Open Access Article
Open Access ArticleHydrophobic nanofibers: a peptide-based functional anti-fouling material†
Kshitish Chandra
Hati‡
a,
Santosh
Kumar‡
a,
Sahabaj
Mondal
a,
Surajit
Singh
a,
Ananda
Shit
a,
Sujay Kumar
Nandi
a and
Debasish
Haldar
 *ab
*ab
aDepartment of Chemical Sciences, Indian Institute of Science Education and Research Kolkata, Mohanpur-741246, West Bengal, India. E-mail: deba_h76@yahoo.com; deba_h76@iiserkol.ac.in
bCentre for Advanced Functional Materials (CAFM), Indian Institute of Science Education and Research, Kolkata, Mohanpur-741246, West Bengal, India
First published on 31st March 2022
Abstract
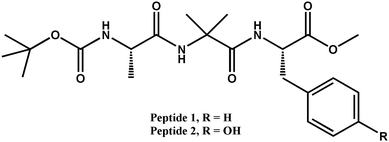
Functional bioinspired materials have been developed by molecular self-assembly of a hydrophobic peptide. Two analogue peptides containing hydrophobic L-alanine, α-aminoisobutyric acid (Aib), L-phenylalanine and L-tyrosine deliver different supramolecular structures and functions. Peptide 1 containing L-phenylalanine adopts a β-turn conformation and self-assembles through intermolecular hydrogen bonds to form a supramolecular hydrophobic sheet-like structure. But peptide 2 with L-tyrosine adopts kink-like conformations and self-assembles to form a supramolecular hydrogen bonded helix-like architecture. Irrespective of the presence of the same peptide backbone conformation, only the side chain hydroxyl functional group has introduced a huge change in self-assembly pattern and function. The hydrophobic peptide 1 further self-assembled to form slippery nanofibers. The resulting surfaces show anti-sticking effects against water and exhibit anti-fouling properties, like inhibiting the growth of Escherichia coli.
Introduction
In nature, plenty of plants, insects and animals have slippery self-cleaning surfaces which are able to repel water and exhibit anti-fouling properties. There are many species that exhibit surface properties such as secretion of wax, gum, oil, toxins or mucus for sustainability.1 Even invertebrates like mollusca and cnidaria exhibit surface properties and clean their bodies from soil, dust and debris by perpetually discharging mucus.2 The lotus leaves exhibit excellent water repellence by making the surface super hydrophobic using wax.3 The fouling of surfaces has wide effects. Various approaches have been developed to design anti-fouling surfaces. The fabrication of novel anti-fouling materials inspired by natural systems, using the self-assembly of synthetic organic moieties is highly interesting due to their potential application in bioorganic chemistry, medicine and material sciences.4,5 Recently Lynn's group have developed an anti-fouling coating that can host and sustain the release of water-soluble agents.6 Hozumi and coworkers have developed self-lubricating organogels with exceptional anti-sticking properties against viscous emulsions.7 The self-assembly process requires a combination of several noncovalent interactions8 such as hydrogen bonding, π–π stacking, and van der Waals interactions between the building blocks and can be tuned by different external stimuli like pH, concentration, heat, light radiation, addition of salts and solvent polarity.9–16Different building blocks such as urea, amides, peptides, nucleobases and carbohydrates have shown spectacular abilities to form supramolecular gels. The gels have wide applications such as removal of toxic dyes, oils and metal ions from water;17–24 as catalysts,25,26 dye sensitized solar cells,27 and matrices for nanoparticle synthesis;28–32 and in drug delivery systems.33–35 In this context, a small peptide scaffold that can serve as a hydrogen bond donor or acceptor is highly interesting.36 In particular, we are interested in tripeptide-based scaffolds due to their structural versatility, biocompatibility, robustness, and accessibility by standard analytical methods.37
Herein, we explore the use of tripeptides as building blocks for the formation of functional bioinspired materials. X-ray crystallography sheds some light on the conformations and self-assembly propensities of the tripeptides. Peptide 1 containing L-Phe forms a ten-member intramolecular hydrogen-bonded β-turn conformation and self-assembles through intermolecular hydrogen bonds to form supramolecular sheet-like structures. Whereas the tyrosine analogue adopts a kink-like conformation and self-assembles to form a supramolecular helical architecture. Moreover, the hydrophobic peptide 1 further self-assembles to form slippery nanofibers. The resulting surfaces show an anti-sticking effect against water and exhibit anti-fouling properties, like inhibiting the growth of Escherichia coli.
Results and discussion
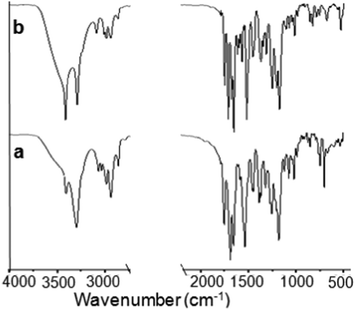
We have designed and synthesized two tripeptides. The peptides contain L-Ala at the N-terminus, a helicogenic Aib residue at the center and Phe/Tyr at the C-terminus (Scheme 1).38 The central helicogenic Aib residue may help to form a turn-like structure. The strong intermolecular hydrogen bonding and π–π stacking interactions between the side chains may dictate the assembly pattern. Target tripeptides 1 and 2 were synthesized by conventional solution phase reaction using DCC/HOBT as the coupling reagent. The final compounds were purified by column chromatography and characterized by 1H-NMR, 13C-NMR, FT-IR spectroscopy and mass spectrometry.First, the self-assembly of the tripeptides was examined by solid FT-IR spectroscopy. In FT-IR, the region 3500–3200 cm−1 is responsible for the N–H stretching vibrations and 1800–1600 cm−1 corresponds to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the amide and ester groups. The FT-IR spectra (Fig. 1) of the peptides 1 and 2 exhibit N–H stretching frequencies at 3270 and 3356 cm−1 respectively, responsible for hydrogen bonded N–H stretching. The band at 3450 cm−1 appeared for non hydrogen-bonded N–H. For tripeptide 1, the ester and amide peaks appeared at 1682 and 1630 cm−1. The tripeptide 2 exhibits peaks at 1690 and 1645 cm−1. We conclude that both tripeptides 1 and 2 have similar hydrogen bonded structures in the solid state.
O stretching vibration of the amide and ester groups. The FT-IR spectra (Fig. 1) of the peptides 1 and 2 exhibit N–H stretching frequencies at 3270 and 3356 cm−1 respectively, responsible for hydrogen bonded N–H stretching. The band at 3450 cm−1 appeared for non hydrogen-bonded N–H. For tripeptide 1, the ester and amide peaks appeared at 1682 and 1630 cm−1. The tripeptide 2 exhibits peaks at 1690 and 1645 cm−1. We conclude that both tripeptides 1 and 2 have similar hydrogen bonded structures in the solid state.
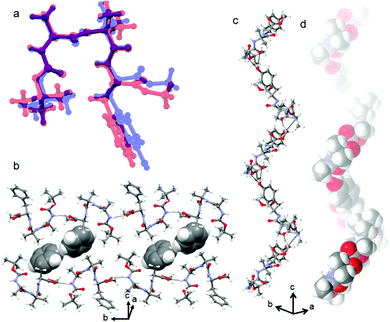
Furthermore, the structure and self-assembly pattern of the tripeptides 1 and 2 have been studied by single-crystal X-ray diffraction analysis. The colorless monoclinic crystals of tripeptides Boc-Ala-Aib-Phe-OMe (tripeptide 1) and Boc-Ala-Aib-Tyr-OMe (tripeptide 2) suitable for crystallography were obtained from methanol-water solution by slow evaporation. The molecular conformations of tripeptide 1 in the crystal state are illustrated in Fig. S1 (ESI†). Most of the backbone dihedral angle ϕ and ψ values (except ϕ1 and ψ3) of the constituent amino acid residues of tripeptide 1 fall within the helical region of the Ramachandran plot. The important backbone dihedral angles of tripeptide 1 are listed in Table S1 (ESI†). The tripeptide 1 forms an intramolecular hydrogen bonded β-turn conformation (N3–H3⋯O2) (Table S2, ESI†). The ORTEP diagram of peptide 2 with the atom numbering scheme is presented in Fig. S2 (ESI†). In the crystal, peptide 2 is unable to form any intramolecularly hydrogen bonded β-turn structure though the ϕ and ψ values of the constituent amino acid residues fall within the type II β-turn region of the Ramachandran map. The important backbone dihedral angles of tripeptide 2 are listed in Table S4 (ESI†). We have compared the molecular conformations of peptide 1 and peptide 2 (Fig. 2a). From Fig. 2a, the backbone conformations of peptides 1 and 2 are almost the same. The β-turn conformations of tripeptide 1 further self-assemble through the intermolecular hydrogen bond (N2–H2⋯O4) along the crystallographic b axis to form an anti-parallel sheet-like structure (Fig. 2b).39 This anti-parallel sheet-like structure further self-assembled through van der interactions and edge to edge π–π stacking interactions (shortest C–C distance 4.29 Å) along the crystallographic b and c directions (Fig. 2b).39 The hydrogen bonding parameters of peptide 1 and 2 are listed in Tables S2 and S3 (ESI†), respectively. The tripeptide 2 forms an intermolecular hydrogen bonded supramolecular helical structure along the crystallographic c axis (Fig. 2c).40 There are three intermolecular hydrogen bonds N2–H2⋯O4 and O5–H5⋯O3 (Table S3, ESI†) connecting individual peptide molecules to form the supramolecular helical columns. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and NH groups of Ala and Aib are engaged in intermolecular hydrogen bonding leaving the same hydrogen bonding functionalities of the Tyr residue uninvolved. However, the Tyr side chain OH is in hydrogen bonding with Ala C
O and NH groups of Ala and Aib are engaged in intermolecular hydrogen bonding leaving the same hydrogen bonding functionalities of the Tyr residue uninvolved. However, the Tyr side chain OH is in hydrogen bonding with Ala C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and helps the supramolecular helix formation. The crystal and data collection parameters of peptides 2 are listed in Table S6 (ESI†). Hence, the peptide 1 forms a supramolecular sheet-like structure and peptide 2 forms a supramolecular helix-like structure in higher order assembly.41
O and helps the supramolecular helix formation. The crystal and data collection parameters of peptides 2 are listed in Table S6 (ESI†). Hence, the peptide 1 forms a supramolecular sheet-like structure and peptide 2 forms a supramolecular helix-like structure in higher order assembly.41
Moreover, tripeptide 1 forms a transparent gel in toluene, xylene, chlorobenzene and 1,2-dichlorobenzene (Fig. S3, ESI†). The stuff was preliminarily categorized as a gel as it did not obey gravitational flow upon turning the tube upside-down at room temperature. The minimum gelation concentration (MGC) is 5 mg mL−1 in 1,2-dichlorobenzene. But, gelation does not occur for tripeptide 2 under the same conditions even at high concentration.
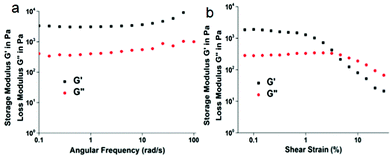
The rheological experiments confirmed the formation of an organogel by tripeptide 1. From angular frequency measurement at fixed shear strain 0.1%, the storage modulus (G′) was found to be larger than the loss modulus (G′′) at room temperature (Fig. 3a). This indicates that an entangled fiber network is formed. From shear strain measurement at 25 °C and constant angular frequency (10 rad s−1) LVER 0.15, G′ and G′′ cross over at 3.9% (Fig. 3b). This indicates the formation of a physically cross-linked organogel through non-covalent interactions.
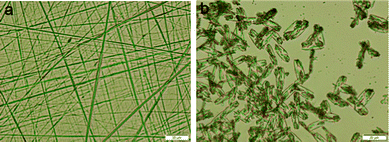
Initially, the morphology of the tri-peptide organogel was studied by polar optical microscope (POM). A thin slice of the peptide 1 gel was placed on a microscopic glass slide and it was allowed to dry under reduced pressure at room temperature for 24 h. The POM image shows that peptide 1 forms a wire-like morphology (Fig. 4a). The POM image shows that peptide 2 forms a flake-like morphology (Fig. 4b).
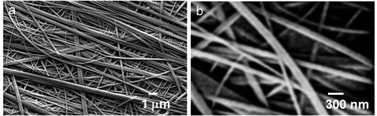
Furthermore, field-emission scanning electron microscopy (FE-SEM) has performed to study the morphology of the tripeptide 1 xerogel. Fig. 5a shows that the peptide forms an entangled fibre-like morphology in gel. The fibers are polydisperse in nature. Fig. 5b shows that the average diameter of peptide 1 fibers is 200 nm and several micrometers long.
The organogel formed by tripeptide 1 in 1,2-dichlorobenzene is so stable that the gel could be suspended by holding one edge and can be shaped into any self-supporting geometrical form.42 Also, a big organogel block of tripeptide 1 can be chopped into multiple pieces.43 The organogel formed by tripeptide 1 showed significant self-healing nature. Self-healing is a process of spontaneous formation of a new self-assembling structure when old structures are deceased within a gel. The optical microscopic image exhibits a gradual microscopic healing of a cut gel surface, although the damaged area was still visible (Fig. S4, ESI†). This self-healing experiment exhibits that the organogel is living and dynamic in nature. The FT-IR spectra (Fig. S5, ESI†) of the peptide 1 xerogel exhibit N–H stretching frequency at 3294 cm−1 responsible for hydrogen bonded N–H stretching. The band at 3412 cm−1 appeared for non hydrogen-bonded N–H. For tripeptide 1, the ester and amide peaks appeared at 1694 and 1654 cm−1. To identify the driving force of peptide 1 self-assembly, concentration dependent NMR studies were performed on peptide 1 dissolved in deuterated benzene. The concentration dependent 1H NMR study in C6D6 exhibits downfield shift of the Ala NH and Aib NH protons with increasing concentration (Fig. S6, ESI†), which suggests that the NH protons are intermolecularly hydrogen bonded. There is no shift for Phe NH, which suggest that the Phe NH proton is intramolecularly hydrogen bonded. This is consistent with the solid state structure. Moreover, we have performed CD spectroscopy. The CD spectrum of peptide 1 (Fig. S7, ESI†) has a positive band at 228 nm and negative bands at 216 and 255 nm.
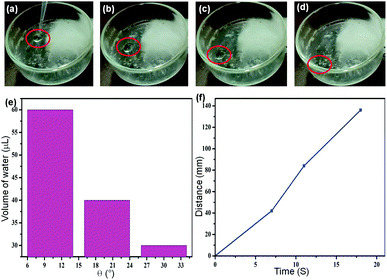
Moreover, the inherent surface properties of the bioinspired tripeptide 1 gel were examined by water adhesion experiment.6 As shown in Fig. 6a–d, the water droplet (10 μL) slid off the organogel surface at the tilt angle of 20°. The organogel consists of mobile and non-crosslinked components (aromatic solvents in the gel matrices) entrapped within a three-dimensional physically crosslinked gelator network (solid phase). The mobile and solid phases are miscible and the free energy of mixing (ΔG = ΔH − TΔS) is negative (ΔG < 0 for homogenious gel).15 But, cross-linking of the gel fibers on gelation enhances ΔG, which may trigger demixing (ΔG becomes >0). Hence the gel surface is dynamic in nature. It has been observed that the velocity of water droplets on inclined organogel surfaces markedly increased with the increasing size of the impregnated water droplet as well as the tilting angle (Fig. 6b). Fig. 6c shows the plot of distance covered by the water droplet with respect to time.
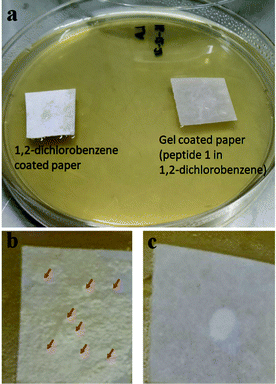
The tripeptide based gel shows an anti-fouling effect against bacteria. The tripeptide 1 and 1,2-dichlorobenzene gel coated paper shows resistance toward bacterial growth (Fig. 7). Fig. 7b shows the growth of Escherichia coli colonies on the paper surface coated with only 1,2-dichlorobenzene under aqueous media. Under the same conditions, the tripeptide 1 and 1,2-dichlorobenzene gel coated paper inhibits the growth of Escherichia coli on the paper surface (Fig. 7c).
Experimental
Materials
L-Amino acids (L-alanine, L-tyrosine, L-phenylalanine) and α-aminoisobutyric acid were supplied by Sigma chemicals. We have purchased HOBt (1-hydroxybenzotriazole) and DCC (N,N′-dicyclohexylcarbodiimide) from Sisco Research Laboratories (SRL). The solvents were supplied by Merck.Peptide synthesis
See ESI.†NMR experiments
NMR spectra of all the intermediate and final compounds were recorded with a JEOL 400 MHz or Bruker 500 MHz NMR spectrometer. We used CDCl3 or DMSO-d6 as the solvent and TMS as an internal standard.Conclusions
In conclusion, irrespective of the presence of the same peptide backbone, only the side chain hydroxyl functional group has introduced a huge change in the self-assembly and function of analogue peptides. From X-ray crystallography, tripeptide 1 with phenylalanine adopts a β-turn conformation and self-assembles through intermolecular hydrogen bonds to form a supramolecular hydrophobic sheet-like structure. Peptide 1 also forms slippery nanofibers and a gel in aromatic solvents. The resulting gel surfaces show anti-sticking effects against water and exhibit anti-fouling properties. The gel surface inhibits the growth of Escherichia coli colonies. Whereas the tyrosine analogue adopts kink like conformations and self-assembles to form a supramolecular helix-like architecture and failed to form a gel. This information may be helpful for fabricating new bioinspired supramolecular functional materials.Author contributions
K. C. H., S. K. and D. H. planned the experiments. K. C. H. and S. K. carried out the synthesis and experiments. S. M., S. S., A. S. and S. K. N. helped to analyse the data. K. C. H., S. K. and D. H. wrote the paper and S. M., S. S., A. S. and S. K. N. reviewed the paper. All the authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
K. C. Hati thanks Indian Institute of Science Education and Research Kolkata, India for the fellowship. S. Kumar, S. Mondal, S. Singh and S. K. Nandi acknowledge CSIR, India for research fellowships. A. Shit thanks UGC, India for fellowship. We thank Indian Institute of Science Education and Research Kolkata, India for funding and analytical facilities.Notes and references
- T. Darmanin and F. Guittard, Mater. Today, 2015, 18, 273 CrossRef CAS.
- J. G. Tidball, in Biology of the Integument, ed. J. Bereiter-Hahn, A. G. Matoltsy and K. S. Richards, Springer, Berlin, 1984, pp. 69–78 Search PubMed.
- C. Neinhuis, K. Koch and W. Barthlott, Planta, 2001, 213, 427 CrossRef CAS PubMed.
- M. Liu, S. Wang and L. Jiang, Nat. Rev. Mater., 2017, 2, 17036 CrossRef CAS.
- (a) P. Zhang, C. Zhao, T. Zhao, M. Liu and L. Jiang, Adv. Sci., 2019, 6, 1900996 CrossRef CAS PubMed; (b) R. Dou, J. Chen, Y. Zhang, X. Wang, D. Cui, Y. Song, L. Jiang and J. Wang, ACS Appl. Mater. Interfaces, 2014, 6, 6998 CrossRef CAS PubMed.
- H. Agarwal, T. J. Polaske, G. Sánchez-Velázquez, H. E. Blackwell and D. M. Lynn, Chem. Commun., 2021, 57, 12691 RSC.
- S. Hecht and I. Huc, Foldamers: Structure, Properties, and Applications, Wiley-VCH, Weinheim, 2007 Search PubMed.
- (a) D. Haldar and C. Schmuck, Chem. Soc. Rev., 2009, 2, 363 RSC; (b) M. Albrecht, Chem. Rev., 2001, 101, 3457 CrossRef CAS PubMed; (c) J. Li, J. A. Wisner and M. C. Jennings, Org. Lett., 2007, 9, 3267 CrossRef CAS PubMed; (d) H. Ito, Y. Furusho, T. Hasegawa and E. Yashima, J. Am. Chem. Soc., 2008, 130, 14008 CrossRef CAS PubMed; (e) D. Haldar, H. Jiang, J. M. Léger and I. Huc, Angew. Chem., Int. Ed., 2006, 45, 5483 CrossRef CAS PubMed; (f) B. Baptiste, J. Zhu, D. Haldar, B. Kauffmann, J. M. Léger and I. Huc, Chem. – Asian J., 2010, 5, 1364 CAS.
- (a) T. Tu, W. Fang, X. Bao, X. Li and K. H. Dotz, Angew. Chem., Int. Ed., 2011, 50, 6601 CrossRef CAS PubMed; (b) N. Fujita, Y. Sakamoto, M. Shirakawa, M. Ojima, A. Fujii, M. Ozaki and S. Shinkai, J. Am. Chem. Soc., 2007, 129, 4134 CrossRef CAS PubMed.
- (a) J. J. D. Jong, L. N. Lucas, R. M. Kellogg and J. H. V. Esch, Science, 2004, 304, 278 CrossRef PubMed; (b) W. Cai, G. T. Wang, Y. X. Xu, K. Jiang and Z. T. Li, J. Am. Chem. Soc., 2008, 130, 6936 CrossRef CAS PubMed.
- O. J. Dautel, M. Robitzer, J. P. L. Porte, F. S. Spirau and J. J. E. Moreau, J. Am. Chem. Soc., 2006, 128, 16213 CrossRef CAS PubMed.
- G. Cravotto and P. Cintas, Chem. Soc. Rev., 2009, 38, 2684 RSC.
- C. Dou, D. Li, H. Gao, C. Wang, H. Zhang and Y. Wang, Langmuir, 2010, 26, 2113 CrossRef CAS PubMed.
- K. Isozaki, H. Takaya and T. Naota, Angew. Chem., Int. Ed., 2007, 46, 2855 CrossRef CAS PubMed.
- A. Ajayaghosh, P. Chithra and R. Varghese, Angew. Chem., Int. Ed., 2007, 46, 230 CrossRef CAS PubMed.
- H. J. Kim, J. H. Lee and M. Lee, Angew. Chem., Int. Ed., 2005, 44, 5810 CrossRef CAS PubMed.
- S. Basak, J. Nanda and A. Banerjee, J. Mater. Chem., 2012, 22, 11658 RSC.
- D. M. Wood, B. W. Greenland, A. L. Acton, F. Rodr′ıguez- Llansola, C. A. Murray, C. J. Cardin, J. F. Miravet, B. Escuder, I. W. Hamley and W. Hayes, Chem. – Eur. J., 2012, 18, 2692 CrossRef CAS PubMed.
- S. Basak, N. Nandi, S. Paul, I. W. Hamley and A. Banerjee, Chem. Commun., 2017, 53, 5910 RSC.
- B. O. Okesola and D. K. Smith, Chem. Commun., 2013, 49, 11164 RSC.
- P. Chakraborty, B. Roy, P. Bairi and A. K. Nandi, J. Mater. Chem., 2012, 22, 20291 RSC.
- S. Sengupta and R. Mondal, J. Mater. Chem. A, 2014, 2, 16373 RSC.
- A. Chakrabarty, U. Maitra and A. D. Das, J. Mater. Chem., 2012, 22, 18268 RSC.
- N. Cheng, Q. Hu, Y. Guo, Y. Wang and L. Yu, ACS Appl. Mater. Interfaces, 2015, 7, 10258 CrossRef CAS PubMed.
- M. Tena-Solsona, J. Nanda, S. D′ıaz-Oltra, A. Chotera, G. Ashkenasy and B. Escuder, Chem. – Eur. J., 2016, 22, 6687 CrossRef CAS PubMed.
- B. Escuder, F. Rodríguez-Llansola and J. F. Miravet, New J. Chem., 2010, 34, 1044 RSC.
- W. Kubo, T. Kitamura, K. Hanabusa, Y. Wada and S. Yanagida, Chem. Commun., 2002, 374 RSC.
- J. H. Jung, H. Kobayashi, M. Masuda, T. Shimizu and S. Shinkai, J. Am. Chem. Soc., 2001, 123, 8785 CrossRef CAS PubMed.
- E. D. Sone, E. R. Zubarev and S. I. Stupp, Angew. Chem., Int. Ed., 2002, 41, 1705 CrossRef CAS.
- S. Kobayashi, N. Hamasaki, M. Suzuki, M. Kimura, H. Shinkai and K. Hanabusa, J. Am. Chem. Soc., 2002, 124, 6550 CrossRef CAS PubMed.
- P. Xue, R. Lu, Y. Huang, M. Jin, C. Tan, C. Bao, Z. Wang and Y. Zhao, Langmuir, 2004, 20, 6470 CrossRef CAS PubMed.
- S. Kobayashi, K. Hanabusa, N. Hamasaki, M. Kimura, H. Shirai and S. Shinkai, Chem. Mater., 2000, 12, 1523 CrossRef CAS.
- Y. Yamada and J. P. Schneider, Biomacromolecules, 2016, 17, 2634 CrossRef CAS PubMed.
- S. R. Jadhav, B. S. Chiou, D. F. Wood, G. DeGrande Hoffman, G. M. Glenn and G. John, Soft Matter, 2011, 7, 864 RSC.
- K. Basu, A. Baral, S. Basak, A. Dehsorkhi, J. Nanda, D. Bhunia, S. Ghosh, V. Castelletto, I. W. Hamley and A. Banerjee, Chem. Commun., 2016, 52, 5045 RSC.
- C. H. Gorbitz, Chem. – Eur. J., 2007, 13, 1022 CrossRef PubMed.
- (a) Y. Liu, Y. Wang, L. Jin, T. Chen and B. Yin, Soft Matter, 2016, 12, 934 RSC; (b) E. Azuma, K. Kuramochi and K. Tsubaki, Chem. Pharm. Bull., 2010, 58, 680 CrossRef CAS PubMed; (c) D. Podder, S. Roy Chowdhury, S. K. Nandi and D. Haldar, New J. Chem., 2019, 43, 3743 RSC; (d) S. Basak, N. Nandi, S. Paul, I. W. Hamley and A. Banerjee, Chem. Commun., 2017, 53, 5910 RSC.
- A. K. Das, A. Banerjee, M. G. B. Drew, S. Ray, D. Haldar and A. Banerjee, Tetrahedron, 2005, 61, 5027 CrossRef CAS.
- A. Banerjee, S. K. Maji, M. G. B. Drew, D. Haldar and A. Banerjee, Tetrahedron Lett., 2003, 44, 6741 CrossRef CAS.
- D. Haldar, S. K. Maji, M. G. B. Drew, A. Banerjee and A. Banerjee, Tetrahedron Lett., 2002, 43, 5465–5468 CrossRef CAS.
- M. A. Spackman and J. J. McKinnon, CrystEngComm, 2002, 4, 378 RSC.
- A. Vidyasagar, K. Handore and K. M. Sureshan, Angew. Chem., Int. Ed., 2011, 50, 8021 CrossRef CAS PubMed.
- T. Das, M. Haring, D. Haldar and D. D. Diaz, Biomater. Sci., 2018, 6, 38 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S9, Table S1–S6, Synthesis, and characterizations (Fig. S10–S16). CCDC 2106154 and 2106153. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2ma00084a |
| ‡ Have equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |