Open Access Article
Open Access ArticleModerate molecular recognitions on ZnO m-plane and their selective capture/release of bio-related phosphoric acids†
Eisuke
Kanao
 *ab,
Katsuya
Nakano
c,
Ryoma
Kamei
d,
Takuro
Hosomi
*ab,
Katsuya
Nakano
c,
Ryoma
Kamei
d,
Takuro
Hosomi
 de,
Yasushi
Ishihama
ab,
Jun
Adachi
ab,
Takuya
Kubo
de,
Yasushi
Ishihama
ab,
Jun
Adachi
ab,
Takuya
Kubo
 *c,
Koji
Otsuka
*c,
Koji
Otsuka
 c and
Takeshi
Yanagida
d
c and
Takeshi
Yanagida
d
aGraduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: kanao.eisuke.7s@kyoto-u.ac.jp; Fax: +81-75-753-4601; Tel: +81-75-753-4565
bNational Institutes of Bio Medical Innovation, Health and Nutrition, Ibaraki, Osaka 567-0085, Japan
cDepartment of Material Chemistry, Graduate School of Engineering, Kyoto University, Katsura, Nishikyo-ku, Kyoto 615-8510, Japan. E-mail: kubo.takuya.6c@kyoto-u.ac.jp; Fax: +81-75-383-2450; Tel: +81-75-383-2448
dDepartment of Applied Chemistry, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8654, Japan
ePrecursory Research for Embryonic Science and Technology (PRESTO), Japan Science and Technology Agency (JST), 4-1-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
First published on 17th February 2022
Abstract
Herein, we explore the hidden molecular recognition abilities of ZnO nanowires uniformly grown on the inner surface of an open tubular fused silica capillary via liquid chromatography. Chromatographic evaluation revealed that ZnO nanowires showed a stronger intermolecular interaction with phenylphosphoric acid than any other monosubstituted benzene. Furthermore, ZnO nanowires specifically recognized the phosphate groups present in nucleotides even in the aqueous mobile phase, and the intermolecular interaction increased with the number of phosphate groups. This discrimination of phosphate groups in nucleotides was unique to the rich (10![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0) m-plane of ZnO nanowires with a moderate hydrophilicity and negative charge. The discrimination could be evidenced by the changes in the infrared bands of the phosphate groups on nucleotides on ZnO nanowires. Finally, as an application of the molecular recognition, nucleotides were separated by the number of phosphate groups, utilizing optimized gradient elution on ZnO nanowire column. Thus, the present results elucidate the unique and versatile molecular selectivity of well-known ZnO nanostructures for the capture and separation of biomolecules.
0) m-plane of ZnO nanowires with a moderate hydrophilicity and negative charge. The discrimination could be evidenced by the changes in the infrared bands of the phosphate groups on nucleotides on ZnO nanowires. Finally, as an application of the molecular recognition, nucleotides were separated by the number of phosphate groups, utilizing optimized gradient elution on ZnO nanowire column. Thus, the present results elucidate the unique and versatile molecular selectivity of well-known ZnO nanostructures for the capture and separation of biomolecules.
Introduction
Metal oxides exhibit thermal and chemical stabilities in harsh environments and diverse functional properties such as ferroelectricity, ferromagnetism, high temperature superconductivity, metal-insulator transitions, and memristors.1–7 Consequently, they have been studied for numerous applications. Since the development of the self-assembly process in the last half of the 20th century, the morphologies of various dimensional nanostructures (e.g., nanoparticles,8,9 nanosheets,10,11 nanorods,12,13 and other characteristic three-dimensional structures14 consisting of metal oxides) have been elucidated. Nanostructures have a large surface, uniform size, perfect crystals, and anisotropy. They have potential to provide programable and integrated properties in various technological fields, especially in molecular recognition and sensing materials.15–17Wire-like one-dimensional nanostructures (nanowires; NWs) are fascinating nanostructures consisting of metal oxides. NWs have been applied to design gas sensors,18–20 biosensors,21,22 optoelectronic devices,23 and photocatalysts.24,25 In particular, ZnO NWs are of great interest from industrial perspectives in the materials field due to their simple syntheses, biocompatibility, biodegradability, and biosafety.26–30 For example, a ZnO NW semiconductor-based electrical sensor was fabricated to detect nonanal, a biomarker for lung cancer.31 In addition, biomolecular analysis systems based on specific interactions with biomolecules have been constructed using ZnO NWs. Examples include a highly sensitive and selective ZnO NW sensor for immunoglobulin G 32 and a ZnO NW microfluidic device to detect cancer-related miRNA biomarkers from urinary extracellular vesicles.33
Despite the large number of applications, few reports have comprehensively investigated the fundamental mechanism of molecular recognition on ZnO NWs. A detailed understanding of molecular recognition on ZnO NWs should provide information on the rational design of a highly functional device. Liquid chromatography (LC), which is a fundamental and highly versatile separation technique, is suitable to investigate the intermolecular interactions on nanomaterials. LC directly reflects the strength of intermolecular interactions with the given stationary and mobile phases. Previously, we developed novel separation media utilizing nanocarbon materials as the stationary phase and investigated their unique molecular recognitions. These included the spherical recognition of C60- or C70-fullerenes,34,35 the difference in the intensities of the CH or CD–π interaction,36,37 the strong CH–π interaction between corannulene and planar polycyclic aromatic hydrocarbons,38 and specific halogen–π interaction of C70-fullerene.39 Furthermore, we have grown spatially uniform ZnO NWs in meter-long microtubes with an aspect ratio up to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (ZnO-NW column) by a new self-assembly strategy called the “flow-assisted method”.40 This microtube can be used as a separation medium for LC. We confirmed the specific retention of the polar functional groups due to the Lewis acidity of their Zn sites. Consequently, the evaluation of the retention behaviors on the ZnO-NW column may shed light on the molecular recognition of ZnO NWs in a particular liquid mobile phase.
000 (ZnO-NW column) by a new self-assembly strategy called the “flow-assisted method”.40 This microtube can be used as a separation medium for LC. We confirmed the specific retention of the polar functional groups due to the Lewis acidity of their Zn sites. Consequently, the evaluation of the retention behaviors on the ZnO-NW column may shed light on the molecular recognition of ZnO NWs in a particular liquid mobile phase.
In this study, we investigate the retention behaviors of various analytes on the ZnO-NW column in LC for a precise understanding of the molecular recognition on ZnO NWs in aqueous solvents. We also demonstrate the potential of nucleotide analysis using ZnO NWs due to the efficient molecular recognition on the (10![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0) m-planes. Finally, we show effective gradient separation of the nucleotides, which require selective sensing to reveal their roles as energy currencies of cells in cellular activities derived from biological processes such as cell motility, intercellular transport, synthesis of biomolecules, and metabolic reactions.41–44 We successfully separated adenosine mono-, di-, and triphosphates (AMP, ADP, and ATP) based on specific intermolecular interactions on ZnO NWs by optimizing the aqueous mobile phase and the column length.
0) m-planes. Finally, we show effective gradient separation of the nucleotides, which require selective sensing to reveal their roles as energy currencies of cells in cellular activities derived from biological processes such as cell motility, intercellular transport, synthesis of biomolecules, and metabolic reactions.41–44 We successfully separated adenosine mono-, di-, and triphosphates (AMP, ADP, and ATP) based on specific intermolecular interactions on ZnO NWs by optimizing the aqueous mobile phase and the column length.
Results and discussion
Retention behaviors of monosubstituted benzenes on the ZnO-NW column
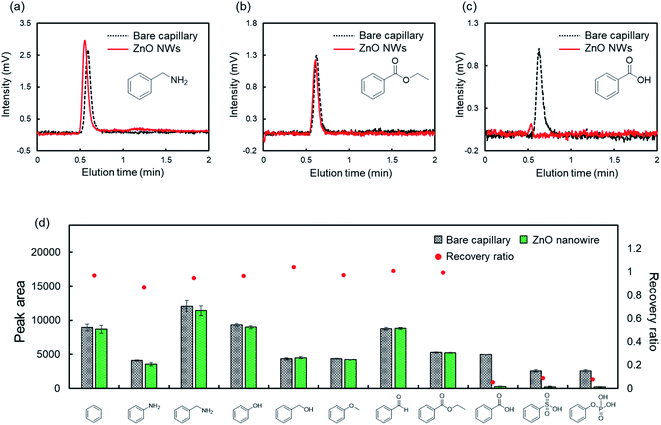
To evaluate the fundamental characteristics of the intermolecular interactions on ZnO NWs, we investigated the retention behavior of monosubstituted benzenes with various functional groups in LC on the ZnO-NW column. For comparison, we also examined the retention behavior on a bare capillary column using acetonitrile as the mobile phase. Acetonitrile is commonly used in LC. Fig. 1a–c show the typical chromatograms of benzyl amine, ethyl benzoate, and benzoic acid by the column type, respectively. Benzyl amine and ethyl benzoate passed through these columns. In contrast, benzoic acid was strongly retained on the ZnO-NW column and was not eluted even after an hour.We then determined the functional groups with a strong intermolecular interaction on ZnO NWs. The recovery ratio of each analyte on the ZnO-NW column was calculated by comparing the peak areas on the ZnO-NW column to those on the bare capillary column (Fig. 1d). The recovery ratios were calculated as
| Recovery ratio = (peak area on the ZnO-NW column)/(peak area on the bare capillary column) |
The recovery ratios of benzoic acid, phenylsulfonic acid, and phenylphosphonic acid were drastically reduced on the ZnO-NW column. ZnO NWs showed a strong and irreversible intermolecular interaction with acidic functional groups in acetonitrile.
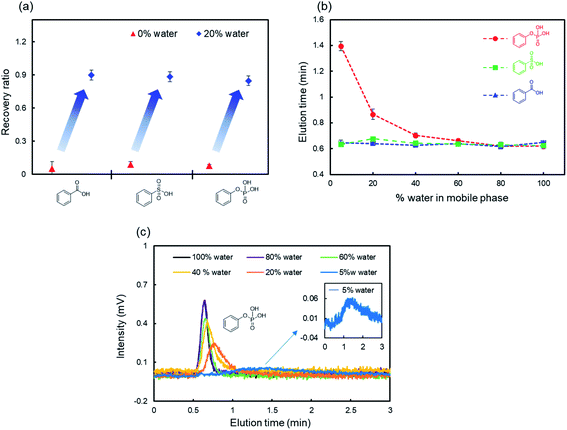
The specific adsorption of acidic functional groups on ZnO NWs may depend on several factors such as hydrophilic interactions45,46 and ionic electrostatic interactions.47–49 We hypothesized that adding water to the mobile phase increases the recovery ratios of these compounds because water competitively coordinates to ZnO NWs and has a high dielectric constant.50,51Fig. 2a shows the recovery ratios of monosubstituted benzenes with acidic functional groups in the mobile phase containing water. As expected, the recovery ratio of each analyte exceeded 80% by adding 20% water to the mobile phase, suggesting that water competitively interacts with ZnO NWs and suppresses the adsorption of monosubstituted benzenes with acidic functional groups. Interestingly, phenylphosphoric acid showed much longer retention times and broader peaks on the ZnO-NW column than the other analytes even in mobile phases containing water (Fig. 2b and c), although the retention time slightly decreased as the content of water in the mobile phase increased. Thus, ZnO NWs interact more strongly with the phosphate groups of the analytes than the other functional groups. The number of negative charges may account for this trend. Zn2+, which is the divalent cation in ZnO NWs, preferred the divalent anion –PO32− in phenylphosphoric acid than the other monovalent anions.
Retention behaviors of phosphorylated nucleotides on the ZnO-NW column
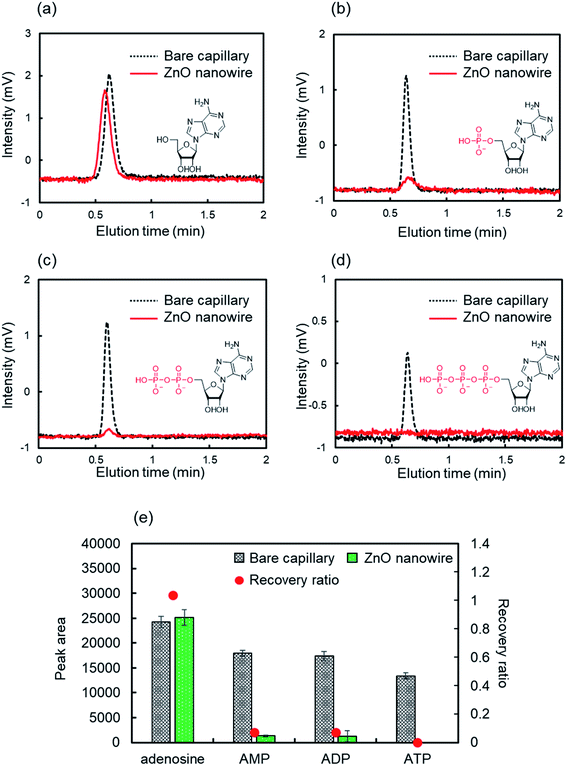
ZnO NWs may interact specifically with the phosphate groups in the mobile phase containing water due to hydrophilic and ionic electrostatic interactions. We evaluated the retention behaviors of nucleotides on the ZnO-NW column to test this hypothesis and to explore the potential of applying ZnO NWs to phosphorylated biomolecular analysis. Nucleotides contribute to many biological processes by regulating the concentration and are important for maintaining the cell health and expression of cellular functions.52–54 Furthermore, they are useful biomarkers for several cancers.55 Consequently, there is a growing demand for detection or measurement techniques to determine these concentration ratios.Firstly, we evaluated the retention behaviors of AMP, ADP, and ATP, which are nucleotides found in RNA, on the ZnO-NW column in acetonitrile as the mobile phase. The mobile phase contained 5% water, considering the solubility of nucleotides. For comparison, adenosine, which has the same structure except for the phosphate group, was also evaluated. Fig. 3 shows the retention behaviors of adenosine, AMP, ADP, and ATP in acetonitrile on ZnO-NWs and bare capillary columns. The retention behavior of adenosine was similar for both the ZnO-NW column and the bare capillary column (Fig. 3a and e). On the other hand, the nucleotides were adsorbed on the ZnO-NW column and exhibited drastically reduced recovery ratios (Fig. 3b–e). These results are consistent with our hypothesis that ZnO NWs show highly selective retention for the phosphate groups. Notably, phosphorylated nucleotides were completely adsorbed on the ZnO-NW column even in the mobile phase containing 5% water but phenylphosphoric acid was eluted (Fig. 2b and c). This stronger interaction of the nucleotides with ZnO NWs than phenylphosphoric acid may be due to the higher hydrophilicity of nucleotides compared to phenylphosphoric acid.
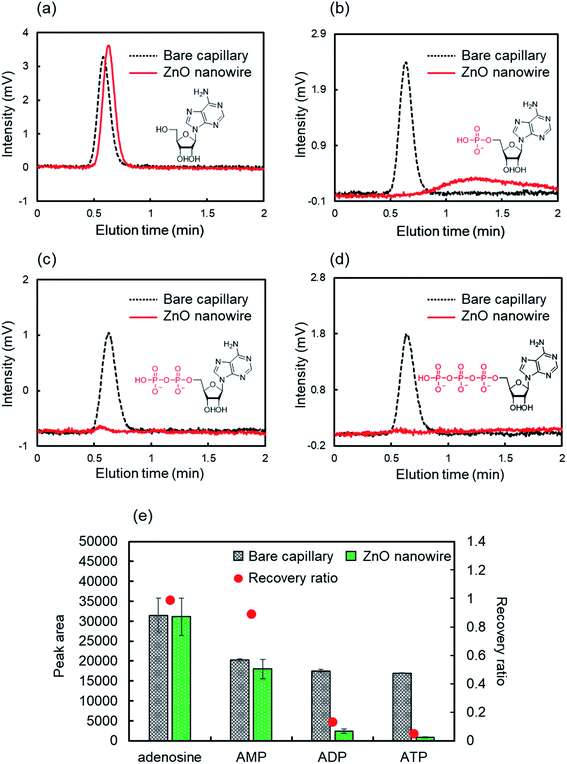
The suppression of the hydrophilic interactions by molecular recognition in aqueous environments is critical for biomolecular analysis.56 We evaluated the retention behaviors of nucleotides in pure water as the mobile phase on the ZnO-NW column (Fig. 4). AMP showed a higher recovery ratio in pure water than in acetonitrile but its retention time was longer than adenosine (Fig. 4a, b, and e). This weakened intermolecular interaction between AMP and ZnO NWs may be due to the competitive hydrophilic interaction with water. Interestingly, ADP and ATP adsorbed on the ZnO-NW column, and their recovery ratios were very low even in water (Fig. 4c–e). The recovery ratios on the ZnO-NW column decreased in the order AMP < ADP < ATP, which coincides with the increase in the number of phosphate groups in the nucleotides. Thus, the strength of the intermolecular interactions between ZnO and nucleotides depends on the number of phosphate groups in the nucleotide.
We then examined whether the ZnO NW nanostructure or the typical properties of ZnO lead to the discrimination of the phosphate groups in the nucleotides. We evaluated the recovery ratios of adenosine and nucleotides on a column fabricated only by the ZnO seed layer, which lacks uniform nanostructures but has the same properties as that of ZnO. Both ADP and ATP were fully eluted on the column fabricated using only the ZnO seed layer (Fig. S1†). The discrimination for the phosphate groups of nucleotides on ZnO NWs in water is attributed to the nanostructures.
IR spectra of adsorbed nucleotides on the ZnO NWs
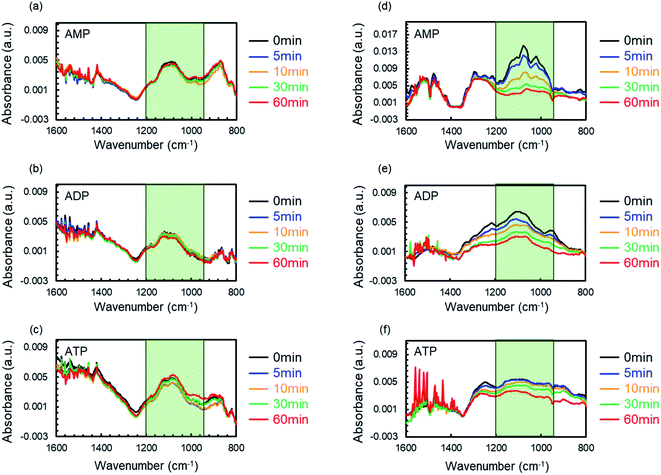
Two fundamental aspects should be considered for molecular recognition since the nanostructure is responsible for the phosphate group discrimination of nucleotides on ZnO NWs: the specific surface area and the morphology. Two types of ZnO nanostructures (ZnO nanowalls and ZnO NWs, Fig. S2†) were fabricated on silicon wafers. IR spectroscopy was used for the molecular recognition of the nucleotide on each nanostructure. The substrate with each nanostructure was immersed into an aqueous solution of AMP, ADP, or ATP (7 mM) and was dried to adsorb the nucleotide on the nanostructure. The substrates were subsequently immersed in water. Then, IR spectroscopy was performed after drying the substrates well.Fig. 5 shows the IR spectra of the nucleotides on each substrate for different immersion times. Each substrate displayed a narrow band in the range of 900–1200 cm−1,57,58 which is derived from the phosphate groups, immediately after immersing the substrates into the nucleotide solution (0 min, Fig. 5, black lines). The signal was not confirmed without immersing ZnO NWs in the nucleotides (Fig. S3,† black line) and the intensity was increased with the immersion time (Fig. S3†). These results suggested that the nucleotide was adsorbed on ZnO nanostructures. On the ZnO nanowalls, the band intensity of the phosphate group in the nucleotide was independent of the immersion time in water, indicating that all the nucleotides are completely adsorbed on the ZnO nanowalls, regardless of the number of phosphate groups (Fig. 5a–c). On the other hand, on the ZnO NWs, the band intensity of the phosphate group in the nucleotides decreased as the immersion time in water increased. More interestingly, although the band intensity in ATP slightly decreased, the decreases in the band intensity were smaller in the order AMP > ADP > ATP (Fig. 5d–f). Briefly, the intermolecular interaction with nucleotides on ZnO NWs was weaker than that on the ZnO nanowalls, whereas the strength varied slightly with the number of phosphate groups in the nucleotides. These results suggest that the discrimination based on the number of phosphate groups in the nucleotides is due to the morphology of ZnO NWs.
The morphology of the ZnO nanostructures could be controlled by competitive nucleation from the (0001) c-planes and m-planes. Thus, ZnO NWs and nanowalls provide unique face-selective electrostatic interactions by anisotropic crystal growth.26,59–63 Fig. S4† shows the SEM image of the top surface of the ZnO NWs. The small hexagonal shape on the top surface and the long columnar shape (Fig. S2†) implied abundant m-planes. The ZnO nanowalls and ZnO NWs exposed the c-planes and the m-planes, respectively. The polar c-plane provides a higher ionicity due to the biased surface to either Zn2+ or O2−. In contrast, Zn2+ and O2− are present in the same proportion on the nonpolar m-plane.64–66 Therefore, the ZnO nanowalls may indiscriminately adsorb the nucleotides on the c-planes via their strong hydrophilic and ionic electrostatic interactions with phosphate groups. On the other hand, ZnO NWs may flexibly recognize the phosphate groups due to their moderate hydrophilic and ionic electrostatic interactions on the m-planes.
Dependence of molecular recognition on the ZnO-NW column on the temperature and flow rate
Ionic electrostatic and hydrophilic interactions may be responsible for the interactions between ZnO NWs and the phosphate groups. The ionic electrostatic interaction is dominant, especially in water, because the hydrophilic interactions are suppressed. The potential energy of the ionic electrostatic interaction is given as| E = z+z−e2/4πεR | (1) |
Here, z+ and z− are the valences of the cation and anion, respectively, e is the elementary charge, ε is the dielectric constant, and R is the distance between the cation and the anion.67Eqn (1) indicates that the ionic electrostatic interaction is not influenced by the temperature. Furthermore, ionic electrostatic interactions rapidly form even in water.68 Here, we hypothesized that the interactions of nucleotides on ZnO NWs are not influenced by the temperature or flow rate in LC analysis. Then, we derived the temperature and flow rate dependencies from the plot of the recovery ratios of nucleotides at different temperatures and flow rates (Fig. S5†). The recovery ratios of each nucleotide were almost the same at different temperatures and flow rates, supporting our hypothesis that molecular recognition for nucleotides on ZnO NWs is due to the ionic electrostatic interaction and is independent of the temperature and flow rate. In addition, the SEM images indicate that the measurement does not corrupt ZnO NWs in the capillary column (Fig. S6†). This physical robustness of ZnO NWs may realize stable molecular recognition in harsh physical conditions.
Separation of phosphorylated nucleotides by gradient elution
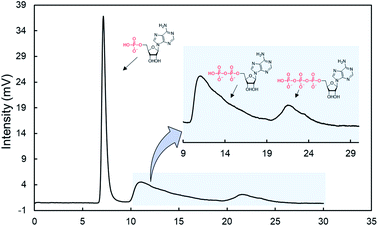
Nucleotides could be adsorbed on ZnO NWs via the ionic electrostatic interaction of their phosphate groups. Eqn (1) indicates that the electrostatic interaction is strongly influenced by the dielectric constant of the solvent. Hence, the competitive coordination of salts or decreasing the dielectric constant in the electrolyte solutions may tune the adsorption of nucleotides. For the collection of adsorbed nucleotides on ZnO NWs, we evaluated the retention behaviors of ATP in different electrolyte solutions such as a NaCl solution, tris(hydroxymethyl)aminomethane-HCl (Tris–HCl buffer), and phosphate buffer. Fig. S7† shows the recovery ratios of ATP in the different electrolyte solutions. For each electrolyte solution, the ionic strength was standardized to 0.2 M. Unexpectedly, the recovery ratios of ATP did not increase in the NaCl solution or the Tris–HCl buffer. On the other hand, the recovery ratio of ATP significantly increased in the phosphate buffer. These observations may be due to the stronger coordination of the phosphate groups to Zn2+ than any other monovalent ions. Thus, the adsorption of nucleotides can be controlled by the competitive coordination of phosphate ions in the mobile phase due to their same charge.Finally, we investigated the effectiveness of the phosphate-gradient condition to separate the nucleotides. As expected, AMP, ADP, and ATP were eluted from the ZnO-NW column in this order as the concentration of the phosphate ions increased in the mobile phase. However, ADP and ATP were not sufficiently separated due to the much lower plate number of the column, which is an index of the column efficiency. Briefly, the 30 cm ZnO-NW column was not long enough for sufficient separation (Fig. S8†). Increasing the length of the ZnO-NW column achieved a more effective separation because a longer column can generate enough plate numbers to distinguish between the neighboring peaks.69
We prepared a 90 cm ZnO-NW column (meter-long ZnO-NW column) using the flow-assisted method.40Fig. 6 shows chromatograms for a mixture of nucleotides. As expected, the nucleotides were successfully separated. Furthermore, the change in the back pressure was negligible on the meter-long ZnO-NW column (3.1 MPa) compared with the shorter column (0.4 MPa) due to the lower packing rate. This value was much lower than that in a typical silica-based monolithic column (15 cm, 21.1 MPa) under the same conditions. Therefore, the meter-long ZnO-NW column may realize a novel separation medium for low back pressure and robust biomolecular analysis with unique phosphate recognition. In the present situation, the gradient separation slightly dissolved the ZnO NWs, and the same column could not provide a reproducible separation (Fig. S9†). The dissolutions of ZnO NWs were not confirmed with NaCl or Tris-buffer (Fig. S10†). Thus, the chemical robustness toward the phosphate buffer may be low due to the high affinity to the phosphoric groups. To realize a practical separation media, the chemical robustness toward the phosphate buffer should be improved in the future. For example, thermal annealing treatment is effective for increasing the crystallization and improving the stability of ZnO nanowires.70,71 Then, we compared the stabilities of the annealed nanowires (350 °C, 1 h) and as-grown nanowires in phosphate buffer. The stability of the annealed nanowires was clearly improved and the morphology is maintained after immersing into phosphate buffer for 5 hours, even though the as-grown nanowires gradually change their morphology and dissolve completely after 5 hours (Fig. S11†).
Conclusion
LC experiments were conducted for monosubstituted benzenes and nucleotides in various aqueous mobile phases to understand the molecular recognition ability on ZnO NWs. ZnO NWs showed an irreversible intermolecular interaction with acidic functional groups in acetonitrile. ZnO NWs strongly retained the phosphate groups even in water containing the mobile phase. Furthermore, ZnO NWs were discriminated by the number of phosphate groups of nucleotides in water, which was not influenced by the temperature and the flow rate in LC analysis. The delicate recognition for the phosphate group was unique to the nanostructure of ZnO NWs via the moderate ionic electrostatic interaction on the (10![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) 0) m-planes of ZnO NWs. In addition, optimizing the phosphate-gradient LC conditions effectively separated the nucleotides. We believe that this report will greatly contribute to the elucidation of the molecular recognition on metal-oxide nanostructures.
0) m-planes of ZnO NWs. In addition, optimizing the phosphate-gradient LC conditions effectively separated the nucleotides. We believe that this report will greatly contribute to the elucidation of the molecular recognition on metal-oxide nanostructures.
Experimental section
Preparation of ZnO-NW columns
ZnO nanowire-decorated microtubes were fabricated via the seed-assisted hydrothermal method, which consists of two steps: seed layer formation and nanowire formation. The seed layer formation process used zinc acetate Zn(OAc)2 in an ethanolamine/EtOH solution. The seed solution was injected into a flexible fused silica capillary column with an inner diameter of 100 μm and an outer diameter of 375 μm (molex 1068150023). Then, the column was annealed at 150 °C for 1 hour to crystallize the ZnO seed layer.Then, the ZnO NW formation process was performed. The growth solution was prepared at room temperature by dissolving zinc nitrate hexahydrate Zn(NO3)2·6H2O (Wako, 99.0% pure), hexamethylenetetramine (HMTA) (CH2)6N4 (Wako, 99.0% pure), and polyethylenimine (PEI) [–CH2CH2NH–]n with an average molecular weight (mol wt.) of 1800 (Aldrich, 50 wt% in H2O) in deionized (DI) water. The ratio of Zn(NO3)2/HMTA/PEI = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1. The pH of the solution was measured prior to nanowire growth. A growth temperature of 75 °C was maintained during the whole growth process, which took five hours.
0.1. The pH of the solution was measured prior to nanowire growth. A growth temperature of 75 °C was maintained during the whole growth process, which took five hours.
Nano-LC analysis
LC analyses were carried out with a nano-LC system consisting of a DiNa S (KYA Technologies, Tokyo, Japan) as the pump, a CE-2070 (JASCO, Tokyo, Japan) as the UV detector, a CHEMINERT (Valco Instruments, Houston, TX) as the sample injector, and a Chemco capillary column conditioner Model 380-b (Chemco, Osaka, Japan) as the column oven. A Nexera Micros (Shimadzu, Kyoto, Japan) series was used for gradient LC separation. This system consisted of LC-Mikros (Shimadzu, Kyoto, Japan) as the pump, MU701 (GL Sciences, Tokyo, Japan) as the UV detector, a CHEMINERT as the sample injector, and CTO-20AC (Shimadzu, Kyoto, Japan) as the column oven. Acetonitrile, DI, sodium phosphate buffer, and Tris–HCl buffer were utilized as the mobile phases. DI was obtained by a Milli-Q Direct-Q 3UV system (Merck Millipore, Tokyo, Japan). Sodium dihydrogenphosphate dihydrate, disodium hydrogenphosphate, and tris(hydroxymethyl)aminomethane were purchased from Tokyo Chemical Industry (Tokyo, Japan) for buffer preparation. As samples, AMP, ADP, ATP, phenylphosphoric acid disodium salt, benzyl alcohol, benzyl amine, aniline, benzene, anisole, acetophenone, ethyl benzoate, benzoic acid, phenol, and sodium benzenesulfonate were purchased from Nacalai Tesque (Kyoto, Japan). A bare capillary column was treated with water and acetone, and then dried.Growth of ZnO NWs on silicon wafers
The hydrothermal method was utilized to grow ZnO NWs arrays on Si(100) wafers. First, a 5 nm-thick Ti buffer layer was sputtered on the Si substrate. Then, a 100 nm-thick ZnO thin film was sputtered as the seed layer. Zinc nitrate hexahydrate (25 mM) (Zn(NO3)2·6H2O, 99.0%), hexane ethylenetetraamine (25 mM, (CH2)6N4, 99.0%), and polyethylenimine (2.5 mM, number-average MW 1800, 50 wt% in H2O) were dissolved in DI water (150 mL) sequentially. The prepared substrate was dipped into the solution and kept at 80 °C for 12 h. After growth, the samples were rinsed with DI water and dried by dry air flow.Growth of ZnO nanowalls on silicon wafers
The hydrothermal method was utilized to grow ZnO nanowalls on Si(100) wafers. A solution of ethanol (25 mL), zinc acetate (Zn(OAc)2, 99.99%) (1.65 g), and ethanolamine (0.76 g) was applied on the Si substrate. The Si substrate was uniformly coated via the spin-coating method. Next, a seed layer was created by sintering at 180 °C for 10 minutes. Zn(OAc)2 (120 mM) and citric acid (C6H8O7·H2O, 99.0%) (257 mM) were dissolved in DI water (150 mL) sequentially. The substrate was dipped into the solution and kept at 95 °C for 12 h. Afterward, the samples were rinsed with DI water and dried by dry air flow. Finally, the as-grown ZnO nanowalls were annealed at 400 °C in air for 1 h.Scanning electron microscopy images and infrared (IR) spectra
The morphology of the ZnO nanostructures was characterized by field emission scanning electron microscopy with energy dispersive X-ray spectroscopy (FESEM-EDS, JEOL JSM-7610F 30 kV). Fourier transform infrared (FT-IR) spectra were recorded using a Nicolet iS50 FT-IR spectrometer (Thermo Fisher Scientific) equipped with a mercury-cadmium-telluride detector. The FT-IR spectra were scanned 300 times for each measurement with a resolution of 4.0 cm−1 and a data point interval of 0.482 cm−1.Author contributions
Experiments were designed by E. Kanao, T. Kubo, T. Hosomi, Y. Ishihama, J. Adachi, K. Otsuka and T. Yanagida. The ZnO-NWs columns were prepared by R. Kamei and T. Hosomi. Chromatographic analyses were operated by E. Kanao and K. Nakano, and SEM and IR measurments by R. Kamei, and T. Hosomi. The manuscript was written by E. Kanao, T. Kubo, K. Otsuka and T. Yanagida.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (KAKENHI; Grants JP21K14652, JP20H05737 JP18H01831, JP18H05243, and JP18KK0112), PRESTO, JST, Japan (Grant JPMJPR19T8).References
- M. Stengel, D. Vanderbilt and N. A. Spaldin, Nat. Mater., 2009, 8, 392–397 CrossRef CAS PubMed.
- H. Y. Hwang, Y. Iwasa, M. Kawasaki, B. Keimer, N. Nagaosa and Y. Tokura, Nat. Mater., 2012, 11, 103–113 CrossRef CAS PubMed.
- N. C. Bristowe, J. Varignon, D. Fontaine, E. Bousquet and P. Ghosez, Nat. Commun., 2015, 6, 6677 CrossRef CAS PubMed.
- W. M. Li, J. F. Zhao, L. P. Cao, Z. Hu, Q. Z. Huang, X. C. Wang, Y. Liu, G. Q. Zhao, J. Zhang, Q. Q. Liu, R. Z. Yu, Y. W. Long, H. Wu, H. J. Lin, C. T. Chen, Z. Li, Z. Z. Gong, Z. Guguchia, J. S. Kim, G. R. Stewart, Y. J. Uemura, S. Uchida and C. Q. Jin, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 12156 CrossRef CAS PubMed.
- S. Zhang, H. Vo and G. Galli, Chem. Mater., 2021, 33, 3187–3195 CrossRef CAS.
- E. J. Braham, D. Sellers, E. Emmons, R. Villarreal, H. Asayesh-Ardakani, N. A. Fleer, K. E. Farley, R. Shahbazian-Yassar, R. Arròyave, P. J. Shamberger and S. Banerjee, Chem. Mater., 2018, 30, 214–224 CrossRef CAS.
- T. Wang, Y. Shi, F. M. Puglisi, S. Chen, K. Zhu, Y. Zuo, X. Li, X. Jing, T. Han, B. Guo, K. Bukvišová, L. Kachtík, M. Kolíbal, C. Wen and M. Lanza, ACS Appl. Mater. Interfaces, 2020, 12, 11806–11814 CrossRef CAS PubMed.
- A. Pucci, M.-G. Willinger, F. Liu, X. Zeng, V. Rebuttini, G. Clavel, X. Bai, G. Ungar and N. Pinna, ACS Nano, 2012, 6, 4382–4391 CrossRef CAS PubMed.
- G. Guo, L. Ji, X. Shen, B. Wang, H. Li, J. Hu, D. Yang and A. Dong, J. Mater. Chem. A, 2016, 4, 16128–16135 RSC.
- F. Ji, X. Ren, X. Zheng, Y. Liu, L. Pang, J. Jiang and S. Liu, Nanoscale, 2016, 8, 8696–8703 RSC.
- X. Liu, Y. Sun, M. Yu, Y. Yin, B. Du, W. Tang, T. Jiang, B. Yang, W. Cao and M. N. R. Ashfold, Sens. Actuators, B, 2018, 255, 3384–3390 CrossRef CAS.
- Z. Zhang, Q. Wu, G. Johnson, Y. Ye, X. Li, N. Li, M. Cui, J. D. Lee, C. Liu, S. Zhao, S. Li, A. Orlov, C. B. Murray, X. Zhang, T. B. Gunnoe, D. Su and S. Zhang, J. Am. Chem. Soc., 2019, 141, 16548–16552 CrossRef CAS PubMed.
- H. Kloust, R. Zierold, J.-P. Merkl, C. Schmidtke, A. Feld, E. Pöselt, A. Kornowski, K. Nielsch and H. Weller, Chem. Mater., 2015, 27, 4914–4917 CrossRef CAS.
- V. Polshettiwar, B. Baruwati and R. S. Varma, ACS Nano, 2009, 3, 728–736 CrossRef CAS PubMed.
- Y.-B. Hahn, R. Ahmad and N. Tripathy, Chem. Commun., 2012, 48, 10369–10385 RSC.
- Z. Li, H. Li, Z. Wu, M. Wang, J. Luo, H. Torun, P. Hu, C. Yang, M. Grundmann, X. Liu and Y. Fu, Mater. Horiz., 2019, 6, 470–506 RSC.
- M. J. Limo, A. Sola-Rabada, E. Boix, V. Thota, Z. C. Westcott, V. Puddu and C. C. Perry, Chem. Rev., 2018, 118, 11118–11193 CrossRef CAS PubMed.
- M. J. Limo, A. Sola-Rabada, E. Boix, V. Thota, Z. C. Westcott, V. Puddu and C. C. Perry, Chem. Rev., 2018, 118, 11118–11193 CrossRef CAS PubMed.
- A. Kolmakov, D. O. Klenov, Y. Lilach, S. Stemmer and M. Moskovits, Nano Lett., 2005, 5, 667–673 CrossRef CAS PubMed.
- W. T. Koo, S. Qiao, A. F. Ogata, G. Jha, J.-S. Jang, V. T. Chen, I.-D. Kim and R. M. Penner, ACS Nano, 2017, 11, 9276–9285 CrossRef CAS PubMed.
- R. Janissen, P. K. Sahoo, C. A. Santos, A. M. da Silva, A. A. G. von Zuben, D. E. P. Souto, A. D. T. Costa, P. Celedon, N. I. T. Zanchin, D. B. Almeida, D. S. Oliveira, L. T. Kubota, C. L. Cesar, A. P. d. Souza and M. A. Cotta, Nano Lett., 2017, 17, 5938–5949 CrossRef CAS PubMed.
- M. Chen, L. Mu, S. Wang, X. Cao, S. Liang, Y. Wang, G. She, J. Yang, Y. Wang and W. Shi, ACS Chem. Neurosci., 2020, 11, 1283–1290 CrossRef CAS PubMed.
- Y. Li, F. Qian, J. Xiang and C. M. Lieber, Mater. Today, 2006, 9, 18–27 CrossRef CAS.
- H. S. Jung, Y. J. Hong, Y. Li, J. Cho, Y.-J. Kim and G.-C. Yi, ACS Nano, 2008, 2, 637–642 CrossRef CAS PubMed.
- P. Tongying, F. Vietmeyer, D. Aleksiuk, G. J. Ferraudi, G. Krylova and M. Kuno, Nanoscale, 2014, 6, 4117–4124 RSC.
- X. Zhao, K. Nagashima, G. Zhang, T. Hosomi, H. Yoshida, Y. Akihiro, M. Kanai, W. Mizukami, Z. Zhu, T. Takahashi, M. Suzuki, B. Samransuksamer, G. Meng, T. Yasui, Y. Aoki, Y. Baba and T. Yanagida, Nano Lett., 2020, 20, 599–605 CrossRef CAS PubMed.
- P. Sharma, H. A. Cho, J.-W. Lee, W. S. Ham, B. C. Park, N.-H. Cho and Y. K. Kim, Nanoscale, 2017, 9, 15371–15378 RSC.
- Z. Li, R. Yang, M. Yu, F. Bai, C. Li and Z. L. Wang, J. Phys. Chem. C, 2008, 112, 20114–20117 CrossRef CAS.
- J. Zhou, N. S. Xu and Z. L. Wang, Adv. Mater., 2006, 18, 2432–2435 CrossRef CAS.
- Y. Wang, Y. Wu, F. Quadri, J. D. Prox and L. Guo, Nanomaterials, 2017, 7(4), 80 CrossRef PubMed.
- C. Wang, T. Hosomi, K. Nagashima, T. Takahashi, G. Zhang, M. Kanai, H. Yoshida and T. Yanagida, ACS Appl. Mater. Interfaces, 2020, 12, 44265–44272 CrossRef CAS PubMed.
- R. Yu, C. Pan and Z. L. Wang, Energy Environ. Sci., 2013, 6, 494–499 RSC.
- T. Yasui, T. Yanagida, S. Ito, Y. Konakade, D. Takeshita, T. Naganawa, K. Nagashima, T. Shimada, N. Kaji, Y. Nakamura, I. A. Thiodorus, Y. He, S. Rahong, M. Kanai, H. Yukawa, T. Ochiya, T. Kawai and Y. Baba, Sci. Adv., 2017, 3, e1701133 CrossRef PubMed.
- T. Kubo, Y. Murakami, M. Tsuzuki, H. Kobayashi, T. Naito, T. Sano, M. Yan and K. Otsuka, Chem.–Eur. J., 2015, 21, 18095–18098 CrossRef CAS PubMed.
- T. Kubo, E. Kanao, T. Matsumoto, T. Naito, T. Sano, M. Yan and K. Otsuka, ChemistrySelect, 2016, 1, 5900–5904 CrossRef CAS.
- E. Kanao, T. Kubo, T. Naito, T. Matsumoto, T. Sano, M. Yan and K. Otsuka, J. Phys. Chem. C, 2018, 122, 15026–15032 CrossRef CAS.
- E. Kanao, T. Kubo, T. Naito, T. Sano, M. Yan, N. Tanaka and K. Otsuka, Anal. Chem., 2020, 92, 4065–4072 CrossRef CAS PubMed.
- E. Kanao, T. Kubo, T. Naito, T. Matsumoto, T. Sano, M. Yan and K. Otsuka, Anal. Chem., 2019, 91, 2439–2446 CrossRef CAS PubMed.
- E. Kanao, T. Morinaga, T. Kubo, T. Naito, T. Matsumoto, T. Sano, H. Maki, M. Yan and K. Otsuka, Chem. Sci., 2020, 11, 409–418 RSC.
- R. Kamei, T. Hosomi, E. Kanao, M. Kanai, K. Nagashima, T. Takahashi, G. Zhang, T. Yasui, J. Terao, K. Otsuka, Y. Baba, T. Kubo and T. Yanagida, ACS Appl. Mater. Interfaces, 2021, 13, 16812–16819 CrossRef CAS PubMed.
- D. G. Hardie, F. A. Ross and S. A. Hawley, Nat. Rev. Mol. Cell Biol., 2012, 13, 251–262 CrossRef CAS PubMed.
- C. Bressan, A. Pecora, D. Gagnon, M. Snapyan, S. Labrecque, P. De Koninck, M. Parent and A. Saghatelyan, eLife, 2020, 9, e56006 CrossRef CAS PubMed.
- B. Li, S.-X. Dou, J.-W. Yuan, Y.-R. Liu, W. Li, F. Ye, P.-Y. Wang and H. Li, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 12118 CrossRef CAS PubMed.
- K. Y. Hara and A. Kondo, Microb. Cell Fact., 2015, 14, 198 CrossRef PubMed.
- H. Ghannam, A. Chahboun and M. Turmine, RSC Adv., 2019, 9, 38289–38297 RSC.
- S. L. Cheng, J. H. Syu, S. Y. Liao, C. F. Lin and P. Y. Yeh, RSC Adv., 2015, 5, 67752–67758 RSC.
- T. Shimada, T. Yasui, A. Yonese, T. Yanagida, N. Kaji, M. Kanai, K. Nagashima, T. Kawai and Y. Baba, Micromachines, 2020, 11, 610 CrossRef PubMed.
- L. Cao, J. Kiely, M. Piano and R. Luxton, Sci. Rep., 2018, 8, 12687 CrossRef PubMed.
- V. Gerbreders, M. Krasovska, I. Mihailova, A. Ogurcovs, E. Sledevskis, A. Gerbreders, E. Tamanis, I. Kokina and I. Plaksenkova, Sens. Bio-Sens. Res., 2019, 23, 100276 CrossRef.
- A. Domínguez, S. grosse Holthaus, S. Köppen, T. Frauenheim and A. L. da Rosa, Phys. Chem. Chem. Phys., 2014, 16, 8509–8514 RSC.
- L. G. Gagliardi, C. B. Castells, C. Ràfols, M. Rosés and E. Bosch, J. Chem. Eng. Data, 2007, 52, 1103–1107 CrossRef CAS.
- L. Liu, G. Duclos, B. Sun, J. Lee, A. Wu, Y. Kam, E. D. Sontag, H. A. Stone, J. C. Sturm, R. A. Gatenby and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 1686 CrossRef CAS PubMed.
- H.-X. Yuan, Y. Xiong and K.-L. Guan, Mol. Cell, 2013, 49, 379–387 CrossRef CAS PubMed.
- U. Olsson and M. Wolf-Watz, Nat. Commun., 2010, 1, 111 CrossRef PubMed.
- C. Zhang, Z. Liu, X. Liu, L. Wei, Y. Liu, J. Yu and L. Sun, Acta Pharm. Sin. B, 2013, 3, 254–262 CrossRef.
- K. Ariga, H. Ito, J. P. Hill and H. Tsukube, Chem. Soc. Rev., 2012, 41, 5800–5835 RSC.
- A. A. Ahmed, S. Gypser, P. Leinweber, D. Freese and O. Kühn, Phys. Chem. Chem. Phys., 2019, 21, 4421–4434 RSC.
- M. Yaguchi, T. Uchida, K. Motobayashi and M. Osawa, J. Phys. Chem. Lett., 2016, 7, 3097–3102 CrossRef CAS PubMed.
- C. Wang, T. Hosomi, K. Nagashima, T. Takahashi, G. Zhang, M. Kanai, H. Zeng, W. Mizukami, N. Shioya, T. Shimoaka, T. Tamaoka, H. Yoshida, S. Takeda, T. Yasui, Y. Baba, Y. Aoki, J. Terao, T. Hasegawa and T. Yanagida, Nano Lett., 2019, 19, 2443–2449 CrossRef CAS PubMed.
- J. Joo, B. Y. Chow, M. Prakash, E. S. Boyden and J. M. Jacobson, Nat. Mater., 2011, 10, 596–601 CrossRef CAS PubMed.
- B. Saravanakumar and S.-J. Kim, J. Phys. Chem. C, 2014, 118, 8831–8836 CrossRef CAS.
- T.-P. Chen, S.-P. Chang, F.-Y. Hung, S.-J. Chang, Z.-S. Hu and K.-J. Chen, Sensors, 2013, 13, 3941–3950 CrossRef CAS PubMed.
- J. Liu, K. Nagashima, H. Yamashita, W. Mizukami, J. Uzuhashi, T. Hosomi, M. Kanai, X. Zhao, Y. Miura, G. Zhang, T. Takahashi, M. Suzuki, D. Sakai, B. Samransuksamer, Y. He, T. Ohkubo, T. Yasui, Y. Aoki, J. C. Ho, Y. Baba and T. Yanagida, Commun. Mater., 2020, 1, 58 CrossRef.
- K. Sun, H.-Y. Su and W.-X. Li, Theor. Chem. Acc., 2013, 133, 1427 Search PubMed.
- S. Akhter, K. Lui and H. H. Kung, J. Phys. Chem., 1985, 89, 1958–1964 CrossRef CAS.
- D. Mora-Fonz, T. Lazauskas, M. R. Farrow, C. R. A. Catlow, S. M. Woodley and A. A. Sokol, Chem. Mater., 2017, 29, 5306–5320 CrossRef CAS.
- T. Enomoto, Y. Nakamori, K. Matsumoto and R. Hagiwara, J. Phys. Chem. C, 2011, 115, 4324–4332 CrossRef CAS.
- M. Kohagen, E. Pluhařová, P. E. Mason and P. Jungwirth, J. Phys. Chem. Lett., 2015, 6, 1563–1567 CrossRef CAS PubMed.
- H. G. Barth, LC GC N. Am., 2018, 36, 830–835 CAS.
- K. Nakamura, T. Takahashi, T. Hosomi, T. Seki, M. Kanai, G. Zhang, K. Nagashima, N. Shibata and T. Yanagida, ACS Appl. Mater. Interfaces, 2019, 11, 40260–40266 CrossRef CAS PubMed.
- O. Lupan, N. Magariu, R. Khaledialidusti, A. K. Mishra, S. Hansen, H. Krüger, V. Postica, H. Heinrich, B. Viana, L. K. Ono, B. R. Cuenya, L. Chow, R. Adelung and T. Pauporté, ACS Appl. Mater. Interfaces, 2021, 13, 10537–10552 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00865j |
| This journal is © The Royal Society of Chemistry 2022 |