Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Ring systems in natural products: structural diversity, physicochemical properties, and coverage by synthetic compounds†
Ya
Chen
 *a,
Cara
Rosenkranz
b,
Steffen
Hirte
*a,
Cara
Rosenkranz
b,
Steffen
Hirte
 ac and
Johannes
Kirchmair
ac and
Johannes
Kirchmair
 a
a
aDepartment of Pharmaceutical Sciences, Division of Pharmaceutical Chemistry, Faculty of Life Sciences, University of Vienna, 1090 Vienna, Austria. E-mail: ya.chen@univie.ac.at
bCenter for Bioinformatics (ZBH), Universität Hamburg, 20146 Hamburg, Germany
cVienna Doctoral School of Pharmaceutical, Nutritional and Sport Sciences (PhaNuSpo), University of Vienna, 1090 Vienna, Austria
First published on 16th June 2022
Abstract
Covering: up to 2021
The structural core of most small-molecule drugs is formed by a ring system, often derived from natural products. However, despite the importance of natural product ring systems in bioactive small molecules, there is still a lack of a comprehensive overview and understanding of natural product ring systems and how their full potential can be harnessed in drug discovery and related fields. Herein, we present a comprehensive cheminformatic analysis of the structural and physicochemical properties of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 natural product ring systems, and the coverage of natural product ring systems by readily purchasable, synthetic compounds that are commonly explored in virtual screening and high-throughput screening. The analysis stands out by the use of comprehensive, curated data sets, the careful consideration of stereochemical information, and a robust analysis of the 3D molecular shape and electrostatic properties of ring systems. Among the key findings of this study are the facts that only about 2% of the ring systems observed in NPs are present in approved drugs but that approximately one in two NP ring systems are represented by ring systems with identical or related 3D shape and electrostatic properties in compounds that are typically used in (high-throughput) screening.
662 natural product ring systems, and the coverage of natural product ring systems by readily purchasable, synthetic compounds that are commonly explored in virtual screening and high-throughput screening. The analysis stands out by the use of comprehensive, curated data sets, the careful consideration of stereochemical information, and a robust analysis of the 3D molecular shape and electrostatic properties of ring systems. Among the key findings of this study are the facts that only about 2% of the ring systems observed in NPs are present in approved drugs but that approximately one in two NP ring systems are represented by ring systems with identical or related 3D shape and electrostatic properties in compounds that are typically used in (high-throughput) screening.
1. Introduction
Natural products (NPs) have a long record of use in traditional medicines. They also remain one of the most prolific sources of inspiration for modern small-molecule drug discovery.1,2 According to the latest survey of Newman and Cragg on the origin of approved drugs,3 68% of all small-molecule drugs approved between 1981 and 2019 are NPs, NP derivatives, NP mimics, or structures containing NP pharmacophores.NPs are, on average, heavier and more hydrophobic than synthetic compounds explored in the context of drug discovery.4–6 They also feature a higher content of oxygen atoms and a lower content of nitrogen atoms.4,5 Most outstanding, however, is their enormous structural diversity and, in part, high molecular complexity.5–7 In particular the stereochemical properties of NPs can pose fundamental challenges to organic chemistry.
Due to the difficulties involved in the sourcing and synthesis of NPs, the availability of materials for experimental testing is limited.8 In a recent survey of more than 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known NPs we found that only approximately 10% are readily obtainable from commercial and non-commercial sources.9 Experimental high-throughput screening (HTS) therefore rarely is an option in NPs research. Instead, a strategy which has been applied very successfully in the search for novel, bioactive NPs is virtual screening.10 The power of virtual screening methods lies in their capacity to cherry-pick the few, most promising compounds for sourcing and testing, thereby enabling researchers to optimise the use of the limited experimental resources. Examples include the identification of influenza neuraminidase inhibitors with docking11 and shape-based approaches,12 the discovery of bioactive triterpenes from Ganoderma lucidum as farnesoid X receptor agonists with a pharmacophore-based approach,13 and the identification of a truxillic acid derivative as a selective activator of the peroxisome proliferator-activated receptor gamma with machine learning methods.14
000 known NPs we found that only approximately 10% are readily obtainable from commercial and non-commercial sources.9 Experimental high-throughput screening (HTS) therefore rarely is an option in NPs research. Instead, a strategy which has been applied very successfully in the search for novel, bioactive NPs is virtual screening.10 The power of virtual screening methods lies in their capacity to cherry-pick the few, most promising compounds for sourcing and testing, thereby enabling researchers to optimise the use of the limited experimental resources. Examples include the identification of influenza neuraminidase inhibitors with docking11 and shape-based approaches,12 the discovery of bioactive triterpenes from Ganoderma lucidum as farnesoid X receptor agonists with a pharmacophore-based approach,13 and the identification of a truxillic acid derivative as a selective activator of the peroxisome proliferator-activated receptor gamma with machine learning methods.14
Virtual screening can substantially reduce the time and costs involved in the identification of bioactive NPs. However, just about 5% of the approved small-molecule drugs are unmodified NPs3 and only a fraction of them are sourced directly from organisms. (Partial) synthesis hence remains an essential component of the research and production of NPs-based drugs. Consequently, the synthetic accessibility of NPs and their derivatives is a major concern.
An established strategy to address the issue of synthetic accessibility is the design of simplified NP derivatives that can be delivered by total synthesis. However, these derivatives often lack key structural features for compounds to exhibit high biological activity.15 Modern synthetic strategies aim at covering the biologically relevant NP space.15 For example, in biology-oriented synthesis (BIOS), focused compound libraries are generated that are inspired by known bioactive NPs.16 Likewise, the pharmacophore-directed retrosynthesis strategy considers the pharmacophore (or “pharmacophoric” elements) of NPs for the elaboration of retrosynthetic routes.17
The vast majority of small-molecule drugs (>90%) contain at least one ring system, regardless of whether they are of natural or synthetic origin or not.18 These ring systems form the structural core of most molecules and determine their shape and conformational flexibility, as well as the orientation of substituents.19 In consequence, they are often essential to biological activity.20 In contrast to molecular scaffolds (or frameworks), which are most commonly defined as the union of a molecule's ring systems and linkers,21 and which represent a substantial subset of atoms of molecules, ring systems present a more fine-graded concept of molecular design (in particular fragment-based design) and compound optimization.22–24 However, despite the importance of ring systems to bioactivity, there is still a lack of a comprehensive overview and understanding of NP ring systems and how their full potential can be harnessed to boost the discovery and design of new drugs. One reason for this knowledge gap is the fact that most reported studies of the physicochemical properties of NPs are focused on complete molecular structures6,25–27 or molecular scaffolds28–32 rather than ring systems.
Among the few studies emphasising on NP ring systems is the pioneering work published by Lee and Schneider in 2001.33 They analysed the ring systems present in a set of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 495 NPs and found that only 17% of these ring systems are represented in a comprehensive collection of drugs. Ertl and Schuffenhauer5 extended this type of analysis to a processed set of 113
495 NPs and found that only 17% of these ring systems are represented in a comprehensive collection of drugs. Ertl and Schuffenhauer5 extended this type of analysis to a processed set of 113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 664 unique molecules extracted from the Dictionary of Natural Products (DNP).34 One of their key messages is that NP ring systems form a highly diverse, feature-rich pool of structural templates for library and compound design. Many of the NP ring systems are of moderate complexity and have benign properties that make them promising starting points for drug discovery.
664 unique molecules extracted from the Dictionary of Natural Products (DNP).34 One of their key messages is that NP ring systems form a highly diverse, feature-rich pool of structural templates for library and compound design. Many of the NP ring systems are of moderate complexity and have benign properties that make them promising starting points for drug discovery.
The existing studies provide valuable insights into the properties of NP ring systems but they are clearly limited with respect to the coverage of the known NP space (which is expanding quickly), and they largely disregard some key molecular properties related to stereochemistry, 3D shape and electrostatics. With this work, we aim to overcome these limitations by building on comprehensive, curated compound libraries and emphasising on 3D molecular properties. This allows us to develop a comprehensive and accurate picture of the diversity and physicochemical properties of NP ring systems, and to determine their coverage by synthetic compounds (SCs).
2. Results and discussion
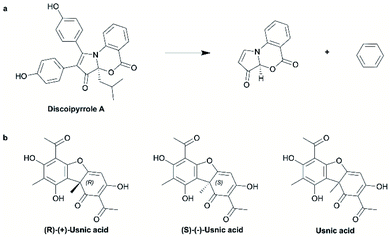
For the purpose of this analysis (and consistent with previous works5,22,35) we define a ring system as the graph composed of all atoms forming one or more rings (i.e., including fused and spiro rings), plus any proximate exocyclic atom connected to the ring atom via any bond other than a single bond (depicted in Fig. 1a; see the ESI Methods section and Table S1† for full detail on all methods employed for this analysis).For the representation of the NP chemical space we referred to the Collection of Open Natural Products (COCONUT) database.36 With over 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 listed compounds, the COCONUT database is the largest public resource of molecular information on NPs. Likewise, for the representation of the SC chemical space, we referred to the “in-stock subset” of the ZINC20 database,37 with more than 9 million readily obtainable compounds (which are typically used in virtual screening and HTS). The ZINC20 database is one of the largest resources of molecular information on purchasable compounds. Both data sets were processed in order to identify and remove any SCs contaminating the COCONUT database and any NPs included in the ZINC20 subset.
000 listed compounds, the COCONUT database is the largest public resource of molecular information on NPs. Likewise, for the representation of the SC chemical space, we referred to the “in-stock subset” of the ZINC20 database,37 with more than 9 million readily obtainable compounds (which are typically used in virtual screening and HTS). The ZINC20 database is one of the largest resources of molecular information on purchasable compounds. Both data sets were processed in order to identify and remove any SCs contaminating the COCONUT database and any NPs included in the ZINC20 subset.
In the context of NPs research, stereochemical information is particularly important. Nevertheless, cheminformatics studies often disregard stereochemical information because it is incomplete and sometimes even wrong.38 However, the question of whether or not to consider stereochemical information can mean the need to decide between a substantial loss of molecular structures (by disregarding any molecular structures with incomplete annotations) and reduced accuracy. In this analysis, we follow a two-pronged approach, depending on the data situation and relevance of stereochemistry to the analysed properties (as indicated at the appropriate locations in the text):
• Analysis disregarding stereochemical information: this approach prioritises data quantity and comprehensiveness over accurate representation. It is primarily used for analysing properties that are not influenced by configurations (e.g. the number of heavy atoms in a ring system).
• Analysis considering stereochemical information: this approach prioritises the correct configuration of tetrahedral atoms over data quantity and comprehensiveness. It is used whenever the stereochemical information adds value and the data situation permits.
We explain the workings of these approaches by the example of different representations of usnic acid depicted in Fig. 1b: the available chemical information indicates that the molecules depicted on the left and in the middle are structurally distinct. For the molecule depicted on the right no conclusion can be drawn as to whether or not it is identical with the molecule depicted on the left or the molecule depicted in the middle (due to a lack of stereochemical annotation). The analysis considering stereochemical information exactly follows this logic whereas the analysis disregarding stereochemical information considers all three structures depicted in Fig. 1b as identical.
2.1. Abundance and structural diversity of ring systems
When considering stereochemical information, the refined set of NPs and the refined set of SCs count 269![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226 and 8
226 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790
790![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 153 unique compounds, respectively (see Table 1 for details, including the respective numbers when disregarding stereochemical information). About 94% of the NPs (and 99% of the synthetic compounds) contain at least one ring. This percentage varies moderately across the NPs from different kingdoms (and marine life) and is 96% for the 63
153 unique compounds, respectively (see Table 1 for details, including the respective numbers when disregarding stereochemical information). About 94% of the NPs (and 99% of the synthetic compounds) contain at least one ring. This percentage varies moderately across the NPs from different kingdoms (and marine life) and is 96% for the 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 658 NPs from plants, 92% for the 15
658 NPs from plants, 92% for the 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 NPs from bacteria, 96% for the 17
662 NPs from bacteria, 96% for the 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 NPs from fungi, and 92% for the 35
937 NPs from fungi, and 92% for the 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 NPs from marine life. For comparison, the percentage of compounds with ring systems among the 2238 approved drugs listed in the “Approved” subset of DrugBank39 (a primary resource for chemical, biological and pharmacological information on drugs and drug candidates) is 89%.
340 NPs from marine life. For comparison, the percentage of compounds with ring systems among the 2238 approved drugs listed in the “Approved” subset of DrugBank39 (a primary resource for chemical, biological and pharmacological information on drugs and drug candidates) is 89%.
| Ring system sets | No. unique compounds | No. unique ring systems | No. compounds/no. ring systemsa | No. singletonsb | Fraction of singletonsc | No. macrocycles | Fraction of macrocyclesc | No. chiral ring systems | Fraction of chiral ring systemsc |
|---|---|---|---|---|---|---|---|---|---|
| a Average number of compounds containing a specific ring system. b Number of ring systems represented by only a single compound. c Among all ring systems. d A substantial number of NPs cannot be assigned to a specific subset due to a lack of annotations of the origin of NPs. | |||||||||
| When considering stereochemical information | |||||||||
| Natural productsd | 269![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226 226 |
38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 662 |
6.96 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 299 299 |
0.60 | 7597 | 0.20 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 063 063 |
0.83 |
| Plants | 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 658 658 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 217 217 |
5.21 | 7772 | 0.64 | 896 | 0.07 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 180 |
0.83 |
| Bacteria | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 662 |
3094 | 5.06 | 1760 | 0.57 | 1184 | 0.38 | 1869 | 0.60 |
| Fungi | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 937 937 |
4938 | 3.63 | 3112 | 0.63 | 525 | 0.11 | 3829 | 0.78 |
| Marine | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 340 |
7204 | 4.91 | 4200 | 0.58 | 1821 | 0.25 | 5552 | 0.77 |
| Synthetic compounds | 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790 790![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 153 153 |
53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 229 229 |
165.14 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
0.49 | 1636 | 0.03 | 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 053 053 |
0.62 |
| Approved drugs | 2238 | 602 | 3.72 | 357 | 0.59 | 52 | 0.09 | 186 | 0.31 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| When disregarding stereochemical information | |||||||||
| Natural productsd | 246![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 003 003 |
7.95 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 450 450 |
0.53 | 6619 | 0.21 | 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 347 347 |
0.79 |
| Plants | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 164 164 |
8418 | 6.43 | 4851 | 0.58 | 677 | 0.08 | 6388 | 0.76 |
| Bacteria | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 329 329 |
2694 | 5.32 | 1408 | 0.52 | 1003 | 0.37 | 1471 | 0.55 |
| Fungi | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 272 272 |
4126 | 3.94 | 2411 | 0.58 | 459 | 0.11 | 3016 | 0.73 |
| Marine | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 506 506 |
6195 | 5.25 | 3431 | 0.55 | 1575 | 0.25 | 4493 | 0.73 |
| Synthetic compounds | 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312 312![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 695 695 |
30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 265 265 |
207.41 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 350 |
0.51 | 1388 | 0.05 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 011 011 |
0.33 |
| Approved drugs | 2225 | 596 | 3.73 | 351 | 0.59 | 52 | 0.09 | 180 | 0.30 |
With a total of 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 658 compounds, the subset of plant NPs is approximately half the size of the subset of plant NPs included in the DNP (the most comprehensive, commercial database of its kind). The lower number of plant NPs results from the fact that some of the largest, non-commercial databases relevant to plant NPs (such as the Universal Natural Products Database (UNPD)40 with more than 220k NPs, and the Traditional Chinese Medicine (TCM) Database@Taiwan41 with more than 60k TCM-related NPs) lack annotations that would allow to identify them as such. For this reason the non-labelled NPs could not be considered in this specific part of the analysis.
658 compounds, the subset of plant NPs is approximately half the size of the subset of plant NPs included in the DNP (the most comprehensive, commercial database of its kind). The lower number of plant NPs results from the fact that some of the largest, non-commercial databases relevant to plant NPs (such as the Universal Natural Products Database (UNPD)40 with more than 220k NPs, and the Traditional Chinese Medicine (TCM) Database@Taiwan41 with more than 60k TCM-related NPs) lack annotations that would allow to identify them as such. For this reason the non-labelled NPs could not be considered in this specific part of the analysis.
The number of unique ring systems in NPs is 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662. Across the subset of NPs from plants (with 63
662. Across the subset of NPs from plants (with 63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 658 compounds the largest NP subset) we count 12
658 compounds the largest NP subset) we count 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 217 unique ring systems (see Table 1 for additional information). For comparison, the numbers of unique ring systems in SCs and approved drugs are 53
217 unique ring systems (see Table 1 for additional information). For comparison, the numbers of unique ring systems in SCs and approved drugs are 53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 229 and 602, respectively. This means that a unique ring system represents, on average, approximately 7 NPs, 165 SCs and 4 approved drugs. Considering the fact that the set of SCs is approximately 32-fold larger than the set of NPs, this corroborates the remarkable diversity of NPs.
229 and 602, respectively. This means that a unique ring system represents, on average, approximately 7 NPs, 165 SCs and 4 approved drugs. Considering the fact that the set of SCs is approximately 32-fold larger than the set of NPs, this corroborates the remarkable diversity of NPs.
When disregarding stereochemical information, the numbers of unique ring systems observed in NPs and SCs are reduced to 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 003 (−20%) and 30
003 (−20%) and 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 265 (−43%), respectively (Table 1). The substantially larger decrease in SCs is related to the fact that NPs result primarily from stereoselective biochemical synthesis while synthetic compounds are products primarily of non-selective chemical reactions (hence, the levels of stereochemical annotations differ accordingly).
265 (−43%), respectively (Table 1). The substantially larger decrease in SCs is related to the fact that NPs result primarily from stereoselective biochemical synthesis while synthetic compounds are products primarily of non-selective chemical reactions (hence, the levels of stereochemical annotations differ accordingly).
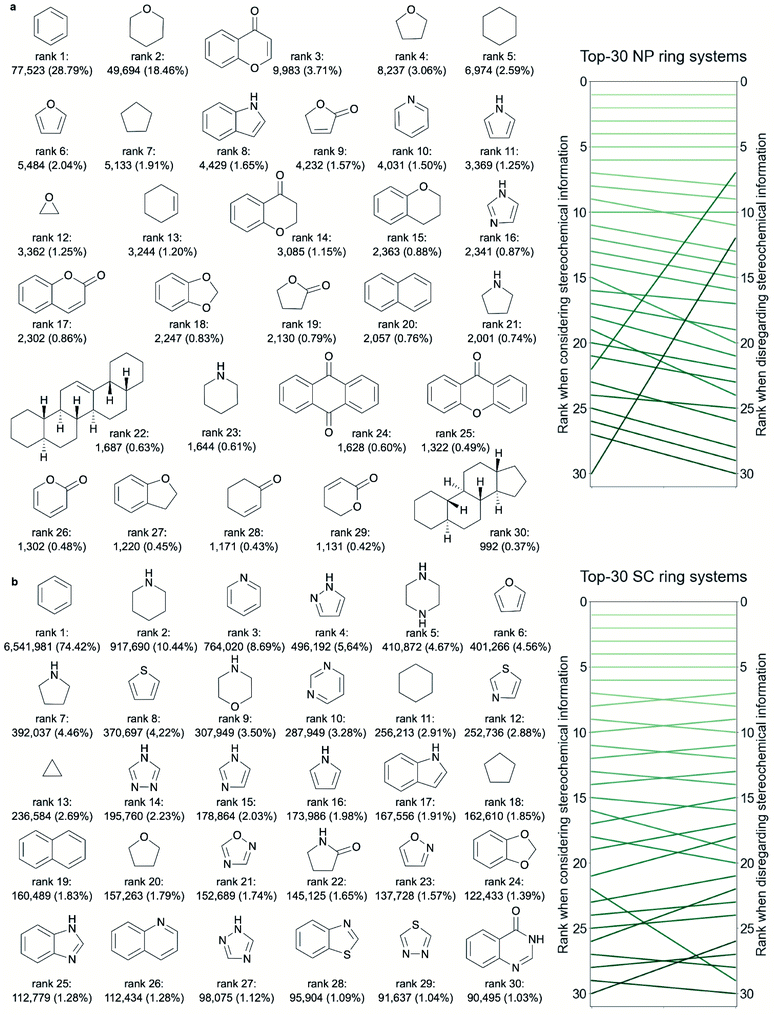
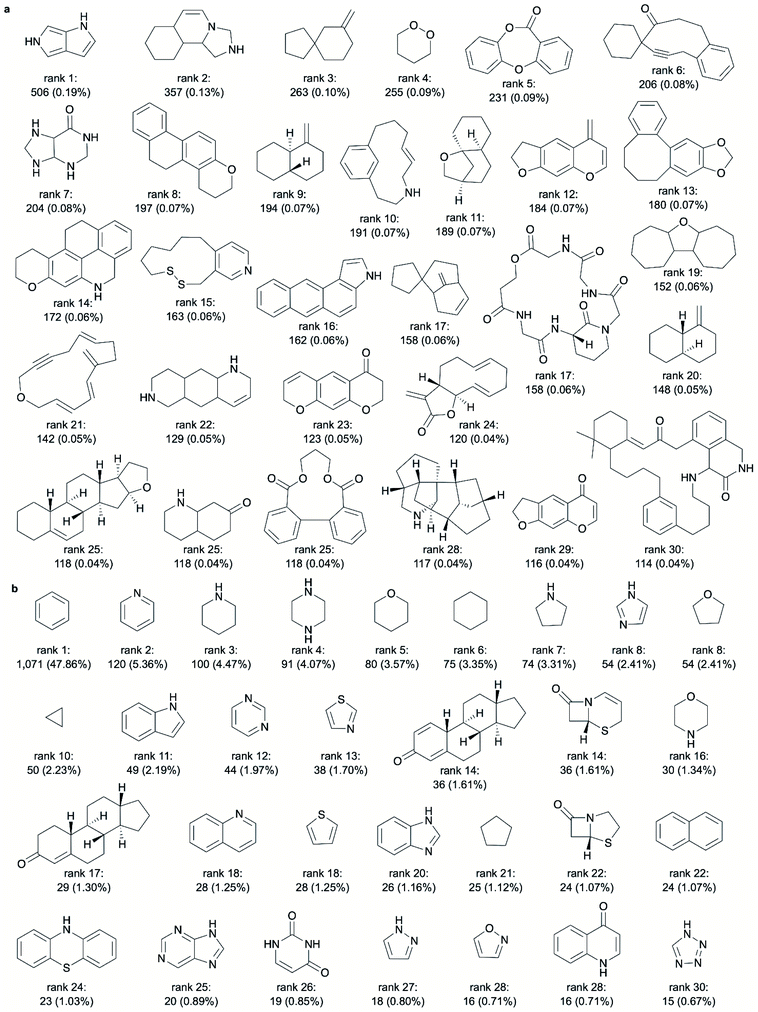
From rank 7 on, the lists of ring systems start to differ, depending on whether or not stereochemical information is considered. For example, ranked no. 7, when disregarding stereochemical information, is the scaffold common to pentacyclic triterpenes (such as ursolic acid), representing 2.05% of all NPs. In contrast, when considering stereochemical information, the first representation of a pentacyclic triterpene is placed on rank 22 only (representing 0.63% of all NPs). This discrepancy stems from the fact that for this scaffold (counting seven tetrahedral atoms) a total of 105 stereoisomers are recorded among the NPs (ESI Fig. S2† shows 30 of these stereoisomers).
Among the 30 most common NP ring systems, 17 ring systems (when considering stereochemical information; 15 when disregarding this information) contain at least one oxygen atom and 6 (in either approach) contain at least one nitrogen atom. Furthermore, in both approaches, 11 out of the 30 top-ranked ring systems (∼37%) are aromatic and further 5 ring systems contain at least one aromatic ring.
Because of the lower proportion of ring systems with chiral centres among SC ring systems compared to NP ring systems (Table 1 and Fig. 6d), the rankings of the most common ring systems are largely unaffected by the consideration or disregard of stereochemical information (Fig. 2b). For this reason we limit our discussion in the following paragraphs to the approach considering stereochemical information.
Like for the NP ring systems, rank 1 of the SC ring systems is held by benzene, although at a much higher percentage of occurrence in molecules (74% vs. 29%). Again, the decline in the frequencies of the individual ring systems is steep (ESI Fig. S1b†). An abundance of nitrogen-containing ring systems (70% vs. 20% among the 30 top-ranked NP ring systems, Table 2), mostly of aromatic character (70% vs. 53% of the ring systems contain at least one aromatic ring), is apparent. Besides benzene, twelve of the most common ring systems in SCs are also common across the 30 top-ranked NP ring systems: tetrahydrofuran, cyclohexane, furan, cyclopentane, 1H-indole, pyridine, 1H-pyrroline, imidazole, 1,3-benzodioxole, naphthalene, pyrrolidine and piperidine. In contrast, sulphur-containing ring systems are observed only among the 30 top-ranked ring systems from SCs and not from NPs.
| Features | NP ring systems | SC ring systems | ||||
|---|---|---|---|---|---|---|
| Top-30a | Top-100a | Alla | Top-30a | Top-100a | Alla | |
| a Considering stereochemical information (disregarding stereochemical information). | ||||||
| Only carbon atoms | 0.23 (0.30) | 0.28 (0.29) | 0.08 (0.07) | 0.17 (0.17) | 0.12 (0.11) | 0.08 (0.05) |
| Only carbon and oxygen atoms | 0.80 (0.80) | 0.75 (0.75) | 0.65 (0.62) | 0.27 (0.23) | 0.22 (0.21) | 0.31 (0.20) |
| At least one oxygen atom | 0.57 (0.50) | 0.55 (0.52) | 0.84 (0.85) | 0.27 (0.27) | 0.46 (0.44) | 0.68 (0.66) |
| Only carbon and nitrogen atoms | 0.43 (0.50) | 0.43 (0.46) | 0.15 (0.14) | 0.60 (0.60) | 0.44 (0.46) | 0.25 (0.24) |
| At least one nitrogen atom | 0.20 (0.20) | 0.24 (0.24) | 0.34 (0.36) | 0.70 (0.73) | 0.74 (0.75) | 0.64 (0.75) |
| At least one sulfur atom | 0.00 (0.00) | 0.02 (0.02) | 0.05 (0.06) | 0.13 (0.17) | 0.17 (0.17) | 0.18 (0.23) |
| Aromatic atoms | 0.53 (0.53) | 0.49 (0.41) | 0.37 (0.41) | 0.70 (0.70) | 0.64 (0.65) | 0.49 (0.62) |
| Heteroaromatic atoms | 0.30 (0.30) | 0.28 (0.23) | 0.15 (0.17) | 0.60 (0.60) | 0.50 (0.53) | 0.28 (0.41) |
| At least one stereo cent (atom tetrahedral) | 0.07 (0.13) | 0.13 (0.30) | 0.83 (0.79) | 0.00 (0.00) | 0.00 (0.00) | 0.62 (0.33) |
At least one non-aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond C bond |
0.17 (0.17) | 0.29 (0.38) | 0.64 (0.63) | 0.00 (0.03) | 0.13 (0.13) | 0.32 (0.26) |
For NPs and SCs alike, a high percentage of ring systems (approximately 50% and 60%, depending on whether or not stereochemical information is considered; see Table 1) are singletons, meaning that they are recorded in a single compound only. Among the approved drugs, the percentage of singletons is close to that calculated for NPs (59%, with and without the consideration of stereochemical information). After removal of singletons we visualised and analysed every 500th ring system, as well as the 30 most distinct ring systems, based on k-means clustering (ESI Fig. S3 and S4†). These additional analyses revealed a higher proportion of macrocyclic structures among the NP ring systems than among the SC ring systems (ESI Fig. S3 and S4,†Table 1 and Section 2.1.5). The fused polycyclic steroid- and terpene-derived ring systems observed in the SC data set are likely related to the semi-synthetic origin of some of the compounds.
Overall, the set of most common SC ring systems is clearly of lower complexity and diversity than the respective set of NP ring systems. Additional statistics on the composition of NP and SC ring systems are reported in Table 2.
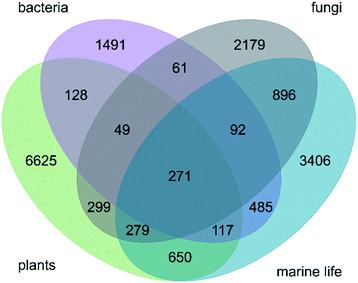
There are 271 ring systems that are represented in NPs from all three investigated kingdoms (plants, bacteria and fungi) and marine life (Fig. 3). Thirty-five of these ring systems are observed in at least 20 NPs of each of these subsets (ESI Fig. S5†). These 35 ring systems are structurally non-complex and include (i) benzene and common 5-membered and 6-membered aromatic and saturated heterocycles (e.g. tetrahydrofuran and furan, pyrrole and pyridine), (ii) bicyclic ring systems such as benzoquinone and indole, and (iii) the tricyclic ring systems anthraquinone and carbazoline. Overall, an accumulation of cyclic ketones is observed, which points to the importance of the polyketide pathway to secondary metabolism across species from different kingdoms. Furthermore, the presence of alkaloids highlights the relevance of amino acid metabolism. The proportions of ring systems exclusive to the individual kingdoms (and marine life) are high: 78.70% (6625) for plants, 55.35% (1491) for bacteria, 52.81% (2179) for fungi and 54.97% (3406) for marine species.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 003 unique ring systems present in NPs, only 2949 (fewer than 10%) are also present in SCs (stereochemical information disregarded). Many of the ring systems common to NPs and SCs (1329 out of 2949, or 45%) are singletons (meaning that they are linked to exactly one NP or SC). Only 815 ring systems out of the 2949 (28%; corresponding to 3% of the unique ring systems in NPs) occur in at least 5 NPs and 5 SCs (meaning that we can state with high confidence that these NP ring systems are covered by SCs). Among these 815 NP ring systems are all of the 30 most frequent NP ring systems.
003 unique ring systems present in NPs, only 2949 (fewer than 10%) are also present in SCs (stereochemical information disregarded). Many of the ring systems common to NPs and SCs (1329 out of 2949, or 45%) are singletons (meaning that they are linked to exactly one NP or SC). Only 815 ring systems out of the 2949 (28%; corresponding to 3% of the unique ring systems in NPs) occur in at least 5 NPs and 5 SCs (meaning that we can state with high confidence that these NP ring systems are covered by SCs). Among these 815 NP ring systems are all of the 30 most frequent NP ring systems.
The 30 most frequent ring systems observed exclusively in NPs are shown in Fig. 4a (stereochemistry considered). They represent 114 to 506 NPs each. A high degree of diversity is observed among these ring systems, although the ring systems ranked 9 and 20 differ only by their stereochemical configuration and those ranked 12 and 29 differ by only one atom.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 941 additional ring systems present in the known NPs (stereochemistry considered) that may be of relevance to drug discovery and are not yet harnessed. In other words, only about 2% of the ring systems observed in NPs (i.e. 721 out of 38
941 additional ring systems present in the known NPs (stereochemistry considered) that may be of relevance to drug discovery and are not yet harnessed. In other words, only about 2% of the ring systems observed in NPs (i.e. 721 out of 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 NP ring systems; stereochemistry considered) are represented in the approved drugs (note that the difference between the 721 and 426 mentioned ring systems results from the fact that, under the consideration of the available stereochemical information, multiple mappings between ring systems are possible in cases where the configuration of at least one tetrahedral atoms is not annotated). Fig. 4b reports the 30 most frequent ring systems present in approved drugs. All of these ring systems (and continuing down to rank 110 without exception) are known to be present in NPs (the ring system ranked 30 is a representative of 3 ring systems representing an identical number of NPs).
662 NP ring systems; stereochemistry considered) are represented in the approved drugs (note that the difference between the 721 and 426 mentioned ring systems results from the fact that, under the consideration of the available stereochemical information, multiple mappings between ring systems are possible in cases where the configuration of at least one tetrahedral atoms is not annotated). Fig. 4b reports the 30 most frequent ring systems present in approved drugs. All of these ring systems (and continuing down to rank 110 without exception) are known to be present in NPs (the ring system ranked 30 is a representative of 3 ring systems representing an identical number of NPs).
2.2. Physicochemical properties of ring systems
The ring systems present in NPs and SCs were analysed and compared (after deduplication) with regard to 14 key physicochemical properties such as molecular weight, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, the number of hydrogen bond donors/acceptors (see the caption of Fig. 5 for a complete list). Since the descriptors for these physicochemical properties are invariant with respect to the configuration of a molecule, stereochemical information was disregarded in this part of the study.
P, the number of hydrogen bond donors/acceptors (see the caption of Fig. 5 for a complete list). Since the descriptors for these physicochemical properties are invariant with respect to the configuration of a molecule, stereochemical information was disregarded in this part of the study.
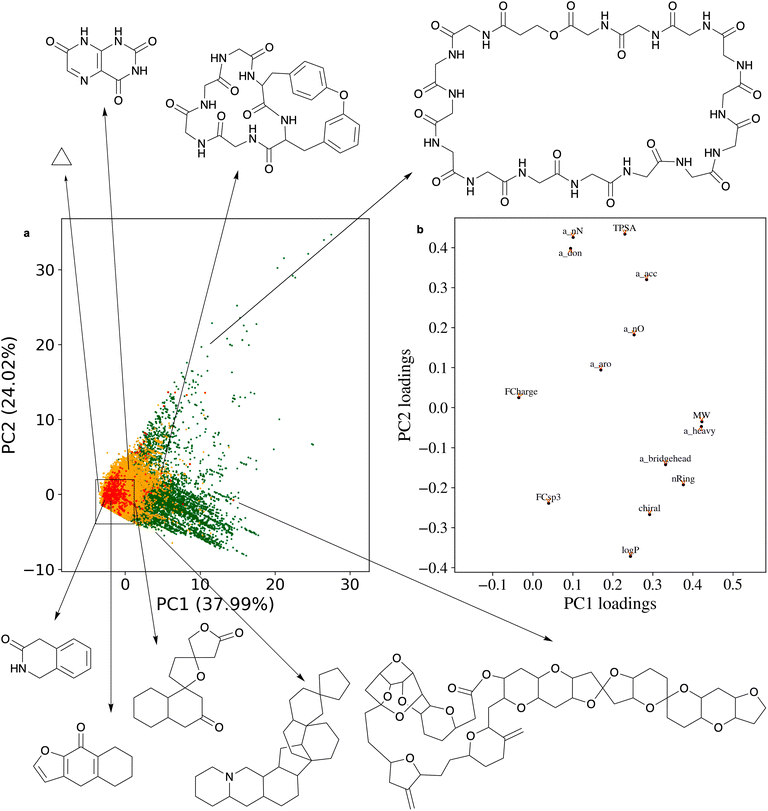
Principal component analysis (PCA) of the 14 physicochemical properties confirms the superior chemical diversity of the ring systems present in NPs in comparison to those present in SCs (Fig. 5; see the ESI† Method section for the exact protocol). PC1 is dominated by molecular weight and the number of heavy atoms: larger ring systems are located towards the right of the plot, and most of them originate from NPs. PC2 primarily describes the polarity of the ring systems: more polar ring systems are located towards the upper part of the plot. The NP ring systems populate a wider chemical space than those derived from SCs. The area most densely populated with NP and SC ring systems alike is also the one that is of primary relevance to small-molecule drug discovery. It envelopes all of the 30 most frequent ring systems observed in NPs.
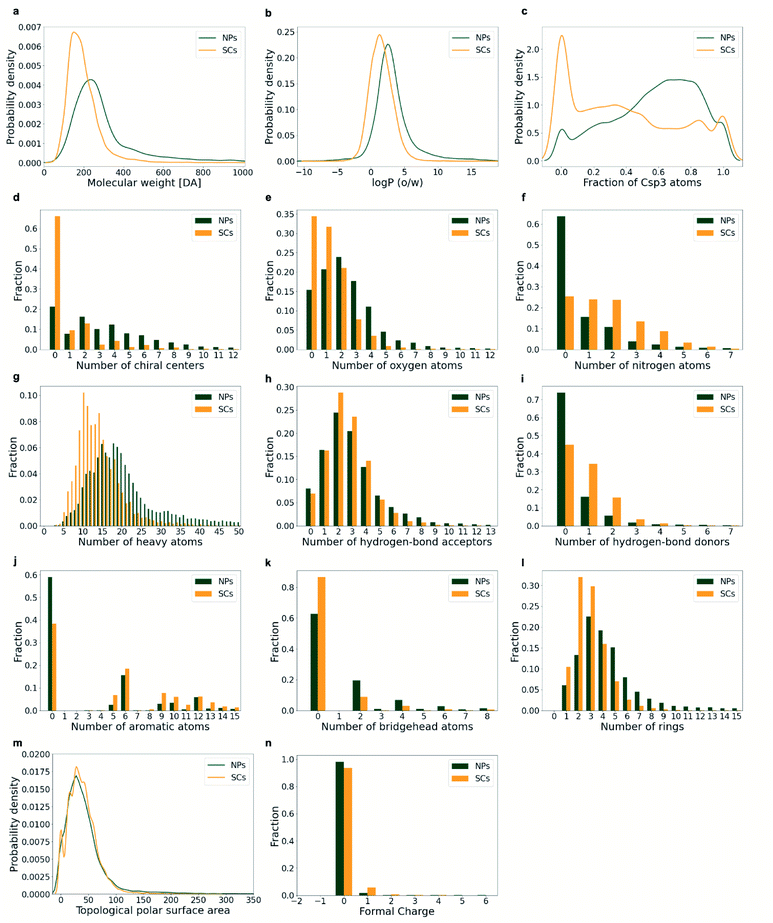
The diversity of the compounds and the trends observed by PCA are reflected by the distributions of the individual physicochemical properties (Fig. 6): the ring systems extracted from NPs are more diverse than those originating from SCs with respect to their size (represented by their molecular weight (MW) and the number of heavy atoms (a_heavy); Figs. 6a and g), the number of rings (Fig. 6l), and the number of oxygen atoms (Fig. 6e). In contrast, SCs show a wider distribution of the number of nitrogen atoms (Fig. 6f) and the hydrogen bond donors (Fig. 6i).
Rings are more frequent in NPs than they are in SCs: on average, NPs contain 4.65 rings whereas SCs contain only 2.98 (Fig. 6l). Moreover, NP ring systems are, on average, larger (heavier) than SC ring systems (MW 302 Da vs. 193 Da, Fig. 6a; a_heavy 22 vs. 14, Fig. 6g) and more lipophilic (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.26 vs. 1.57, Fig. 6b). NP ring systems are often also more complex with regard to their 3D molecular shape: they have a higher fraction of sp3 hybridised carbon atoms (0.57 vs. 0.39; Fig. 6c) and a higher number of bridgehead atoms (1.59 vs. 0.42; Fig. 6k). Related to these characteristics, NP ring systems are more likely to contain chiral tetrahedral atoms (76% in ring systems derived from plant NPs; 55% derived from bacterial NPs; 73% derived from fungi NPs; 73% derived from marine life NPs; Table 1; when disregarding stereochemical information) compared to both SCs (33%) and approved drugs (30%). Also, the average number of chiral centres in ring systems from NPs is 3.75 compared to 0.94 for ring systems from SCs (Fig. 6d).
P 3.26 vs. 1.57, Fig. 6b). NP ring systems are often also more complex with regard to their 3D molecular shape: they have a higher fraction of sp3 hybridised carbon atoms (0.57 vs. 0.39; Fig. 6c) and a higher number of bridgehead atoms (1.59 vs. 0.42; Fig. 6k). Related to these characteristics, NP ring systems are more likely to contain chiral tetrahedral atoms (76% in ring systems derived from plant NPs; 55% derived from bacterial NPs; 73% derived from fungi NPs; 73% derived from marine life NPs; Table 1; when disregarding stereochemical information) compared to both SCs (33%) and approved drugs (30%). Also, the average number of chiral centres in ring systems from NPs is 3.75 compared to 0.94 for ring systems from SCs (Fig. 6d).
NP ring systems further stand out by a higher average number of oxygen atoms (2.43 vs. 1.21; Fig. 6e), a lower average number of nitrogen atoms (0.87 vs. 1.74; Fig. 6f), and a lower degree of aromaticity (i.e. average number of aromatic ring atoms 4.42 vs. 6.03; Fig. 6j) compared to SC ring systems. With respect to the topological polar surface area (TPSA) and formal charges, the ring systems observed in NPs and SCs are comparable (Fig. 6m and n). Among the NP ring systems, 99.8% have a formal charge of 0 or 1. Among the SC ring systems, this percentage is 99.3.
In general, the trends observed for the property distributions among ring systems are consistent with those observed for the complete molecules.4,5 One exception is lipophilicity, which is, on average, higher for NP than for SC ring systems (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.26 vs. 1.57) but comparable for the complete molecules (log
P 3.26 vs. 1.57) but comparable for the complete molecules (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P 3.25 vs. 3.31; data from ref. 4).
P 3.25 vs. 3.31; data from ref. 4).
2.3. Three-dimensional shape and electrostatic properties of ring systems
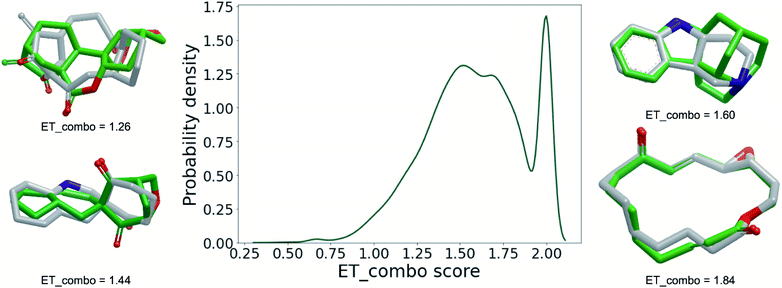
The interaction of small molecules with macromolecules is determined by the compatibility of the 3D molecular shapes and electrostatic potentials of the binding partners. Therefore, in the last part of this analysis, we employ powerful, established 3D approaches (ROCS47,48 and EON49) to ascertain to what extent synthetic organic chemistry covers the ring systems present in NPs. More specifically, we quantified the 3D molecular shape and electrostatic similarity of each NP ring system and its nearest neighbour in the data set of SC ring systems (only the 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 721 NPs ring systems with fully defined tetrahedral atoms were considered; SC ring systems were allowed to have undefined tetrahedral atoms because with respect to synthesis, enantiomerically pure compounds generally pose higher challenges than racemic mixtures; the configuration of all undefined tetrahedral atoms of SC ring systems was hence enumerated). The results of this approach are visualised in Fig. 7 as a density distribution plot derived from the maximum pairwise similarities of the NP and SC ring systems, quantified by the “ET_combo score” scoring function. ET_combo score ranges from 0 to 2 and puts equal weights on shape similarity and electrostatic similarity. A value of 2 indicates a perfect match of the 3D shape and electrostatics between a pair of structures. Accordingly the peak in the probability density distribution at an ET_combo score value close to 2.00 indicates that there are a substantial number of NP ring systems (∼15%) represented by identical or closely related ring systems in the SC data set.
721 NPs ring systems with fully defined tetrahedral atoms were considered; SC ring systems were allowed to have undefined tetrahedral atoms because with respect to synthesis, enantiomerically pure compounds generally pose higher challenges than racemic mixtures; the configuration of all undefined tetrahedral atoms of SC ring systems was hence enumerated). The results of this approach are visualised in Fig. 7 as a density distribution plot derived from the maximum pairwise similarities of the NP and SC ring systems, quantified by the “ET_combo score” scoring function. ET_combo score ranges from 0 to 2 and puts equal weights on shape similarity and electrostatic similarity. A value of 2 indicates a perfect match of the 3D shape and electrostatics between a pair of structures. Accordingly the peak in the probability density distribution at an ET_combo score value close to 2.00 indicates that there are a substantial number of NP ring systems (∼15%) represented by identical or closely related ring systems in the SC data set.
Approximately one out of two NP ring systems (∼13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) is matched by a ring system of the SC data set with ET_combo scores of at least 1.60, a threshold above which ring systems can be typically considered as structurally closely related or “covered” (Fig. 7; data for alternative ET_combo score thresholds is provided in ESI Table S2†). This means that roughly half of the recorded NP ring systems are accessible to synthetic organic chemistry. It also means, however, that there is another half, in other words a large number of NP ring systems, that is clearly still underexplored as potentially relevant structural templates for drug discovery.
500) is matched by a ring system of the SC data set with ET_combo scores of at least 1.60, a threshold above which ring systems can be typically considered as structurally closely related or “covered” (Fig. 7; data for alternative ET_combo score thresholds is provided in ESI Table S2†). This means that roughly half of the recorded NP ring systems are accessible to synthetic organic chemistry. It also means, however, that there is another half, in other words a large number of NP ring systems, that is clearly still underexplored as potentially relevant structural templates for drug discovery.
3. Conclusions and outlook
Ring systems form the core of most small-molecule drugs. They determine the molecular shape, flexibility and orientation of substituents and, hence, are key to the bioactivity and specificity of compounds. NPs are recognized in drug discovery as the richest source of chemical inspiration and much of their significance can be attributed to the vast diversity and feature-richness of NP ring systems.In this comprehensive analysis of the ring systems present in NPs we show that about one in two NP ring systems (∼13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) are represented by ring systems with identical or related 3D shape and electrostatic properties in readily obtainable, synthetic compounds (which are typically used for virtual screening and HTS). At the same time, only about 2% of the 38
500) are represented by ring systems with identical or related 3D shape and electrostatic properties in readily obtainable, synthetic compounds (which are typically used for virtual screening and HTS). At the same time, only about 2% of the 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 662 unique ring systems observed in NPs (stereochemistry considered) are components of approved drugs, leaving a huge pool of potentially relevant ring systems yet to be exploited in small-molecule drug discovery. One particular area of interest are macrocycles, which are represented by significant numbers among NPs (7597 unique macrocycles, representing 20% of the total number of unique ring systems; stereochemistry considered) but not among SCs (1636 unique macrocycles, representing 3%).
662 unique ring systems observed in NPs (stereochemistry considered) are components of approved drugs, leaving a huge pool of potentially relevant ring systems yet to be exploited in small-molecule drug discovery. One particular area of interest are macrocycles, which are represented by significant numbers among NPs (7597 unique macrocycles, representing 20% of the total number of unique ring systems; stereochemistry considered) but not among SCs (1636 unique macrocycles, representing 3%).
While it will take time for synthetic and biotechnological approaches to advance the exploration of NP ring systems, the full wealth of information on the existing ring systems can be exploited already today, by rapidly advancing artificial intelligence (AI) technologies for compound design.7,50–53 These technologies utilise various types of biological, chemical and structural information to train models that can generate NP-inspired compounds which have a high likelihood of being synthetically accessible and active on the target(s) of interest. The most promising way forward certainly is the integration of these in silico approaches with the advanced experimental techniques that are at our disposal today. The synergy generated from this effort will boost NPs research and small-molecule drug discovery.
4. Conflicts of interest
There are no conflicts to declare.5. Acknowledgements
We thank Judith M. Rollinger from the University of Vienna for fruitful discussions, the anonymous Reviewers for their diligent work and valuable comments and suggestions, and OpenEye for providing a free academic license for their software collection.6. References
- C. F. Stratton, D. J. Newman and D. S. Tan, Bioorg. Med. Chem. Lett., 2015, 25, 4802–4807 CrossRef CAS PubMed.
- A. L. Harvey, R. Edrada-Ebel and R. J. Quinn, Nat. Rev. Drug Discovery, 2015, 14, 111–129 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef CAS PubMed.
- Y. Chen, C. Stork, S. Hirte and J. Kirchmair, Biomolecules, 2019, 9, 43 CrossRef PubMed.
- P. Ertl and A. Schuffenhauer, in Natural Compounds as Drugs, ed. F. Petersen and R. Amstutz, Birkhäuser, Basel, 1st edn, 2008, vol. 66, pp. 217–235 Search PubMed.
- Y. Chen, M. G. de Lomana, N.-O. Friedrich and J. Kirchmair, J. Chem. Inf. Model., 2018, 58, 1518–1532 CrossRef CAS PubMed.
- T. Rodrigues, Org. Biomol. Chem., 2017, 15, 9275–9282 RSC.
- A. G. Atanasov, B. Waltenberger, E.-M. Pferschy-Wenzig, T. Linder, C. Wawrosch, P. Uhrin, V. Temml, L. Wang, S. Schwaiger, E. H. Heiss, J. M. Rollinger, D. Schuster, J. M. Breuss, V. Bochkov, M. D. Mihovilovic, B. Kopp, R. Bauer, V. M. Dirsch and H. Stuppner, Biotechnol. Adv., 2015, 33, 1582–1614 CrossRef CAS PubMed.
- Y. Chen, C. de Bruyn Kops and J. Kirchmair, J. Chem. Inf. Model., 2017, 57, 2099–2111 CrossRef CAS PubMed.
- Y. Chen and J. Kirchmair, Mol. Inform., 2020, 39, e2000171 CrossRef PubMed.
- N. K. K. Ikram, J. D. Durrant, M. Muchtaridi, A. S. Zalaludin, N. Purwitasari, N. Mohamed, A. S. A. Rahim, C. K. Lam, Y. M. Normi, N. A. Rahman, R. E. Amaro and H. A. Wahab, J. Chem. Inf. Model., 2015, 55, 308–316 CrossRef CAS PubMed.
- J. Kirchmair, J. M. Rollinger, K. R. Liedl, N. Seidel, A. Krumbholz and M. Schmidtke, Future Med. Chem., 2011, 3, 437–450 CrossRef CAS PubMed.
- U. Grienke, J. Mihály-Bison, D. Schuster, T. Afonyushkin, M. Binder, S.-H. Guan, C.-R. Cheng, G. Wolber, H. Stuppner, D.-A. Guo, V. N. Bochkov and J. M. Rollinger, Bioorg. Med. Chem., 2011, 19, 6779–6791 CrossRef CAS PubMed.
- M. Rupp, T. Schroeter, R. Steri, H. Zettl, E. Proschak, K. Hansen, O. Rau, O. Schwarz, L. Müller-Kuhrt, M. Schubert-Zsilavecz, K.-R. Müller and G. Schneider, ChemMedChem, 2010, 5, 191–194 CrossRef CAS PubMed.
- N. J. Truax and D. Romo, Nat. Prod. Rep., 2020, 37, 1436–1453 RSC.
- S. Basu, B. Ellinger, S. Rizzo, C. Deraeve, M. Schürmann, H. Preut, H.-D. Arndt and H. Waldmann, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6805–6810 CrossRef CAS PubMed.
- M. E. Abbasov, R. Alvariño, C. M. Chaheine, E. Alonso, J. A. Sánchez, M. L. Conner, A. Alfonso, M. Jaspars, L. M. Botana and D. Romo, Nat. Chem., 2019, 11, 342–350 CrossRef CAS PubMed.
- R. D. Taylor, M. MacCoss and A. D. G. Lawson, J. Med. Chem., 2014, 57, 5845–5859 CrossRef CAS PubMed.
- P. Ertl, S. Jelfs, J. Mühlbacher, A. Schuffenhauer and P. Selzer, J. Med. Chem., 2006, 49, 4568–4573 CrossRef CAS PubMed.
- P. Ertl, J. Chem. Inf. Model., 2022, 62(9), 2164–2170 CrossRef CAS PubMed.
- G. W. Bemis and M. A. Murcko, J. Med. Chem., 1996, 39, 2887–2893 CrossRef CAS PubMed.
- M. Aldeghi, S. Malhotra, D. L. Selwood and A. W. E. Chan, Chem. Biol. Drug Des., 2014, 83, 450–461 CrossRef CAS PubMed.
- G. Karageorgis, D. J. Foley, L. Laraia and H. Waldmann, Nat. Chem., 2020, 12, 227–235 CrossRef CAS PubMed.
- A. Thakkar, N. Selmi, J.-L. Reymond, O. Engkvist and E. J. Bjerrum, J. Med. Chem., 2020, 63, 8791–8808 CrossRef CAS PubMed.
- H. Chen, O. Engkvist, N. Blomberg and J. Li, MedChemComm, 2012, 3, 312–321 RSC.
- T. El-Elimat, X. Zhang, D. Jarjoura, F. J. Moy, J. Orjala, A. D. Kinghorn, C. J. Pearce and N. H. Oberlies, ACS Med. Chem. Lett., 2012, 3, 645–649 CrossRef CAS PubMed.
- N. Singh, R. Guha, M. A. Giulianotti, C. Pinilla, R. A. Houghten and J. L. Medina-Franco, J. Chem. Inf. Model., 2009, 49, 1010–1024 CrossRef CAS PubMed.
- K. Grabowski, K.-H. Baringhaus and G. Schneider, Nat. Prod. Rep., 2008, 25, 892–904 RSC.
- D. Reker, in Progress in the Chemistry of Organic Natural Products, ed. A. D. Kinghorn, H. Falk, S. Gibbons, J. ichi Kobayashi, Y. Asakawa and J.-K. Liu, Springer, Cham, 1st edn, 2019, vol. 110, pp. 143–175 Search PubMed.
- M. A. Koch, A. Schuffenhauer, M. Scheck, S. Wetzel, M. Casaulta, A. Odermatt, P. Ertl and H. Waldmann, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 17272–17277 CrossRef CAS PubMed.
- P. Ertl and T. Schuhmann, Mol. Inform., 2020, 39, e2000017 CrossRef PubMed.
- A. L. Chávez-Hernández, N. Sánchez-Cruz and J. L. Medina-Franco, Mol. Inform., 2020, 39, e2000050 CrossRef PubMed.
- M. L. Lee and G. Schneider, J. Comb. Chem., 2001, 3, 284–289 CrossRef CAS PubMed.
- Dictionary of Natural Products, https://dnp.chemnetbase.com, accessed 21 December 2021 Search PubMed.
- J. Hert, J. J. Irwin, C. Laggner, M. J. Keiser and B. K. Shoichet, Nat. Chem. Biol., 2009, 5, 479–483 CrossRef CAS PubMed.
- M. Sorokina, P. Merseburger, K. Rajan, M. A. Yirik and C. Steinbeck, J. Cheminform., 2021, 13, 2 CrossRef PubMed.
- J. J. Irwin, K. G. Tang, J. Young, C. Dandarchuluun, B. R. Wong, M. Khurelbaatar, Y. S. Moroz, J. Mayfield and R. A. Sayle, J. Chem. Inf. Model., 2020, 60, 6065–6073 CrossRef CAS PubMed.
- C. Kramer, M. Podewitz, P. Ertl and K. R. Liedl, Planta Med., 2015, 81, 459–466 CrossRef CAS PubMed.
- D. S. Wishart, Y. D. Feunang, A. C. Guo, E. J. Lo, A. Marcu, J. R. Grant, T. Sajed, D. Johnson, C. Li, Z. Sayeeda, N. Assempour, I. Iynkkaran, Y. Liu, A. Maciejewski, N. Gale, A. Wilson, L. Chin, R. Cummings, D. Le, A. Pon, C. Knox and M. Wilson, Nucleic Acids Res., 2018, 46, D1074–D1082 CrossRef CAS PubMed.
- J. Gu, Y. Gui, L. Chen, G. Yuan, H.-Z. Lu and X. Xu, PLoS One, 2013, 8, e62839 CrossRef CAS PubMed.
- C. Y.-C. Chen, PLoS One, 2011, 6, e15939 CrossRef CAS PubMed.
- P. Ertl, Bioorg. Med. Chem., 2022, 54, 116562 CrossRef CAS PubMed.
- P. Ermert, Chimia, 2017, 71, 678–702 CrossRef CAS PubMed.
- E. Marsault and M. L. Peterson, J. Med. Chem., 2011, 54, 1961–2004 CrossRef CAS PubMed.
- P. G. Dougherty, Z. Qian and D. Pei, Biochem. J., 2017, 474, 1109–1125 CrossRef CAS PubMed.
- S. Stone, D. J. Newman, S. L. Colletti and D. S. Tan, Nat. Prod. Rep., 2022, 39, 20 RSC.
- P. C. D. Hawkins, A. G. Skillman and A. Nicholls, J. Med. Chem., 2007, 50, 74–82 CrossRef CAS PubMed.
- ROCS 3.4.1.0: OpenEye Scientific Software, Santa Fe, NM, https://www.eyesopen.com, accessed 17 September 2021 Search PubMed.
- EON 2.3.4.0: OpenEye Scientific Software, Santa Fe, NM, https://www.eyesopen.com, accessed 17 September 2021 Search PubMed.
- D. Merk, F. Grisoni, L. Friedrich and G. Schneider, Commun. Chem., 2018, 68 Search PubMed.
- F. I. Saldívar-González, V. D. Aldas-Bulos, J. L. Medina-Franco and F. Plisson, Chem. Sci., 2022, 13, 1526 RSC.
- L. Friedrich, T. Rodrigues, C. S. Neuhaus, P. Schneider and G. Schneider, Angew. Chem., Int. Ed., 2016, 55, 6789–6792 CrossRef CAS PubMed.
- T. Rodrigues, D. Reker, P. Schneider and G. Schneider, Nat. Chem., 2016, 8, 531–541 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Details on the computational methods and how to access the source code; Table S1, reporting the full names of the data sources of the COCONUT database; Table S2, reporting the numbers and percentages of NP ring systems that are matched by a ring system in the SC data set at different cutoffs of the ET_combo score. Fig. S1, showing the occurrences (in percent) of the 30 most frequent ring systems in (a) NPs and (b) SCs when considering stereochemical information; Fig. S2, showing the 30 most frequent stereoisomers of the pentacyclic triterpene ranked no. 7 of the NP ring system set when disregarding stereochemical information; Fig. S3, showing every 500th (a) NP ring system and (b) SC ring system (stereochemical information considered; singletons omitted); Fig. S4, showing the 30 most diverse (a) NP ring systems and (b) SC ring systems (identified by a k-means clustering method implemented using scikit-learn and RDKit that takes Morgan2 fingerprints with a length of 1024 bits as input; singletons removed prior to clustering); Fig. S5, showing the 35 ring systems recorded for at least 20 times in each of the subsets of NPs from plants, bacteria, fungi and marine life. See https://doi.org/10.1039/d2np00001f |
| This journal is © The Royal Society of Chemistry 2022 |