A colloidal gold immunochromatographic strip assay for the rapid detection of Shigella in milk and meat products
Beijun
Ang
ab,
Xinxin
Xu
ab,
Liqiang
Liu
ab,
Liguang
Xu
 *ab,
Hua
Kuang
*ab,
Hua
Kuang
 ab and
Chuanlai
Xu
ab and
Chuanlai
Xu
 *ab
*ab
aState Key Laboratory of Food Science and Technology, Jiangnan University, Wuxi, People's Republic of China
bInternational Joint Research Laboratory for Biointerface and Biodetection, and School of Food Science and Technology, Jiangnan University, Wuxi, People's Republic of China. E-mail: xlg@jiangnan.edu.cn; xcl@jiangnan.edu.cn
First published on 22nd November 2021
Abstract
Shigella is the most common foodborne pathogen and can cause bacillary dysentery. Therefore, it is necessary to detect Shigella rapidly and accurately, but it is difficult for traditional methods to meet these requirements. In this study, monoclonal antibodies (mAbs) were prepared using a recombinant invasive plasmid antigen C (IpaC) protein, which is specifically located on the outer membrane of Shigella. Based on our mAbs 2G11 and 5A11, a colloidal gold lateral flow immunochromatographic (LFIA) test strip method for the detection of several serotypes of Shigella was produced. The sensitivity of the LFIA strip was 1 × 105 CFU mL−1 for four groups, and seven serotypes of Shigella. The applicability of the strip was tested using spiked milk and meat samples and the results showed that Shigella could be detected in these matrices.
1. Introduction
Shigella is a type of Gram-negative short bacilli, with several natural hosts including humans.1 The strain is mainly found in dairy products, meat, and vegetables. Ingesting food contaminated with Shigella can cause clinical symptoms such as diarrhea, fever, vomiting and dehydration, and even death.2,3Shigella mainly causes intestinal inflammation by infecting human colonic epithelial cells4 and the infection triggers innate immune responses in the host which the bacterium combats by translocating into the host cell cytosol via type III secretion system bacterial effector proteins that interfere with host processes.5,6 According to the antigenic structure of Shigella, it can be divided into four groups, and 48 serotypes: Shigella dysenteriae (S. dysenteriae, group A) has 12 serotypes, among which the eighth serotype are divided into three subtypes. Shigella flexneri (S. flexneri, group B) has 15 serotypes, Shigella boydii (S. boydii, group C) has 18 serotypes and Shigella sonnei (S. sonnei, group D) has one serotype.7,8 They all exhibit different degrees of pathogenicity in different populations and approximately 60% of worldwide bacterially-induced diarrhea is caused by this bacterial infection, representing around 160 million Shigella infections every year, and a death toll of 1.1 million.9,10 Therefore, a rapid, sensitive, simple and efficient method for the detection of Shigella in food is urgently needed.Invasive plasmid antigen (Ipa) B, C and D are three types of secretory proteins encoded by the virulent plasmids of Shigella. These proteins represent important factors affecting processes such as cell invasion and serve as important immunogens.11 Some studies have shown that they are the main protein antigens responsible for the serum immune response during infection in humans and monkeys, among which, the peptides IpaB and IpaC have particular immunogenicity.12 Moreover, studies have also shown that IpaC is located on the outer membrane of bacteria,12,13 and is therefore exposed to recognition by antibodies raised against the bacteria.
With the development of detection technologies for Shigella, these methods can be divided into traditional culture methods, immunological methods, molecular biological methods and biosensor methods.14–16 Results from traditional biochemical culture methods are accurate and do not require complex equipment, but the process is tedious, time-consuming and laborious, and cannot meet the needs required for rapid on the spot detection. Molecular biological methods can be divided into variable temperature amplification techniques such as the polymerase chain reaction (PCR) and its derivatives17 or isothermal amplification techniques such as rolling circle amplification (RCA),18 or loop-mediated isothermal amplification (LAMP).19 These methods are highly sensitive, specific, simple and rapid, and are widely used in the detection of foodborne pathogens. However, these methods require specific instrumentation, which is expensive and needs skilled operators.
The classical immunological analytical methods have advantages of high sensitivity, good specificity, short cycle time, are simple to operate and are low cost. Therefore, they are widely used in the detection of toxic and harmful substances in many foods.20 Among these methods, the lateral flow immunoassay (LFIA) is widely used for the detection of bacteria due to its rapid and convenient procedure, as results can be obtained within 20 min with the naked eye, and thus is suitable for onsite analysis.21 In this study, we expressed the recombinant protein IpaC of Shigella to produce mAbs and then constructed a LFIA strip using the mAbs to recognize several serotypes of Shigella. Our strip was rapid and sensitive and was able to detect Shigella in both milk and meat samples.
2. Materials and methods
2.1. Bacterial strains and culture conditions
Fifteen bacterial strains were used in this study, included seven different serotypes of Shigella and seven other bacteria. The bacterial strains were as follows: S. dysenteriae (CICC 23829), S. dysenteriae (CICC 10983), S. flexneri (CICC 10865), S. flexneri (CICC 21534), S. soonei (CICC 21535), S. soonei (CICC 21679), S. boydii (CICC 21680), Escherichia coli (E. coli) O157:H7 (CICC 21530), Salmonella typhimurium (S. typhimurium) (ATCC 13311), Enterobacter sakazakii (E. sakazakii) (ATCC 29544), Yersinia enterocolitica (Y. enterocolitica) (CICC 21565), Listeria monocytogenes (L. monocytogenes) (ATCC 19115), Staphylococcus aureus (S. aureus) (ATCC 29213), and Vibrio parahaemolyticus (V. parahaemolyticus) (CICC 21617).Seven strains of Shigella, E. coli, S. typhimurium, and E. sakazakii were grown in Luria-Bertani (LB) broth at 37 °C. Y. enterocolitica was grown in LB broth at 26 °C. L. monocytogenes, S. aureus were grown in TSB at 37 °C. V. parahaemolyticus was grown in LB broth containing 3.5% NaCl at 30 °C. All the bacteria were cultured in an incubator and shaken at 180 rpm overnight.
2.2. Reagents and apparatus
Tryptone, yeast extract, TSB and agar powder were purchased from Beijing Solarbio Science & Tech Co., Ltd (Beijing, China). the pET-32a(+)-IpaC gene was synthesized by Talen-Bio Scientific Co., Ltd (Wuxi, China). BL21 (DE3) competent cells, Super Optimal broth with catabolite repression (SOC) medium, and Blue Plus® II Protein Markers (14–120 kDa) were purchased from Beijing TransGen Biotech Co., Ltd (Beijing, China). Isopropyl-β-D-thiogalactoside (IPTG), kanamycin sulfate, and imidazole were obtained from Shanghai Sangon Biotech Co., Ltd (Shanghai, China). Freund's complete adjuvant (FCA) and Freund's incomplete adjuvant (FIA), bovine serum albumin (BSA), horseradish peroxidase (HRP), HRP-conjugated goat anti-mouse IgG and 3,3′,5,5′-tetramethylbenzidine (TMB) were obtained from Sigma Aldrich (Shanghai, China). Sodium periodate, ethylene glycol and sodium cyanoborohydride were purchased from J & K Scientific Co., Ltd (Shanghai, China). All other solvents and chemicals were purchased from the National Pharmaceutical Group Chemical Reagent Co., Ltd (Shanghai, China) at analytical purity. All solutions were prepared using ultrapure water (Milli-Q) ultrapure system, Millipore Co., Ltd (Bedford, MA, USA).BALB/c mice were purchased from Henan Skbex Biotechnology Co., Ltd (Anyang, China). SP2/0 myeloma cells were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Syringe filters (0.2 μm) were obtained from Merck Millipore Ltd (Billerica, USA). Ni-NTA 6FF (Pre-Packed Gravity Column) was obtained from Shanghai Sangon Biotech Co., Ltd (Shanghai, China). A constant temperature shaker (THZ-420) was purchased from Shanghai Jinghong Experimental Equipment Co., Ltd (Shanghai, China), an ultrasonic cell disruptor was purchased from Ningbo Scientz Biotech Co., Ltd (Ningbo, China). Finally, a NanoDrop One Microvolume UV-Vis spectrophotometer was purchased from Thermo Fisher Scientific. (Waltham, MA, USA).
The materials for the LFIA strip, including polyvinylchloride (PVC) backing cards, sample pads (glass fiber membrane, CB-SB08) and absorption pads (SX18) were purchased from JieYi Biotechnology Co., Ltd (Shanghai, China). Nitrocellulose (NC) membranes were purchased from Whatman-Xinhua Filter Paper Co., Ltd (Hangzhou, China). Finally, a CM 4000 guillotine cutting module was obtained from Shanghai Kinbio Tech Co., Ltd (Shanghai, China).
2.3. Expression and purification of recombinant IpaC protein
The full-length gene of IpaC was cloned into pET-32a(+) vector at BamHI and SacI restriction sites with a thioredoxin (Trx) tag and a 6× His tag at the N-terminal for downstream purification, as shown in Fig. 1. The constructed recombinant plasmid was expressed as a fusion protein using prokaryotic expression methods. The pET-32a(+)-IpaC was transformed into E. coli strain BL21 (DE3) competent cells as described previously with slight modification.22 Plasmid (80 ng) was mixed with 50 μL of competent cells and bathed in ice for 30 min. Then, the mixture was heat-shocked at 42 °C for 45 s and then placed back on ice for 2 min. Next, 500 μL of SOC medium was added and the treated cells were placed at 37 °C to recover for 1 h. The 100 μL mixture was then spread onto solid Luria Bertani broth (LB) media containing kanamycin and grown overnight at 37 °C. After this, a monoclonal transformant was selected to pre-amplify in 5 mL LB broth (Kan) on a shaker at 37 °C, and 220 rpm for 12 h, then inoculated into 200 mL LB medium (Kan 1%), and shaken at 37 °C, and 220 rpm. When the optical density (OD 600) reached 0.6–0.8, protein expression was induced with the addition of 0.2 mM IPTG, at 16 °C, and 220 rpm for 20 h. The induced bacterial solution was then centrifuged at 5000 × g for 20 min, and then resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 8.0) and sonicated on ice for 10 min (sonicated for 3 s with 5 s intervals at 35 W). The soluble fraction of the lysate was then separated by ultracentrifugation (20 min at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 4 °C).23
000 × g, 4 °C).23
The resulting clear lysate supernatant containing soluble IpaC protein was then passed through 0.2 μm syringe filters and purified by Ni-NTA affinity chromatography24,25 using a prepacked and equilibrated (with lysis buffer) gravity column. The lysate was loaded onto the column and only the target protein with the 6× His tag was bound to the resin. Then the protein was gradient eluted by washing the resin with elution buffer (50 mM NaH2PO4, 300 mM NaCl, with 10 mM, 20 mM, 50 mM, 250 mM, 500 mM of increasing content of imidazole, pH 8.0). Finally, the gradient eluent was collected and analyzed by SDS-PAGE and target proteins were dialyzed in 0.01 M PBS for 48 h and their final concentrations were determined using the NanoDrop One Microvolume UV-Vis spectrophotometer.
2.4. Immunization of mice and production of monoclonal antibodies
All animal procedures in this study were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Jiangnan University and approved by the Animal Ethics Committee of Jiangnan University. The immunization process was performed as previously described.26 Five female BALB/c mice (6–8 weeks-old) were immunized by subcutaneous injection with 100 μg of purified IpaC emulsified with an equivalent volume of FCA. Then, 18–21 days later, a similar second immunization was given using FIA instead of FCA. Then, booster injections were performed twice at 3 week intervals with half the dose of the original two immunizations emulsified in FIA.For the mAb production, 1 week after the last injection, an optimal spleen was select, using screened mouse serum by indirect ELISA. The IpaC protein was coated on flat-bottomed 96-well plates at concentrations of 1.0 μg mL−1, 0.3 μg mL−1 and 0.1 μg mL−1, respectively and the coating concentrations of the bacterial strains used for testing specificity were 1.0 × 108 CFU mL−1, 3.0 × 107 CFU mL−1 and 1.0 × 107 CFU mL−1, respectively.27 Splenocytes were collected from the mouse with the highest serum titer specific for IpaC protein and Shigella, and then fused with SP2/0 myeloma cells and cultured as described previously.28 Supernatants from the hybridomas were first screened by indirect ELISA on 96-well plates coated with IpaC (0.3 μg mL−1). Then, positive wells were selected using OD450s higher than 1.4, then, the positive wells were tested with both Shigella and other bacteria to select the wells specific for the different strains. The selected hybridomas were finally cloned four times using the limited dilution method, and at each time the screening procedure was performed in the same way. After four rounds of cloning, hybridoma cells were injected into the abdominal cavity of paraffin primed mice to produce antibodies. Ten mAbs were purified from mouse ascites by the caprylic acid-ammonium sulfate precipitation method and the antibody concentrations were determined using a Nanodrop One (Thermo Fisher). And the affinity constant, which means the the equilibrium dissociation constant between the antibody and its antigen, were measured using non-competitive ELISA to work out the KD value. As previously reported,29 briefly, the coating antigen (seven strains of Shigella) was serially diluted three times from 3 × 107 to 3× 106 CFU mL−1 and the antibody was also three times diluted from 1.0 μg mL−1 to 0.1 μg mL−1. Then an S-shaped curve was drawn using logarithms of antibody concentrations as the abscissa and the corresponding absorbance as the ordinate. The KD was calculated according to the following eqn (1):
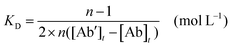 | (1) |
2.5. Construction of the LFIA strip
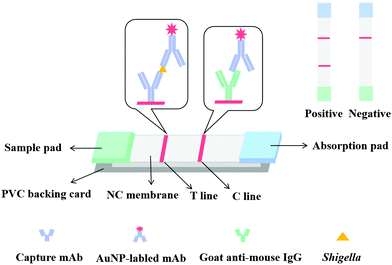
An LFIA strip was constructed as previously reported30 as a sandwich format (Fig. 2). LFIA strips consist of a sample pad, an NC membrane with a test line (T line) and a control line (C line), an absorbent pad, and a polyvinylchloride (PVC) backing card. The T and C lines were formed by spraying anti-IpaC capture antibody and goat anti-mouse IgG antibody. The NC membrane was then dried at 37 °C for 6 h and then pasted onto the backing card, and at the two ends of the membrane, the sample pad and absorption pad were overlapped. This was then cut into 4.0 mm wide by 5 cm long strips and stored in a desiccator.2.6. Detection of Shigella in food samples
Chicken, pork, and milk purchased from a local supermarket were used as test food samples in this study. They were confirmed as Shigella negative using the test procedure according to GB 4789.2–2016. Meat samples were ground and minced but the milk samples were used unprocessed.Shigella strains were grown in LB overnight and centrifuged at 5000 × g for 20 min, then the bacterial precipitates were resuspended in PBS and diluted to the target inoculation number. Samples were spiked with different amounts of Shigella and tested with the strips. For milk samples, 1 mL of Shigella at dilutions of 1.0 × 109, 2.0 × 108, 4.0 × 107, 8.0 × 106, 1.6 × 106, and 3.2 × 105 CFU mL−1 were separately added to 9 mL of the milk solution, and the target inoculation numbers were 1.0 × 108, 2.0 × 107, 4.0 × 106, 8.0 × 105, 1.6 × 105, and 3.2 × 104 CFU mL−1. For the chicken and pork samples, 10 g of meat paste was placed into a sterile homogenizer and then 1 mL of the diluted bacteria solution was added. The samples were homogenized with 90 mL of PBS in a stomacher blender for 1 min and then the solution was poured into a sterile centrifuge tube and centrifuged at 4500 × g for 5 min, and the pellet was resuspended in 10 mL of PBS and tested by the strip and noninoculated samples were used as negative controls. For all three types of samples, after the inoculation procedure, 150 μL of sample solution was reacted with 50 μL of GNP-labeled mAb for 5 min, then the test strip was inserted into the mixture, a few minutes later, the presence/absence of a red band on the T line was recorded.
3. Results and discussion
3.1. Expression and purification of recombinant IpaC protein
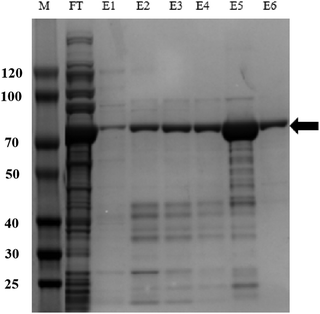
IpaC was expressed as a fusion protein with a Trx tag and a 6× His tag in E. coli BL21 (DE3) cells. Protein was purified by IMAC and verified using SDS-PAGE and the results revealed that the recombinant fusion protein was expressed as a band of approximately 70 kDa (Fig. 1), which is consistent with the predicted molecular mass of IpaC (about 45 kDa) plus the Trx and His tags (approximately 25 kDa). The recombinant protein was used directly for mouse immunization and screening of mAbs produced by the hybridoma cell lines.3.2. Principle of the LFIA strip
The principle of the LFIA strip was based on a double antibody sandwich, where150 μL of sample solution was first mixed with 50 μL of AuNP-labeled detection antibody, then the test strip was inserted into the mixture and the solution was allowed to move onto the sample pad and migrate to the NC membrane, and then finally arrived at the absorption pad by capillary action. If the sample solution contained target bacteria, they will react with the AuNP-labeled mAb first, then the capture antibody fixed on the T line of the NC membrane and then react with the other recognition site of the antigen to form a AuNP-labeled detection antibody-antigen-capture antibody sandwich complex. Excess AuNP-labeled mAb was captured by the goat anti-mouse IgG antibody at the C line, eventually forming a red T and C line. The color intensity of the T line was proportional to the bacteria concentration. In the absence of target bacteria (negative sample), the AuNP-labeled mAb will just bind to the goat anti-mouse IgG antibody at the C line, thus only a red C line was observed. Importantly a red C line should always be observed regardless of whether the sample was negative or positive, otherwise the strip was deemed to be invalid.3.3. Optimization of the strip
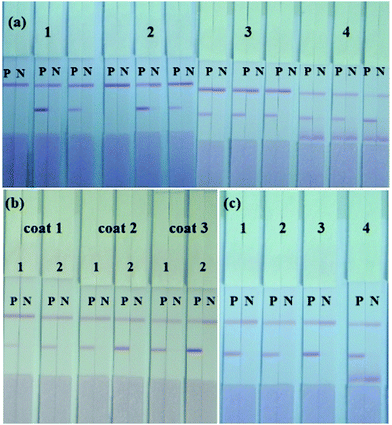
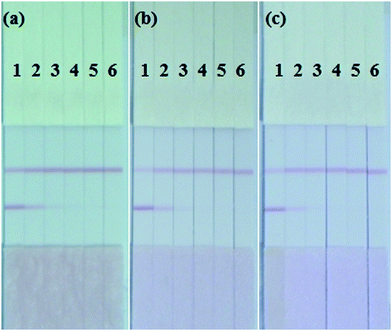
The sensitivity of the ICA strip was greatly affected by the different combinations of capture antibody and detection antibody. An optimal combination should exhibit a strong red T line when testing positive samples and have the weakest background when testing negative samples. Based on this principle, four mAbs (2G11, 3H10, 5A11 and 8A1) with high affinity constant for S. dysenteriae (CICC 23829) were used, while the KD value of them were 5.96 × 109, 5.62 × 109, 6.12 × 109, and 5.37 × 109 L mol−1 as calculated. Each of these four mAbs was used as immobilized mAbs and AuNP-labeled mAbs respectively, making 1.0 × 106 CFU mL−1 of S. dysenteriae (CICC 23829) as positive control and PBS as negative control. As shown in Fig. 3(a), the immobilized mAb 5A11 on the T line and the AuNP-labeled mAb 2G11 exhibited the best results.The concentration of immobilized mAb and AuNP-labeled mAb was also optimized to generate the best color intensity on the T line. Concentrations of 0.3 mg mL−1, 0.5 mg mL−1 and 0.8 mg mL−1 for mAb 5A11 immobilized on the T line were compared using 1.0 × 106 CFU mL−1 of S. dysenteriae (CICC 23829) and PBS. The color intensity of the T line increased with an increase in the concentration of mAb, 0.8 mg mL−1 of mAb 5A11 gave a deeper color on the T line (Fig. 3(b)). Similarly, the concentration of AuNP-labeled antibody was optimized and 10 μg mL−1 of 2G11 showed a better color effect when compared to a concentration of 5 μg mL−1 (Fig. 3(b)).
The type of resuspension buffer was also an important optimization requirement influencing the migration of the AuNP-labeled mAb and the hydrophilicity of the NC membrane.31 Four different resuspension buffers, BSA, 5% Tween-20, 5% Triton and ON-870 were analyzed by comparing the color intensity of the T and C lines using mAbs 2G11 and 5A11. As shown in Fig. 3(c), based on the chromogenic results, ON-870 generated the clearest color when compared to the other buffers.
3.4. Sensitivity and specificity of the LFIA strip
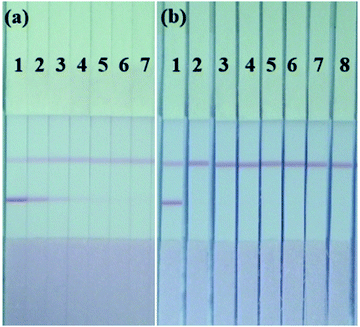
The sensitivity of the LFIA strip was determined based on the optimized conditions above. Shigella dysenteriae (CICC 23829) was diluted to different concentrations 1 × 106, 5 × 105, 2 × 105, 1 × 105, 5 × 104, 2 × 104, and 1 × 104 CFU mL−1, and each was tested on the strip. In this assay the lowest concentration of the target analyte that was able to generate a visually weaker red T line when compared to a negative sample, represented the visual limit of detection (vLOD)32 and the cut-off value was defined as the threshold concentration of the target analyte that generated a colorless T line.33 Results presented in Fig. 4(a), show that the vLOD of the LFIA strip was 1 × 105 CFU mL−1 while the cut-off value was 5 × 104 CFU mL−1 when Shigella dysenteriae (CICC 23829) was tested. There was no significant difference between the strip test results for the four groups, or the seven different serotypes of Shigella (seven aforementioned strains were tested, but the results are not shown), but the strip needed further validation for the remaining serotypes of the strains.To evaluate the specificity of the strip, seven other bacterial strains at a concentration of 1.0 × 107 CFU mL−1 were tested and the results are displayed in Fig. 4(b). No cross-reactivity was observed with E. coli O157: H7 (CICC 21530), S. typhimurium (ATCC 13311), E. sakazakii (ATCC 29544), Y. enterocolitica (CICC 21565), L. monocytogenes (ATCC 19115), S. aureus (ATCC 29213), or V. parahaemolyticus (CICC 21617) at this concentration, but for E. coli O157: H7 (CICC 21530), E. sakazakii (ATCC 29544) and S. typhimurium(ATCC 13311), the red T line on the strip appeared gradually with an increase in biomass concentration from 1.0 × 107 CFU mL−1. Like Shigella, the above three bacteria are all typical enterobacteria with high homology, even at the level of the outer membrane proteins.
3.5. Shigella detection in milk and meat samples
To determine the detection performance of the LFIA strip, milk, chicken and pork samples spiked with different concentrations of Shigella were analyzed. Blank samples were confirmed with no bacteria using the plate count method. The milk, chicken and pork samples spiked with different concentrations of Shigella (CICC 23829) were separately tested and are shown in Fig. 5(a)–(c). In the milk sample, a weaker T line was generated at 4 × 106 CFU mL−1, and the cut-off value was therefore 8 × 105 CFU mL−1. However, for chicken and pork samples, due to the influence of their complex matrices, the cut-off value for both was reduced to 4 × 106 CFU mL−1. The above test results indicated that the LFIA strip construction could be used to detect Shigella in common food samples, but its sensitivity varied with the matrix conditions of the samples.Compared with traditional culture methods, which requires 2–3 days to attain a result, this strip method proved to be a much more rapid method for the detection of Shigella. The entire testing process could be accomplished within 10 h including bacterial enrichment and strip test assay, and the results were clearly visible as a T line simply using the naked eye.
4. Conclusions
In this study, the method used to prepare monoclonal antibodies against pathogenic bacteria was based on their outer membrane proteins. The sensitivity of our strip was 1 × 105 CFU mL−1 and was capable of detecting multiple serotypes of Shigella in common foods such as milk and meat. However, although IpaC is an outer membrane protein of Shigella, it is highly homologous in many Enterobacteria, like E. coli and Salmonella. Therefore, during our mouse and antibody screening procedure, we avoided those mAbs with high cross-reactivity, but some cross-reactivity towards many other bacteria still existed in the final strip test. For future developments, we will analyze the specific epitopes of the outer membrane proteins of Shigella and prepare mAbs with higher specificity through the selection and optimization of the immunogenic protein, enabling better materials for the development of immunoassay products for the detection of Shigella.Conflicts of interest
All the authors declare no competing financial interest.Acknowledgements
This work is financially supported by National Key R&D Program (2018YFC1602906), Suzhou Science and Technology Committee Program (SS202038), and Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_1841).References
- C. J. Kuehl, J. D. D’Gama, A. R. Warr and M. K. Waldor, An Oral Inoculation Infant Rabbit Model for Shigella Infection, mBio, 2020, 11(1), e03105-19 CrossRef.
- R. Gharpure, Z. A. Marsh, D. M. Tack, S. A. Collier, J. Strysko, L. Ray, D. C. Payne and A. G. Garcia-Williams, Disparities in Incidence and Severity of Shigella Infections Among Children-Foodborne Diseases Active Surveillance Network (FoodNet), 2009–2018, J. Pediatr. Infect. Dis. Soc., 2021, 782–788 CrossRef.
- D. P. M. Sethuvel, S. Anandan, J. S. Michael, D. Murugan, A. Neeravi, V. P. Verghese, K. Walia and B. Veeraraghavan, Virulence gene profiles of Shigella species isolated from stool specimens in India: its association with clinical manifestation and antimicrobial resistance, Pathog. Global Health, 2019, 113(4), 173–179 CrossRef CAS PubMed.
- E. Mattock and A. J. Blocker, How Do the Virulence Factors of Shigella Work Together to Cause Disease?, Front. Cell. Infect. Microbiol., 2017, 7, 64 CrossRef.
- K. A. Miller, A. C. Garza-Mayers, Y. Leung and M. B. Goldberg, Identification of interactions among host and bacterial proteins and evaluation of their role early during Shigella flexneri infection, Microbiology, 2018, 164(4), 540–550 CrossRef CAS.
- S. Tachiyama, R. Skaar, Y. Chang, B. L. Carroll, M. Muthuramalingam, S. K. Whittier, M. L. Barta, W. L. Picking, J. Liu and W. D. Picking, Composition and Biophysical Properties of the Sorting Platform Pods in the Shigella Type III Secretion System, Front. Cell. Infect. Microbiol., 2021, 11, 682635 CrossRef CAS.
- L. Zhao, Y. Xiong, D. Meng, J. Guo, Y. Li, L. Liang, R. Han, Y. Wang, X. Guo, R. Wang, L. Zhang, L. Gao and J. Wang, An 11 year study of shigellosis and Shigella species in Taiyuan, China: Active surveillance, epidemic characteristics, and molecular serotyping, J. Infect. Public Health, 2017, 10(6), 794–798 CrossRef.
- D. P. Muthuirulandi Sethuvel, N. K. Devanga Ragupathi, S. Anandan and B. Veeraraghavan, Update on: Shigella new serogroups/serotypes and their antimicrobial resistance, Lett. Appl. Microbiol., 2017, 64(1), 8–18 CrossRef CAS.
- D. Hosangadi, P. G. Smith, D. C. Kaslow, B. K. Giersing and E. Who, Shigella Vaccine Consultation Expert, G., WHO consultation on ETEC and Shigella burden of disease, Geneva, 6-7th April 2017: Meeting report, Vaccine, 2019, 37(50), 7381–7390 CrossRef PubMed.
- R. Walker, R. W. Kaminski, C. Porter, R. K. M. Choy, J. A. White, J. M. Fleckenstein, F. Cassels and L. Bourgeois, Vaccines for Protecting Infants from Bacterial Causes of Diarrheal Disease, Microorganisms, 2021, 9(7), 1382 CrossRef PubMed.
- M. Karimi-Yazdi, Z. Ghalavand, M. Shabani, H. Houri, M. Sadredinamin, M. Taheri and G. Eslami, High Rates of Antimicrobial Resistance and Virulence Gene Distribution Among Shigella spp. Isolated from Pediatric Patients in Tehran, Iran, Infect. Drug Resist., 2020, 13, 485–492 CrossRef CAS PubMed.
- M. L. Barta, S. Tachiyama, M. Muthuramalingam, O. Arizmendi, C. E. Villanueva, K. X. Ramyar, B. V. Geisbrecht, S. Lovell, K. P. Battaile, W. L. Picking and W. D. Picking, Using disruptive insertional mutagenesis to identify the in situ structure-function landscape of the Shigella translocator protein IpaB, Protein Sci., 2018, 27(8), 1392–1406 CrossRef CAS PubMed.
- J. K. Duncan-Lowey, A. L. Wiscovitch, T. E. Wood, M. B. Goldberg and B. C. Russo, Shigella flexneri Disruption of Cellular Tension Promotes Intercellular Spread, Cell Rep., 2020, 33(8), 108409 CrossRef CAS PubMed.
- J. Feng, Q. Shen, J. Wu, Z. Dai and Y. Wang, Naked-eyes detection of Shigella flexneri in food samples based on a novel gold nanoparticle-based colorimetric aptasensor, Food Control., 2019, 98, 333–341 CrossRef CAS.
- N. T. Nguyen, S. Y. Kim, J. H. Wee, Y. H. Kim and J. Min, Cell blocking: an enhancement of foodborne pathogen detection by fluorescent signals of recombinant yeasts, Anal. Biochem., 2020, 606, 113856 CrossRef CAS PubMed.
- P. B. Pavlinac, J. A. Platts-Mills, K. D. Tickell, J. Liu, J. Juma, F. Kabir, J. Nkeze, C. Okoi, D. J. Operario, J. Uddin, S. Ahmed, P. L. Alonso, M. Antonio, S. M. Becker, R. F. Breiman, A. S. G. Faruque, B. Fields, J. Gratz, R. Haque, A. Hossain, M. J. Hossain, S. Jarju, F. Qamar, N. T. Iqbal, B. Kwambana, I. Mandomando, T. L. McMurry, C. Ochieng, J. B. Ochieng, M. Ochieng, C. Onyango, S. Panchalingam, A. Kalam, F. Aziz, S. Qureshi, T. Ramamurthy, J. H. Roberts, D. Saha, S. O. Sow, S. E. Stroup, D. Sur, B. Tamboura, M. Taniuchi, S. M. Tennant, A. Roose, D. Toema, Y. Wu, A. Zaidi, J. P. Nataro, M. M. Levine, E. R. Houpt and K. L. Kotloff, The Clinical Presentation of Culture-positive and Culture-negative, Quantitative Polymerase Chain Reaction (qPCR)-Attributable Shigellosis in the Global Enteric Multicenter Study and Derivation of a Shigella Severity Score: Implications for Pediatric Shigella Vaccine Trials, Clin. Infect. Dis., 2021, 73(3), e569–e579 CrossRef.
- J. Yang, N. Zhang, J. Lv, P. Zhu, X. Pan, J. Hu, W. Wu, S. Li and H. Li, Comparing the performance of conventional PCR, RTQ-PCR, and droplet digital PCR assays in detection of Shigella, Mol. Cell. Probes, 2020, 51, 101531 CrossRef CAS PubMed.
- X. Xu, Y. Su, Y. Zhang, X. Wang, H. Tian, X. Ma, H. Chu and W. Xu, Novel rolling circle amplification biosensors for food-borne microorganism detection, TrAC, Trends Anal. Chem., 2021, 141 CAS.
- L. Zhang, Q. Wei, Q. Han, Q. Chen, W. Tai, J. Zhang, Y. Song and X. Xia, Detection of Shigella in Milk and Clinical Samples by Magnetic Immunocaptured-Loop-Mediated Isothermal Amplification Assay, Front. Microbiol., 2018, 9, 94 CrossRef PubMed.
- Y. Li, G. Jin, L. Liu, J. Xiao and H. Kuang, Fast determination of citreoviridin residues in rice using a monoclonal antibody-based immunochromatographic strip assay, Food Agric. Immunol., 2020, 31(1), 893–906 CrossRef CAS.
- M. L. Yahaya, N. D. Zakaria, R. Noordin and K. A. Razak, The Effect of Nitrocellulose Membrane Pore Size of Lateral Flow Immunoassay on Sensitivity for Detection of Shigella sp. in Milk Sample, Mater. Today: Proc., 2019, 17, 878–883 CAS.
- D. Madi-Moussa, F. Coucheney and D. Drider, Expression of five class II bacteriocins with activity against Escherichia coli in Lacticaseibacillus paracasei CNCM I-5369, and in a heterologous host, Biotechnol. Rep., 2021, 30, e00632 CrossRef.
- A. Li, K. Sun, J. Wang, S. Wang, X. Zhao, R. Liu and Y. Lu, Recombinant expression, purification and characterization of human soluble tumor necrosis factor receptor 2, Protein Expression Purif., 2021, 182, 105857 CrossRef CAS PubMed.
- N. N. Mohanty, N. Chacko, S. K. Biswas, K. Chand, A. B. Pandey, B. Mondal, D. Hemadri and S. B. Shivachandra, Production of recombinant non-structural protein-3 hydrophobic domain deletion (NS3DeltaHD) protein of bluetongue virus from prokaryotic expression system as an efficient diagnostic reagent, Biologicals, 2016, 44(5), 352–359 CrossRef CAS.
- T. Djukic, M. Mladenovic, D. Stanic-Vucinic, J. Radosavljevic, K. Smiljanic, L. Sabljic, M. Devic, D. Cujic, T. Vasovic, A. Simovic, M. Radomirovic and T. Cirkovic Velickovic, Expression, purification and immunological characterization of recombinant nucleocapsid protein fragment from SARS-CoV-2, Virology, 2021, 557, 15–22 CrossRef CAS PubMed.
- Q. Fu, Z. Wei, Y. Chen, J. Xie, X. Zhang, T. He and X. Chen, Development of monoclonal antibody against IgT of a perciform fish, large yellow croaker (Larimichthys crocea) and characterization of IgT(+) B cells, Dev. Comp. Immunol., 2021, 119, 104027 CrossRef CAS.
- L. Zeng, L. Guo, Z. Wang, X. Xu, H. Ding, S. Song, L. Xu, H. Kuang and C. Xu, Gold nanoparticle-based immunochromatographic assay for detection Pseudomonas aeruginosa in water and food samples, Food Chem. X, 2021, 9, 100117 CrossRef CAS PubMed.
- L. Ye, X. Wu, L. Xu, Q. Zheng and H. Kuang, Preparation of an anti-thiamethoxam monoclonal antibody for development of an indirect competitive enzyme-linked immunosorbent assay and a colloidal gold immunoassay, Food Agric. Immunol., 2018, 29(1), 1173–1183 CrossRef CAS.
- Y. Li, L. Liu, H. Kuang and C. Xu, Preparing monoclonal antibodies and developing immunochromatographic strips for paraquat determination in water, Food Chem., 2020, 311, 125897 CrossRef CAS PubMed.
- X. Lei, X. Xu, L. Liu, H. Kuang, L. Xu and C. Hao, Immunochromatographic test strip for the rapid detection of tricaine in fish samples, Food Agric. Immunol., 2020, 31(1), 687–699 CrossRef CAS.
- Z. Wang, J. Zhang, L. Liu, X. Wu, H. Kuang, C. Xu and L. Xu, A colorimetric paper-based sensor for toltrazuril and its metabolites in feed, chicken, and egg samples, Food Chem., 2019, 276, 707–713 CrossRef CAS.
- Z. Chen, K. Zhou, W. Ha, P. Chen, H. Fu, Y. Shen, Y. Sun and Z. Xu, Development of a low-cost, simple, fast and quantitative lateral-flow immunochromatographic assay (ICA) strip for melatonin in health foods, Food Agric. Immunol., 2019, 30(1), 497–509 CrossRef CAS.
- L. Zeng, W. Jiang, L. Liu, S. Song and H. Kuang, Development of ic-ELISA and lateral-flow immunochromatographic strip for detection of vitamin B2 in an energy drink and vitamin tablets, Food Agric. Immunol., 2017, 29(1), 121–132 CrossRef.
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2022 |