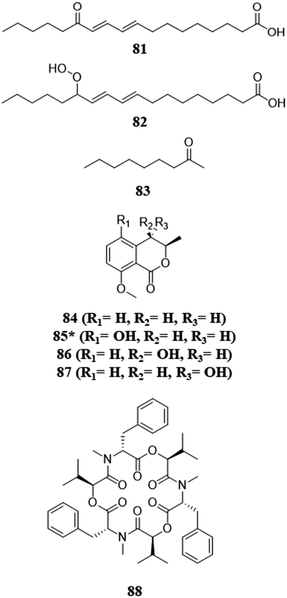 | ||
| Fig. 6 Structures of compounds isolated from the pairings with or between pathogens.79–82 The new secondary metabolites are indicated by an asterisk (*). See Table S1 for more details (ESI†). | ||
Paraconiothyrium variabile is an endophyte from the host plant Cephalotaxus harringtonia, and it has been shown to possess the ability to inhibit common phytopathogens, such as Fusarium oxysporum.81 When these fungi were co-cultured, a strong antagonism was observed via transmission electron microscopy. Even macroscopically, it was observed that the phytopathogen's mycelium became disjointed, and the media was pigmented due to metabolites from the endophyte. Upon LC-MS/MS analysis of the confrontation zone, the known metabolites biosynthesized by P. variabile, 13-oxo-9,11-octadecadienoic acid (13-oxo-ODE; 81) and 13-hydroperoxy-9,11-octadecadienoic acid (82) were induced. This analysis also showed the downregulation of beauvericin (88), which is biosynthesized by F. oxysporum. This downregulation was also observed when 81 was added to a 10 day old monoculture of F. oxysporum. By using LC-MS and RT-qPCR, the authors were able to determine that F. oxysporum was upregulating the production of beauvericin (88) during the co-culture.80 However, P. variabile was biotransforming beauvericin (88), thus causing the concentration of this molecule to decrease in co-culture.80,81 It was hypothesized that P. variabile was biotransforming beauvericin to protect itself from the mycotoxin, and this may have the added benefit of protecting the plant host.80 This is a very interesting line of reasoning, pairing a phytopathogen against endophytes from the same host. The challenge is that there are potentially scores of endophytic fungi within a host plant, and thus some level of screening must be undertaken to determine the best one to pair with the phytopathogen. In addition, the caveats noted above regarding the propagation of endophytes in the lab also apply. Regardless, understanding how phytopathogens can be modulated via co-culture may present ideas for future biocontrol agents.
2.5 Pairings due to the biosynthesis of antagonistic secondary metabolites
This co-culturing rationale is not based so much on from where the fungi reside, but rather, by the antagonistic properties of the compounds they are known to biosynthesize. By utilizing fungi that biosynthesize antagonistic metabolites, there may be a broad range of fungal species that could be explored. In a co-culturing experiment where one or more of the interacting fungi produce antagonistic metabolites, such as compounds with well documented antifungal and/or cytotoxic properties, if the other species does not find a way to mitigate the threat, then it will lose the ability to survive. This antagonistic competition has shown the potential to activate silent biosynthetic gene clusters, revealing the biosynthesis of new scaffolds and the activation of metabolites with antagonistic capabilities (Fig. 7).31,112,113,117,118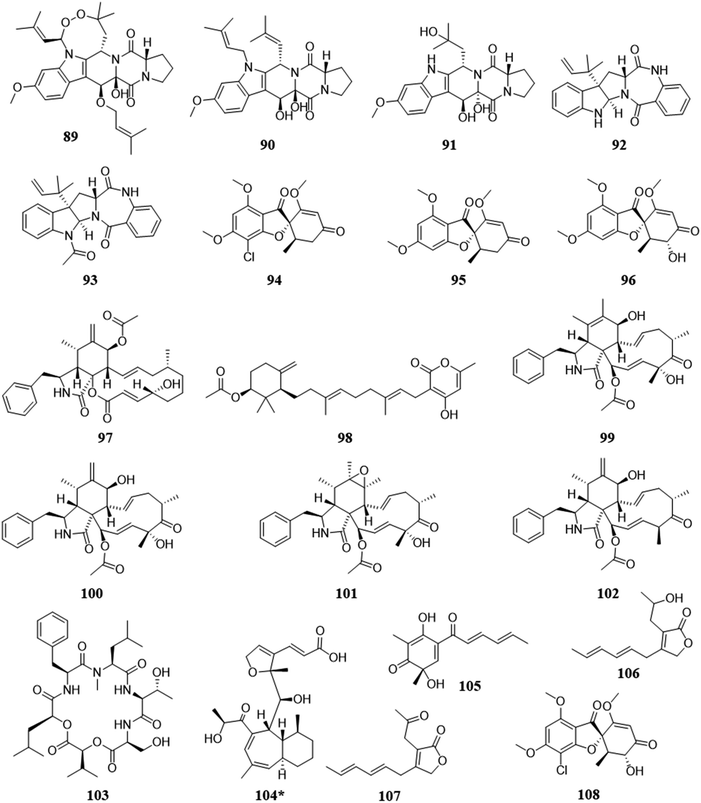 | ||
| Fig. 7 Structures of compounds isolated from the pairings due to biosynthesis of antagonistic secondary metabolites.31,112,113,117 The new secondary metabolites are indicated by an asterisk (*). See Table S1 for more details (ESI†). | ||
3 Ways to grow fungi for co-culturing
There are several ways that fungal co-cultures can be grown. The most common media types are grain-based media,31,71,117,120,121 solid agar,77,83,104,122,123 and liquid media.28,72,73,76,114 In addition, there are essentially two ways to start a fungal co-culture experiment. The most straight forward is to start both monocultures simultaneously. Obviously, this technique is used most often when the fungal strains that are being paired have similar growth rates. Thus, the challenge with this approach is that it may not work well if the fungi have significantly different growth rates. However, since a number of slow growing fungal cultures are antagonistic and exhibit competition,58 this leads to another way to carry out the experiment, which is to stagger the fungal growths before they are combined. This allows for fungi that have a slower growth rate to still be investigated in the context of co-culturing. These growths are typically carried out by either inoculating the co-culture with fungi that have different growth rates, or by using a greater amount of inoculum for the slower growing fungus.31,54,67,69,80,81,92,112,113,117,120A key insight is that one needs to think about the growth rate of the fungal cultures. As we discuss later, empirically we had better “success” with fungi that grow at a relatively fast rate (i.e., two to four weeks to confluency). However, since we often find interesting chemistry when working with monocultures of fungi that grow at a slower growth rate,124–126 we have been pondering how best to use these slower growing fungi in co-culture experiments. While it may be a pragmatic challenge to figure out how to time the growth of starter cultures for fungal co-culture experiments, such experimentation may be extremely important to tap the full biosynthetic potential of the interspecific interactions.
4 Screening of co-cultures for chemical diversity
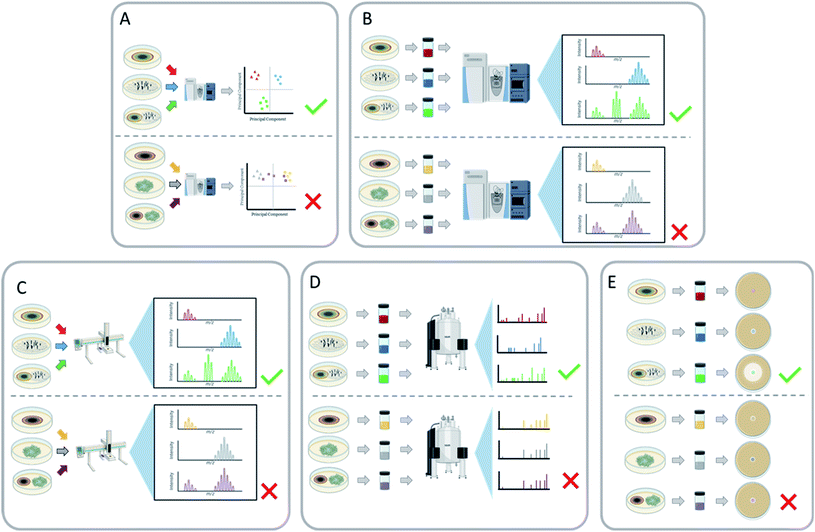
One of the goals of co-culturing is to characterize new fungal secondary metabolites, and this leads to a prevailing question: how does one screen a suite of co-culture conditions for new chemistry? The process of isolating and fully characterizing a secondary metabolite is rather extensive, as is likely well known to this audience, and this process is only complicated by the inclusion of two fungal cultures. If that effort results in the identification of ‘known’ chemistry, then valuable human and financial resources are expended unnecessarily. Techniques to rapidly evaluate co-cultures, in search of conditions that trigger the biosynthesis of new secondary metabolites, are needed, especially to screen for new chemical diversity. Ideally, a screening program of various co-cultures will indicate which specific co-cultures resulted in the activation of silent biosynthetic gene clusters. These interspecific interactions can be assessed by a variety of endpoints, including: an inhibition assay or observing a zone of inhibition,127–130 analyzing the mass spectrometry profiles of the monoculture vs. the co-culture,123,127–129,131,132 NMR spectroscopy,28 and by the observation of certain color pigments only present during co-culture conditions.105,133,1344.1 Mass spectrometry
There are several ways in which mass spectrometry can be utilized to screen co-cultures for new chemistry, and of all analytical techniques, it has probably been used the most. Mass spectrometry metabolomics can be employed for untargeted comparisons between co-culture and corresponding monocultures (Fig. 8A),69,127,130 and this allows for the identification of metabolites that are unique to the co-culture. Principal component analysis (PCA) has been used to visualize these data, to identify how chemically similar or dissimilar a co-culture is to its respective monocultures. This can also be used to identify which secondary metabolites are different between the cultures to aid in the isolation of these newly biosynthesized metabolites.117,130 Another way to identify unique metabolites is by producing a scatter plot from the retention time and m/z ratio of all the features present in the mass spectrum.127 The unique features are identified by comparing the scatter plots of the co-culture with the corresponding monocultures to identify secondary metabolites that are only produced during this interaction. Aside from metabolomics, mass spectrometry has other benefits as a quick screening tool, such as in situ analysis,85,117 dereplication,127,129 and LC-MS chromatogram comparisons (Fig. 8B and C).69,123,132 These approaches can be used to rapidly assess fungal–fungal pairings for the induction of new metabolites.4.2 Nuclear magnetic resonance
Nuclear Magnetic Resonance (NMR) has been used to screen co-culture extracts for chemical shifts that were not present under monoculture conditions (Fig. 8D).28 These resonances can also indicate interesting chemical motifs and structural complexity of the products of cryptic biosynthetic gene clusters. In interesting examples, color changes of the co-culture, which were hypothesized to be indicative of changes in the chemical profile, were then followed up by structure elucidation studies via NMR. This led to the discovery of bioactive secondary metabolites [64, 67–69, 79, 80, 121–126, 178 (121–178 are in the ESI†)], including antifungals and a suite of new cytotoxic compounds.105,133,1344.3 Bioassays
Another example of a screening method to quickly examine the viability of a co-culture is through biological assessment (Fig. 8E). This can be helpful when the point of the co-culture is to isolate and characterize bioactive metabolites, as it can quickly rule out pairings that don't induce the activity of interest.28,130,134 For example, Shen et al.130 paired 16 different fungi to have a total of 110 different co-culture pairings. The broths of those co-cultures were evaluated against C. albicans and C. neoformans. Twenty-nine of the co-culture pairings showed antifungal activity. Notably, the co-culture between Trametes robiniophila and Pleurotus ostreatus exhibited the greatest antifungal activity, while their monocultures were inactive. This co-culture produced three new sesterterpenes (127–129, ESI†) with antifungal activity.130 Overall, a bioassay screening method can increase the likelihood of finding a pairing that will activate secondary metabolites that display a bioactivity of interest, with the caveat that this then focuses only on that particular bioactivity, possibly overlooking beneficial co-culture pairings that generate compounds with very different biological activities.5 Determining the secondary metabolite producer strain
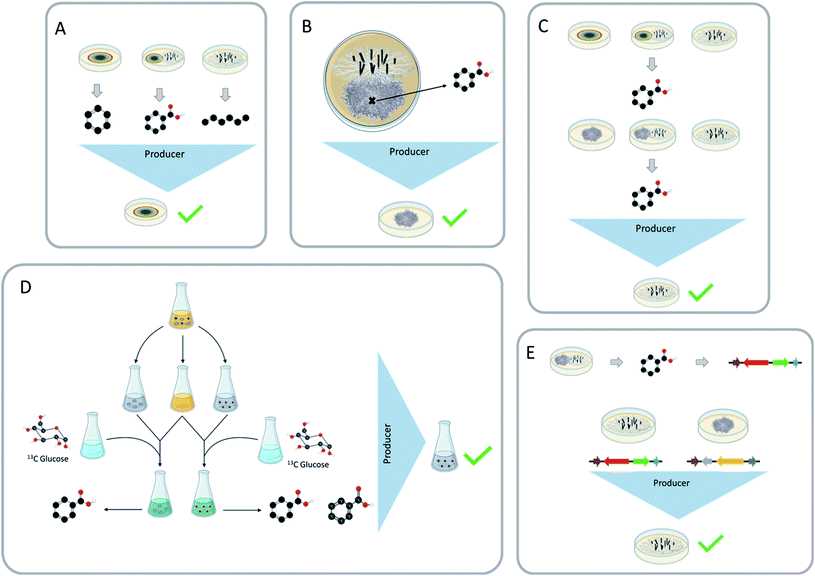
There are a few common questions that arise when discussing fungal–fungal co-culture experiments. Do you know which strain generated the new chemistry? Is it possible that the isolated compound results from the production via one strain and/or the modification (i.e., biotransformation) from the other strain? Would the new chemical diversity be the same if the producing strain was paired with a different fungus in co-culture?Being able to identify which strain is biosynthesizing the secondary metabolites that have been stimulated during the co-culture experiment is both important and beneficial. It could help to address ecological questions, such as how fungi chemically respond to their ever-changing environment and under what circumstances do certain chemical classes get activated. It could also help with drug discovery efforts. For example, if there is a silent biosynthetic gene cluster that was activated during these interspecific interactions, and it resulted in a compound of interest, knowing which fungus has the chemical machinery to produce this compound could be important. This knowledge could then be used to set up similar conditions to isolate other structural analogues, or it could be used to pinpoint the biosynthetic gene cluster, which could open the possibility for further genetic studies, such as heterologous expression.135 Being able to ascertain the producing strain can also lead to a targeted approach for increasing the structural diversity of certain secondary metabolites. The producing strain has the potential to undergo other inducing conditions and activate more structural diversity. Towards these ends, there are several ways that have been used in the literature to identify the producing strain, including: (a) structural similarities, (b) the presence of these metabolites during the monoculture, (c) location of the secondary metabolites during the co-culture, (d) exposing the paired fungi to different conditions, (e) 13C media doping to trace the biosynthesis, and (f) genetic approaches.
5.1 Structural similarities and monoculture presence
The first two methods most commonly used to identify the producing strain are structural similarities and the presence of these metabolites in the monoculture (Fig. 9A). Secondary metabolites identified during these interspecific interactions may be analogous to the monoculture metabolites or other known metabolites, which would lead to the identification of the producer strain by structural similarities. Co-culturing can also increase the abundance of secondary metabolites; since these metabolites were also present in monoculture conditions, the producing strain would remain the same.79,105,133The producer strain can be identified by analyzing the structure of the newly characterized metabolites and comparing them to secondary metabolites isolated from the monocultures66,74,127 as well as literature comparisons.113,117,136 Structural similarities can be assessed through literature comparisons, for example, tremulane sesquiterpenes (23–27) were isolated from the co-culture of Nigrospora oryzae and Irpex lacteus. The literature indicated that I. lacteus has the ability to produce this class of secondary metabolites, indicating that the most likely producer strain of nigrosirpexin A (27) was I. lacteus.72 Isolating secondary metabolites that are building blocks is another way to use structural similarities. When Fusarium tricinctum and F. begonia were co-cultured, they produced enniatins (131–132, ESI†) and secondary metabolites that were building blocks of the enniatins. Due to F. tricinctum producing the enniatins in monoculture, it was deduced that F. tricinctum was the producer of the new linear depsipeptides, which incorporated a nearly identical suite of amino acid building blocks.121
5.2 Spatial location during co-culture
By using imaging mass spectrometry, the location where the metabolites accumulate during the co-culture can indicate which fungus produced the secondary metabolites (Fig. 9B).77,113,129 For example, when Pseudoxylaria sp. and Coriolopsis sp. were co-cultured, several new secondary metabolites were characterized (181–186, ESI†). MALDI-MS imaging showed that four (181–184) of these newly activated metabolites were only present on the mycelium of Pseudoxylaria sp.1295.3 Different pairing/growing in presence of an extract
Fungi have been shown to activate the biosynthesis of the same secondary metabolites when co-cultured with different strains or in the presence of a fungal extract,31,104,128,134 and this approach can be used to identify the producer strain. For example, Shang et al.104 grew Trichoderma hamatum with the monoculture extract of Chaunopycnis sp.; the opposite experiment was also implemented (i.e., Chaunopycnis sp. was grown with the monoculture extract of Trichoderma hamatum). In doing so, they found that T. hamatum made 65, both when paired with Chaunopycnis sp. and the extract thereof.104 An alternate version of this approach is to pair the two fungi with a different fungal species and see if the biosynthesis of the unique compound can be triggered (Fig. 9C). For instance, Aspergillus fischeri and Xylaria flabelliformis were co-cultured, leading to the biosynthesis of wheldone (104).31 In trying to determine which of these two fungi produced this compound, X. flabelliformis was co-cultured with Nectria pseudotrichia where 10431 was observed again, indicating that the producer strain of 104 was X. flabelliformis.Biosynthetic gene clusters that encode for secondary metabolites may have self-protecting genes, which, as the name indicates, allows them to protect themselves from their own potentially harmful secondary metabolites.137 This evolutionary protective trait can be used to ascertain the producer strain. If the co-culture elicits a toxic secondary metabolite, the producer strain will not be as affected from this metabolite as other strains. The co-culture of Talaromyces siamensis and Phomopsis sp. induced the production of the antifungal metabolite BE-31405 (126, ESI†).134 The producer strain of 126 was identified by pairing the co-cultured fungi with other fungi to see if the induced secondary metabolite would be produced, and in this case, when T. siamensis was co-cultured with another fungus, 126 was again produced. The paired fungi were also grown in the presence of 126, and in this case the Phomopsis sp. was unable to grow but T. siamensis successfully grew out, likely because it had a way to protect itself from the antifungal activity of 126. This set of experiments strongly suggests that T. siamensis was the producer strain of compound BE-31405 (126).
5.4 Media doping
Incorporating modified building blocks into the media has been shown to aid in the determination of the producer strain, by identifying which strain was able to produce the modified metabolite. This has been successfully shown by Xu et al.29 and Shen et al.130 The method works by co-culturing two fungi in liquid media, then separating the fungi back into monoculture growths. The media used for the new monoculture growths is a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of the co-culture broth and 13C-labeled glucose media. These cultures are then analyzed by LC-MS to determine which strain was able to uptake the 13C label and incorporate it into the induced metabolites (Fig. 9D).29,130 The caveat is that this seems to work best on liquid media with fungi that have very different morphologies. If the co-culture is grown on grain-based media, there would be no plausible way to separate the co-culture back into monocultures since they grow intertwined on a solid substrate.
1 mixture of the co-culture broth and 13C-labeled glucose media. These cultures are then analyzed by LC-MS to determine which strain was able to uptake the 13C label and incorporate it into the induced metabolites (Fig. 9D).29,130 The caveat is that this seems to work best on liquid media with fungi that have very different morphologies. If the co-culture is grown on grain-based media, there would be no plausible way to separate the co-culture back into monocultures since they grow intertwined on a solid substrate.
5.5 Genomics
In the post genomics era, it is possible to use genomics to identify the producer strain (Fig. 9E). When Aspergillus fischeri and Xylaria flabelliformis were co-cultured, they produced several secondary metabolites that were not present during either monoculture growth.117 Both fungi were genetically tractable, and the secondary metabolites were analogues of known secondary metabolites that are linked to biosynthetic gene clusters.56,138 This made it possible to identify which fungus had the machinery to biosynthesize at least some of the activated secondary metabolites (Fig. 9C). This method is very useful at identifying the producer strain, but it has several limitations. Both fungi would need to have genomes that were made publicly available (i.e., genetically tractable), or they would have to be highly related to a species with a publicly available genome. The identified secondary metabolites would also have to be linked to a biosynthetic gene cluster, and that is likely more challenging for a novel metabolite than realized. Another example of this is using RT-qPCR to identify which genes were responsible for the production of the newly activated secondary metabolites. When Pleurotus ostreatus and Trametes robiniophila Murr were co-cultured, they produce three new sesterterpenes (127–129). RT-qPCR showed the transcription level of expressed genes encoding the new chemistry was much higher in co-culture than in the monoculture. Thus, the biosynthetic gene cluster responsible for these secondary metabolites was identified by RT-qPCR. During this analysis the producer strain was identified through other means, but this concept is a potential way to use gene activation in the identification of the producer strains.1306 Final thoughts
There are many research groups, around the world, interested in discovering new chemical diversity from fungal cultures. Yet, based upon what we are learning through studying the genomes of fungi, it is apparent that there are many more fungal metabolites to be discovered. In fact, given how the cost of genome sequencing will diminish over time, and as tools to annotate the biosynthetic gene clusters in those genomes will improve, it is likely that this gap between the discovered chemical diversity of fungi (i.e. compounds that have been isolated/characterized) vs. their potential chemical diversity (i.e. compounds that can be inferred from genome sequencing) will only grow.If you accept the above to be true, then a clear challenge is determining ways to turn on the biosynthesis of those structurally unique molecules. Co-culturing of fungi, forcing them to fight for resources, is one approach to stimulate the biosynthesis of new chemical diversity, essentially forcing one or both fungi to utilize some of their cryptic biosynthetic gene clusters. While such experiments have produced exciting results, we still seem to be in an early stage of investigation. Over the last two decades, there are fewer than 60 publications that report discoveries from fungal–fungal co-cultures, and that should be contrasted with the approximately 2000 publications, annually, on the isolation of fungal metabolites from monocultures. Hence, there are many opportunities for future improvements in this approach, especially if successful pairings could be developed in a more predictable manner.
Obvious steps to address this challenge include more collaborations between natural products chemists and taxonomic mycologists. For example, their collective knowledge may help us understand how interspecific interactions occur in natural substrates and how competition plays a role in organizing fungal communities. This, in turn, could lead to better choices when selecting fungal species to pair in co-cultures in the laboratory. We anticipate that as more fungal species are isolated and identified, and as analytic chemistry tools improve, particularly those that can be used to scout the chemistry of fungal–fungal co-cultures in situ, fungal metabolites with new scaffolds will be identified. As this research area matures, it is our hope that fungal–fungal co-culturing will become a routine weapon in the arsenal used to identify fungal metabolites that benefit the industrial, agrochemical, and pharmaceutical sectors.
7 Author contributions
SLK was responsible for organizing the data, writing the paper, and making the figures. HAR was responsible for writing the paper and organizing the data. CDR was responsible for writing the paper and organizing the figures. NHO was responsible for overseeing the project and writing/editing the manuscript.8 Conflicts of interest
Nicholas Oberlies is a member of the Scientific Advisory Board of Mycosynthetix, Inc.9 Acknowledgements
Research on fungi in the Oberlies Lab is supported in part by the National Institutes of Health via the National Cancer Institute (P01 CA125066) and the National Institute of Allergy and Infectious Diseases (R56 AI146096 and R01 AI153356), and S.L.K. and C.D.R. were supported in part by the National Center for Complementary and Integrative Health (F31 AT010558) and the National Institute of General Medical Sciences (T34 GM113860), respectively. The authors owe profound gratitude to Tyler N. Graf for helpful edits.10 Notes and references
- J. Houbraken, J. C. Frisvad and R. A. Samson, IMA Fungus, 2011, 2, 87–95 CrossRef PubMed.
- R. Gaynes, Emerging Infect. Dis., 2017, 23, 849 CrossRef.
- A. Fleming, Br. J. Exp. Pathol., 1929, 10, 226 CAS.
- E. Lax, The mold in Dr Florey's coat: the story of the penicillin miracle, Macmillan, 2005 Search PubMed.
- V. J. Cirillo, Perspect. Biol. Med., 2008, 51, 121–133 CrossRef PubMed.
- D. Goyette, Surg. Technol., 2017, 49, 256–264 Search PubMed.
- The Nobel Prize in Physiology or Medicine, Nobel Prize:€ Nobel Prize, 1945, https://www.nobelprize.org/prizes/medicine/1945/summary/ Search PubMed.
- M. Blackwell, Am. J. Bot., 2011, 98, 426–438 CrossRef PubMed.
- D. L. Hawksworth and R. Lücking, Microbiol. Spectrum, 2017, 5, 79–95 Search PubMed.
- M. Cheek, E. Nic Lughadha, P. Kirk, H. Lindon, J. Carretero, B. Looney, B. Douglas, D. Haelewaters, E. Gaya, T. Llewellyn, A. M. Ainsworth, Y. Gafforov, K. Hyde, P. Crous, M. Hughes, B. E. Walker, R. Campostrini Forzza, K. M. Wong and T. Niskanen, Plants, People, Planet, 2020, 2, 371–388 CrossRef.
- D. Hibbett, K. Abarenkov, U. Kõljalg, M. Öpik, B. Chai, J. Cole, Q. Wang, P. Crous, V. Robert and T. Helgason, Mycologia, 2016, 108, 1049–1068 Search PubMed.
- D. S. Hibbett, M. Binder, J. F. Bischoff, M. Blackwell, P. F. Cannon, O. E. Eriksson, S. Huhndorf, T. James, P. M. Kirk and R. Lücking, Mycol. Res., 2007, 111, 509–547 CrossRef PubMed.
- G. F. Bills and J. B. Gloer, Microbiol. Spectrum, 2016, 4, 1087–1119 Search PubMed.
- J. H. Wisecaver, J. C. Slot and A. Rokas, PLoS Genet., 2014, 10, e1004816 CrossRef PubMed.
- H. Zähner, Folia Microbiol., 1979, 24, 435–443 CrossRef PubMed.
- E. Bignell, T. C. Cairns, K. Throckmorton, W. C. Nierman and N. P. Keller, Philos. Trans. R. Soc., B, 2016, 371, 20160023 CrossRef PubMed.
- L. C. Vining, Annu. Rev. Microbiol., 1990, 44, 395–427 CrossRef CAS PubMed.
- A. D. Kinghorn, E. J. C. De Blanco, D. M. Lucas, H. L. Rakotondraibe, J. Orjala, D. D. Soejarto, N. H. Oberlies, C. J. Pearce, M. C. Wani and B. R. Stockwell, Anticancer Res., 2016, 36, 5623–5637 CrossRef CAS PubMed.
- M. Buckley, American Society for Microbiology, 2008, 1–44 Search PubMed.
- N. P. Keller, Nat. Rev. Microbiol., 2019, 17, 167–180 CrossRef CAS PubMed.
- S. Bertrand, O. Schumpp, N. Bohni, A. Bujard, A. Azzollini, M. Monod, K. Gindro and J.-L. Wolfender, J. Chromatogr. A, 2013, 1292, 219–228 CrossRef CAS PubMed.
- G. M. Mueller, Biodiversity of fungi: inventory and monitoring methods, Elsevier, 2011 Search PubMed.
- L. Losada, O. Ajayi, J. C. Frisvad, J. Yu and W. C. Nierman, Med. Mycol., 2009, 47, S88–S96 CrossRef CAS PubMed.
- S. Chatterjee, Y. Kuang, R. Splivallo, P. Chatterjee and P. Karlovsky, BMC Microbiol., 2016, 16, 83 CrossRef PubMed.
- S. Huang, W. Ding, C. Li and D. G. Cox, Pharmacogn. Mag., 2014, 10, 410–414 CrossRef PubMed.
- F. Kuhar, V. Castiglia and L. Levin, Int. Biodeterior. Biodegrad., 2015, 104, 238–243 CrossRef CAS.
- M. Kettering, O. Sterner and T. Anke, Zeitschrift für Naturforschung C, 2004, 59, 816–823 CrossRef CAS PubMed.
- A. A. Stierle, D. B. Stierle, D. Decato, N. D. Priestley, J. B. Alverson, J. Hoody, K. McGrath and D. Klepacki, J. Nat. Prod., 2017, 80, 1150–1160 CrossRef CAS PubMed.
- X.-Y. Xu, X.-T. Shen, X.-J. Yuan, Y.-M. Zhou, H. Fan, L.-P. Zhu, F.-Y. Du, M. Sadilek, J. Yang and B. Qiao, Front. Microbiol., 2018, 8, 2647 CrossRef PubMed.
- N. Adnani, M. G. Chevrette, S. N. Adibhatla, F. Zhang, Q. Yu, D. R. Braun, J. Nelson, S. W. Simpkins, B. R. McDonald and C. L. Myers, ACS Chem. Biol., 2017, 12, 3093–3102 CrossRef CAS PubMed.
- S. L. Knowles, H. A. Raja, I. H. Isawi, L. Flores-Bocanegra, P. H. Reggio, C. J. Pearce, J. E. Burdette, A. Rokas and N. H. Oberlies, Org. Lett., 2020, 22, 1878–1882 CrossRef CAS PubMed.
- K. Scherlach and C. Hertweck, Org. Biomol. Chem., 2009, 7, 1753–1760 RSC.
- C. Nai and V. Meyer, Trends Microbiol., 2018, 26, 538–554 CrossRef CAS PubMed.
- H.-W. Nützmann, V. Schroeckh and A. A. Brakhage, Regulatory cross talk and microbial induction of fungal secondary metabolite gene clusters, in Methods Enzymol, ed. D. A. Hopwood, Elsevier, 2012, vol. 517, pp. 325–341 Search PubMed.
- S. Bertrand, N. Bohni, S. Schnee, O. Schumpp, K. Gindro and J.-L. Wolfender, Biotechnol. Adv., 2014, 32, 1180–1204 CrossRef CAS PubMed.
- A. Rokas, M. E. Mead, J. L. Steenwyk, H. A. Raja and N. H. Oberlies, Nat. Prod. Rep., 2020, 37, 868–878 RSC.
- A. Osbourn, Trends Genet., 2010, 26, 449–457 CrossRef CAS PubMed.
- J. Stajich, 1000 fungal genomes project, in 1kfg Updates – Another 100 genomes, April 26, 2017, 2017 Search PubMed.
- A. A. Brakhage and V. Schroeckh, Fungal Genet. Biol., 2011, 48, 15–22 CrossRef CAS PubMed.
- B. C. Covington, F. Xu and M. R. Seyedsayamdost, Annu. Rev. Biochem., 2021, 90, 763–788 CrossRef CAS PubMed.
- As of April 22, 2021, there were 8091 genome assemblies for fungi available in NCBI.
- I. V. Grigoriev, R. Nikitin, S. Haridas, A. Kuo, R. Ohm, R. Otillar, R. Riley, A. Salamov, X. Zhao and F. Korzeniewski, Nucleic Acids Res., 2013, 42, D699–D704 CrossRef PubMed.
- M. T. Robey, L. K. Caesar, M. T. Drott, N. P. Keller and N. L. Kelleher, Proc. Natl. Acad. Sci. U. S. A., 2021, 118 CrossRef CAS PubMed.
- N. P. Keller and T. M. Hohn, Fungal Genet. Biol., 1997, 21, 17–29 CrossRef CAS PubMed.
- J.-H. Yu and N. Keller, Annu. Rev. Phytopathol., 2005, 43, 437–458 CrossRef CAS PubMed.
- N. P. Keller, G. Turner and J. W. Bennett, Nat. Rev. Microbiol., 2005, 3, 937–947 CrossRef CAS PubMed.
- S. L. Salzberg, Next-generation genome annotation: we still struggle to get it right, BioMed Central, 2019 Search PubMed.
- H. J. Pel, J. H. de Winde, D. B. Archer, P. S. Dyer, G. Hofmann, P. J. Schaap, G. Turner, R. P. de Vries, R. Albang, K. Albermann, M. R. Andersen, J. D. Bendtsen, J. A. Benen, M. van den Berg, S. Breestraat, M. X. Caddick, R. Contreras, M. Cornell, P. M. Coutinho, E. G. Danchin, A. J. Debets, P. Dekker, P. W. van Dijck, A. van Dijk, L. Dijkhuizen, A. J. Driessen, C. d'Enfert, S. Geysens, C. Goosen, G. S. Groot, P. W. de Groot, T. Guillemette, B. Henrissat, M. Herweijer, J. P. van den Hombergh, C. A. van den Hondel, R. T. van der Heijden, R. M. van der Kaaij, F. M. Klis, H. J. Kools, C. P. Kubicek, P. A. van Kuyk, J. Lauber, X. Lu, M. J. van der Maarel, R. Meulenberg, H. Menke, M. A. Mortimer, J. Nielsen, S. G. Oliver, M. Olsthoorn, K. Pal, N. N. van Peij, A. F. Ram, U. Rinas, J. A. Roubos, C. M. Sagt, M. Schmoll, J. Sun, D. Ussery, J. Varga, W. Vervecken, P. J. van de Vondervoort, H. Wedler, H. A. Wosten, A. P. Zeng, A. J. van Ooyen, J. Visser and H. Stam, Nat. Biotechnol., 2007, 25, 221–231 CrossRef PubMed.
- S. E. Helaly, B. Thongbai and M. Stadler, Nat. Prod. Rep., 2018, 35, 992–1014 RSC.
- J. E. Galagan, S. E. Calvo, C. Cuomo, L.-J. Ma, J. R. Wortman, S. Batzoglou, S.-I. Lee, M. Baştürkmen, C. C. Spevak and J. Clutterbuck, Nature, 2005, 438, 1105–1115 CrossRef CAS PubMed.
- H. Gross, Appl. Microbiol. Biotechnol., 2007, 75, 267–277 CrossRef CAS PubMed.
- G. L. Challis, J. Med. Chem., 2008, 51, 2618–2628 CrossRef CAS PubMed.
- A. A. Brakhage, Nat. Rev. Microbiol., 2013, 11, 21–32 CrossRef CAS PubMed.
- T. Degenkolb, S. Heinze, B. Schlegel, G. Strobel and U. Gräfe, Biosci., Biotechnol., Biochem., 2002, 66, 883–886 CrossRef CAS PubMed.
- J. L. Steenwyk, M. E. Mead, S. L. Knowles, H. A. Raja, C. D. Roberts, O. Bader, J. Houbraken, G. H. Goldman, N. H. Oberlies and A. Rokas, Genetics, 2020, 216, 481–497 CrossRef CAS PubMed.
- M. E. Mead, S. L. Knowles, H. A. Raja, S. R. Beattie, C. H. Kowalski, J. L. Steenwyk, L. P. Silva, J. Chiaratto, L. N. Ries, G. H. Goldman, N. H. Oberlies and A. Rokas, mSphere, 2019, 4, e00018-19 CrossRef PubMed.
- J. C. Slot and E. Gluck-Thaler, Curr. Opin. Genet. Dev., 2019, 58, 17–24 CrossRef PubMed.
- C. Shearer, Can. J. Bot., 1995, 73, 1259–1264 CrossRef.
- G. F. Bills, M. Christensen, M. Powell and G. Thorn, Saprobic soil fungi, Biodiversity of fungi: Inventory and monitoring methods, Elsevier, 2004 Search PubMed.
- A. E. Arnold, Fungal Biology Reviews, 2007, 21, 51–66 CrossRef.
- A. E. Arnold, Plant Press, 2008, 32, 13–15 Search PubMed.
- A. E. Arnold, Z. Maynard, G. S. Gilbert, P. D. Coley and T. A. Kursar, Ecol. Lett., 2000, 3, 267–274 CrossRef.
- M. A. Gamboa, S. Laureano and P. Bayman, Mycopathologia, 2003, 156, 41–45 CrossRef PubMed.
- R. X. Tan and W. X. Zou, Nat. Prod. Rep., 2001, 18, 448–459 RSC.
- S. K. Verma, S. K. Gond, A. Mishra, V. K. Sharma, J. Kumar, D. K. Singh, A. Kumar and R. N. Kharwar, Fungal endophytes representing diverse habitats and their role in plant protection, in Developments in fungal biology and applied mycology, Springer, 2017, pp. 135–157 Search PubMed.
- F. O. Chagas, L. G. Dias and M. T. Pupo, J. Chem. Ecol., 2013, 39, 1335–1342 CrossRef CAS PubMed.
- H.-T. Li, H. Zhou, R.-T. Duan, H.-Y. Li, L.-H. Tang, X.-Q. Yang, Y.-B. Yang and Z.-T. Ding, J. Nat. Prod., 2019, 82, 1009–1013 CrossRef CAS PubMed.
- F. Zhu, G. Chen, X. Chen, M. Huang and X. Wan, Chem. Nat. Compd., 2011, 47, 767–769 CrossRef CAS.
- H.-T. Li, T. Liu, R. Yang, F. Xie, Z. Yang, Y. Yang, H. Zhou and Z.-T. Ding, RSC Adv., 2020, 10, 18384–18389 RSC.
- P. Mandelare, D. Adpressa, E. Kaweesa, L. Zakharov and S. Loesgen, J. Nat. Prod., 2018, 81, 1014–1022 CrossRef CAS PubMed.
- S.-Q. Yang, X.-M. Li, X. Li, H.-L. Li, L.-H. Meng and B.-G. Wang, Phytochem. Lett., 2018, 25, 191–195 CrossRef CAS.
- Q.-Y. Zhou, X.-Q. Yang, Z.-X. Zhang, B.-Y. Wang, M. Hu, Y.-B. Yang, H. Zhou and Z.-T. Ding, Fitoterapia, 2018, 130, 26–30 CrossRef CAS PubMed.
- Z.-X. Zhang, X.-Q. Yang, Q.-Y. Zhou, B.-Y. Wang, M. Hu, Y.-B. Yang, H. Zhou and Z.-T. Ding, Molecules, 2018, 23, 1816 CrossRef PubMed.
- C. Li, A. M. Sarotti, B. Yang, J. Turkson and S. Cao, Molecules, 2017, 22, 1166 CrossRef PubMed.
- F. Zhu and Y. Lin, Chin. Sci. Bull., 2006, 51, 1426 CAS.
- F. Zhu, G. Chen, J. Wu and J. Pan, Nat. Prod. Res., 2013, 27, 1960–1964 CrossRef CAS PubMed.
- Y. Sadahiro, H. Kato, R. M. Williams and S. Tsukamoto, J. Nat. Prod., 2020 Search PubMed.
- A. E. Arnold, Z. Maynard and G. S. Gilbert, Mycol. Res., 2001, 105, 1502–1507 CrossRef.
- A. Azzollini, L. Boggia, J. Boccard, B. Sgorbini, N. Lecoultre, P.-M. Allard, P. Rubiolo, S. Rudaz, K. Gindro and C. Bicchi, Front. Microbiol., 2018, 9, 72 CrossRef PubMed.
- M. Bärenstrauch, S. Mann, C. Jacquemin, S. Bibi, O.-K. Sylla, E. Baudouin, D. Buisson, S. Prado and C. Kunz, Fungal Genet. Biol., 2020, 103383 CrossRef PubMed.
- A. Combès, I. Ndoye, C. Bance, J. Bruzaud, C. Djediat, J. Dupont, B. Nay and S. Prado, PLoS One, 2012, 7, e47313 CrossRef PubMed.
- G. Glauser, K. Gindro, J. Fringeli, J.-P. De Joffrey, S. Rudaz and J.-L. Wolfender, J. Agric. Food Chem., 2009, 57, 1127–1134 CrossRef CAS PubMed.
- S. Halecker, J.-P. Wennrich, S. Rodrigo, N. Andrée, L. Rabsch, C. Baschien, M. Steinert, M. Stadler, F. Surup and B. Schulz, Fungal Ecology, 2020, 45, 100918 CrossRef.
- S. S. Soliman and M. N. Raizada, Front. Microbiol., 2013, 4, 3 CAS.
- V. P. Sica, E. R. Rees, E. Tchegnon, R. H. Bardsley, H. A. Raja and N. H. Oberlies, Front. Microbiol., 2016, 7, 544 Search PubMed.
- S. J. Higginbotham, A. E. Arnold, A. Ibañez, C. Spadafora, P. D. Coley and T. A. Kursar, PLoS One, 2013, 8, e73192 CrossRef CAS PubMed.
- I. Bodinaku, J. Shaffer, A. B. Connors, J. L. Steenwyk, M. N. Biango-Daniels, E. K. Kastman, A. Rokas, A. Robbat and B. E. Wolfe, mBio, 2019, 10, e02445-19 CrossRef PubMed.
- T. Sudhakar, S. Dash, R. Rao, R. Srinivasan, S. Zacharia, M. Atmanand, B. Subramaniam and S. Nayak, Curr. Sci., 2013, 104, 178 Search PubMed.
- J. Ludwig-Müller, Biotechnol. Lett., 2015, 37, 1325–1334 CrossRef PubMed.
- T. N. Graf, D. Kao, J. Rivera-Chávez, J. M. Gallagher, H. A. Raja and N. H. Oberlies, Planta Med., 2020, 86, 988–996 CrossRef CAS PubMed.
- W. Ding, Y. Lu, Z. Feng, S. Luo and C. Li, Chem. Nat. Compd., 2017, 53, 691–693 CrossRef CAS.
- S. S. Afiyatullov, O. I. Zhuravleva, A. S. Antonov, D. V. Berdyshev, M. V. Pivkin, V. A. Denisenko, R. S. Popov, A. V. Gerasimenko, G. von Amsberg and S. A. Dyshlovoy, J. Antibiot., 2018, 71, 846–853 CrossRef CAS PubMed.
- C.-Y. Li, W.-J. Ding, C.-L. Shao, Z.-G. She and Y.-C. Lin, J. Asian Nat. Prod. Res., 2010, 12, 809–813 CrossRef CAS PubMed.
- C. Li, J. Wang, C. Luo, W. Ding and D. G. Cox, Nat. Prod. Res., 2014, 28, 616–621 CrossRef CAS PubMed.
- C. Li, J. Zhang, C. Shao, W. Ding, Z. She and Y. Lin, Chem. Nat. Compd., 2011, 47, 382–384 CrossRef CAS.
- J. Wang, S. Huang, C. Li, W. Ding, Z. She and C. Li, Chem. Nat. Compd., 2015, 51, 239–241 CrossRef CAS.
- J. Wang, W. Ding, C. Li, S. Huang, Z. She and Y. Lin, Chem. Nat. Compd., 2013, 49, 799–802 CrossRef CAS.
- A. S. Gladfelter, T. Y. James and A. S. Amend, Curr. Biol., 2019, 29, R191–R195 CrossRef CAS PubMed.
- T. El-Elimat, H. A. Raja, M. Figueroa, A. H. Al Sharie, R. L. Bunch and N. H. Oberlies, J. Nat. Prod., 2021, 84, 898–916 CrossRef CAS PubMed.
- E. Oppong-Danquah, D. Parrot, M. Blümel, A. Labes and D. Tasdemir, Front. Microbiol., 2018, 9, 2072 CrossRef PubMed.
- C. Panebianco, W. Tam and E. Jones, Fungal Diversity, 2002, 10, 77–88 Search PubMed.
- D. P. Overy, T. Rämä, R. Oosterhuis, A. K. Walker and K.-L. Pang, Mar. Drugs, 2019, 17, 42 CrossRef CAS PubMed.
- K.-L. Pang, D. P. Overy, E. G. Jones, M. da Luz Calado, G. Burgaud, A. K. Walker, J. A. Johnson, R. G. Kerr, H.-J. Cha and G. F. Bills, Fungal Biology Reviews, 2016, 30, 163–175 CrossRef.
- Z. Shang, A. A. Salim and R. J. Capon, J. Nat. Prod., 2017, 80, 1167–1172 CrossRef CAS PubMed.
- J. Bao, J. Wang, X. Y. Zhang, X. H. Nong and S. H. Qi, Chem. Biodiversity, 2017, 14, e1600327 CrossRef PubMed.
- A. Medina, M. Schmidt-Heydt, A. Rodriguez, R. Parra, R. Geisen and N. Magan, Curr. Genet., 2015, 61, 325–334 CrossRef CAS PubMed.
- Y. Zhang, W. Wu and L. Cai, Fungal Biol., 2017, 121, 21–43 CrossRef PubMed.
- Y. Zhang, W.-P. Wu, D.-M. Hu, Y.-Y. Su and L. Cai, Mycological Progress, 2014, 13, 165–170 CrossRef.
- R. Chávez, F. Fierro, R. O. García-Rico and I. Vaca, Front. Microbiol., 2015, 6, 903 Search PubMed.
- J. Houbraken, H. Spierenburg and J. C. Frisvad, Antonie van Leeuwenhoek, 2012, 101, 403–421 CrossRef CAS PubMed.
- W. Hüttel and D. Hoffmeister, Fungal biotransformations in pharmaceutical sciences, in Industrial applications, Springer, 2011, pp. 293–317 Search PubMed.
- P. R. Tamandegani, T. Marik, D. Zafari, D. Balázs, C. Vágvölgyi, A. Szekeres and L. Kredics, Biomolecules, 2020, 10, 730 CrossRef CAS PubMed.
- A. Tata, C. Perez, M. L. Campos, M. A. Bayfield, M. N. Eberlin and D. R. Ifa, Anal. Chem., 2015, 87, 12298–12305 CrossRef CAS PubMed.
- L.-J. Shi, Y.-M. Wu, X.-Q. Yang, T.-T. Xu, S. Yang, X.-Y. Wang, Y.-B. Yang and Z.-T. Ding, J. Nat. Prod., 2020, 1374–1382 CrossRef CAS PubMed.
- G. Surico, L. Mugnai and G. Marchi, The esca disease complex, in Integrated management of diseases caused by fungi, phytoplasma and bacteria, Springer, 2008, pp. 119–136 Search PubMed.
- A. Spagnolo, G. Marchi, F. Peduto, A. J. Phillips and G. Surico, Eur. J. Plant Pathol., 2011, 129, 485–500 CrossRef.
- S. L. Knowles, H. A. Raja, A. J. Wright, A. M. L. Lee, L. K. Caesar, N. B. Cech, M. E. Mead, J. L. Steenwyk, L. Ries, G. H. Goldman, A. Rokas and N. H. Oberlies, Front. Microbiol., 2019, 10, 285 CrossRef PubMed.
- J. B. Gloer, Can. J. Bot., 1995, 73, 1265–1274 CrossRef.
- V. P. Sica, H. A. Raja, T. El-Elimat and N. H. Oberlies, RSC Adv., 2014, 4, 63221–63227 RSC.
- W. Wang, J. Gong, X. Liu, C. Dai, Y. Wang, X.-N. Li, J. Wang, Z. Luo, Y. Zhou, Y. Xue, H. Zhu, C. Chen and Y. Zhang, J. Nat. Prod., 2018, 81, 1578–1587 CrossRef CAS PubMed.
- J.-p. Wang, W. Lin, V. Wray, D. Lai and P. Proksch, Tetrahedron Lett., 2013, 54, 2492–2496 CrossRef CAS.
- G. Yu, Z. Sun, J. Peng, M. Zhu, Q. Che, G. Zhang, T. Zhu, Q. Gu and D. Li, J. Nat. Prod., 2019, 82, 2013–2017 CrossRef CAS PubMed.
- S. Murakami, N. Hayashi, T. Inomata, H. Kato, Y. Hitora and S. Tsukamoto, J. Nat. Med., 2020, 1–5 CAS.
- T. El-Elimat, H. A. Raja, C. S. Day, W.-L. Chen, S. M. Swanson and N. H. Oberlies, J. Nat. Prod., 2014, 77, 2088–2098 CrossRef CAS PubMed.
- N. D. Paguigan, H. A. Raja, C. S. Day and N. H. Oberlies, Phytochemistry, 2016, 126, 59–65 CrossRef CAS PubMed.
- N. D. Paguigan, J. Rivera-Chávez, J. J. Stempin, M. Augustinović, A. I. Noras, H. A. Raja, D. A. Todd, K. D. Triplett, C. Day and M. Figueroa, J. Nat. Prod., 2019, 82, 550–558 CrossRef CAS PubMed.
- S. Bertrand, O. Schumpp, N. Bohni, M. Monod, K. Gindro and J.-L. Wolfender, J. Nat. Prod., 2013, 76, 1157–1165 CrossRef CAS PubMed.
- L. Yao, L.-P. Zhu, X.-Y. Xu, L.-L. Tan, M. Sadilek, H. Fan, B. Hu, X.-T. Shen, J. Yang, B. Qiao and S. Yang, Sci. Rep., 2016, 6, 1–13 CrossRef CAS PubMed.
- H. Guo, N. B. Kreuzenbeck, S. Otani, M. Garcia-Altares, H.-M. Dahse, C. Weigel, D. K. Aanen, C. Hertweck, M. Poulsen and C. Beemelmanns, Org. Lett., 2016, 18, 3338–3341 CrossRef CAS PubMed.
- X.-T. Shen, X.-H. Mo, L.-P. Zhu, L.-L. Tan, F.-Y. Du, Q.-W. Wang, Y.-M. Zhou, X.-J. Yuan, B. Qiao and S. Yang, Appl. Environ. Microbiol., 2019, 85, e00293-19 CrossRef PubMed.
- M. H. Kossuga, A. G. Ferreira, L. D. Sette and R. G. Berlinck, Nat. Prod. Commun., 2013, 8, 1934578X1300800610 CrossRef.
- L.-H. Meng, Y. Liu, X.-M. Li, G.-M. Xu, N.-Y. Ji and B.-G. Wang, J. Nat. Prod., 2015, 78, 2301–2305 CrossRef CAS PubMed.
- K. Nonaka, T. Abe, M. Iwatsuki, M. Mori, T. Yamamoto, K. Shiomi, S. Ômura and R. Masuma, J. Antibiot., 2011, 64, 769–774 CrossRef CAS PubMed.
- K. Nonaka, M. Iwatsuki, S. Horiuchi, K. Shiomi, S. Ōmura and R. Masuma, J. Antibiot., 2015, 68, 573–578 CrossRef CAS PubMed.
- C. J. Harvey, M. Tang, U. Schlecht, J. Horecka, C. R. Fischer, H.-C. Lin, J. Li, B. Naughton, J. Cherry and M. Miranda, Sci. Adv., 2018, 4, eaar5459 CrossRef PubMed.
- N. Bohni, V. Hofstetter, K. Gindro, B. Buyck, O. Schumpp, S. Bertrand, M. Monod and J.-L. Wolfender, Molecules, 2016, 21, 370 CrossRef PubMed.
- N. P. Keller, Nat. Chem. Biol., 2015, 11, 671–677 CrossRef CAS PubMed.
- M. E. Mead, H. A. Raja, J. L. Steenwyk, S. L. Knowles, N. H. Oberlies and A. Rokas, Microbiol. Resour. Announce., 2019, 8, e00890-19 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1np00070e |
| This journal is © The Royal Society of Chemistry 2022 |