Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Deep generative models for peptide design
Fangping
Wan†
 abc,
Daphne
Kontogiorgos-Heintz†
abcd and
Cesar
de la Fuente-Nunez
abc,
Daphne
Kontogiorgos-Heintz†
abcd and
Cesar
de la Fuente-Nunez
 *abc
*abc
aMachine Biology Group, Departments of Psychiatry and Microbiology, Institute for Biomedical Informatics, Institute for Translational Medicine and Therapeutics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA. E-mail: cfuente@upenn.edu
bDepartments of Bioengineering and Chemical and Biomolecular Engineering, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania, USA
cPenn Institute for Computational Science, University of Pennsylvania, Philadelphia, Pennsylvania, USA
dDepartment of Computer and Information Science, School of Engineering and Applied Science, University of Pennsylvania, Philadelphia, Pennsylvania, USA
First published on 31st March 2022
Abstract
Computers can already be programmed for superhuman pattern recognition of images and text. For machines to discover novel molecules, they must first be trained to sort through the many characteristics of molecules and determine which properties should be retained, suppressed, or enhanced to optimize functions of interest. Machines need to be able to understand, read, write, and eventually create new molecules. Today, this creative process relies on deep generative models, which have gained popularity since powerful deep neural networks were introduced to generative model frameworks. In recent years, they have demonstrated excellent ability to model complex distribution of real-word data (e.g., images, audio, text, molecules, and biological sequences). Deep generative models can generate data beyond those provided in training samples, thus yielding an efficient and rapid tool for exploring the massive search space of high-dimensional data such as DNA/protein sequences and facilitating the design of biomolecules with desired functions. Here, we review the emerging field of deep generative models applied to peptide science. In particular, we discuss several popular deep generative model frameworks as well as their applications to generate peptides with various kinds of properties (e.g., antimicrobial, anticancer, cell penetration, etc). We conclude our review with a discussion of current limitations and future perspectives in this emerging field.
Introduction
Defined as short chains of amino acids with lengths ranging from 2 to 50, peptides can act as hormones, antimicrobials, or delivery vehicles1,2 and have gained great interest due to their potential as therapeutic drugs.1,3 Indeed, we have witnessed an increasing number of approvals of peptide therapeutics over the past 60 years, with 52 peptide drugs approved in this century.4 Furthermore, over 150 and 500 peptides are in clinical trials and under pre-clinical development,4 respectively. Despite the potential therapeutic value of peptides,5–7 designing them to achieve specific properties or functions remains a challenge. The exponential search space of peptides (e.g., 20L possible peptides taking into account the 20 naturally occurring, canonical amino acids and length L) in addition to the high cost and time-consuming process associated with experimental validation8–12 pose serious issues when developing peptide drugs.In recent years, a data-driven paradigm has emerged by combining deep learning and advanced computational resources [i.e., graphics processing units (GPUs)], revolutionizing a number of fields, including computer vision,13 natural language processing (NLP),14 game playing,15 and computational biology.16–19 As universal approximators,20 deep neural networks have demonstrated superior abilities in modelling complex real-world data, including extracting high-level features from raw inputs,21,22 making predictions,13 fitting data distributions, and generating novel data.23–25 Among various kinds of deep learning frameworks, deep generative models differ from classification and regression models (i.e., discriminative models) in their ability to model the distribution of data using deep neural networks. Consequently, properly trained deep generative models can be used to (1) assign a likelihood/probability to measure if a novel data point is drawn from the data distribution, (2) sample and/or generate novel data points that possess similar properties to those present in the training data, and (3) extract expressive data representations23,24 (i.e., feature learning) or perform casual inference26 (e.g., determine the casual factor that lead to the generation of data) by specifying the generation process of data. In a number of application fields, deep generative models have exhibited superior performance in generating complicated and high-dimensional data, including realistic images,27,28 syntactically correct programming language codes,29 and drug-like molecules.30 Deep generative models could potentially serve as a new tool to efficiently explore the vast peptide sequence space and facilitate the peptide design process by prioritizing promising peptides for experimental validation.
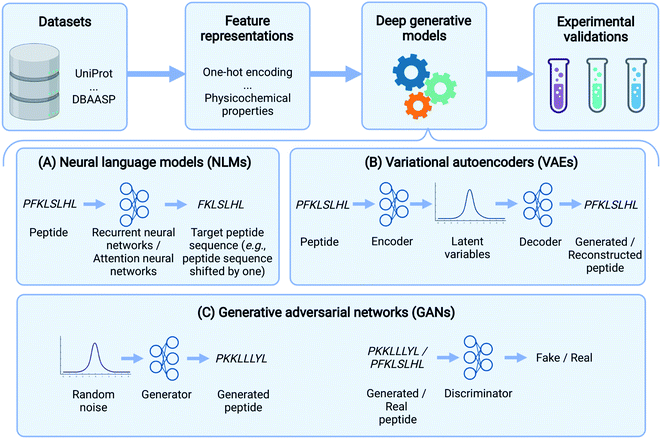
In the past few years, deep learning methods have also been widely used in peptide science to perform various tasks like peptide identification, property prediction, and peptide generation.31 Specifically, for peptide generation, deep generative models have been used to generate peptides with a range of activities, including the following: antimicrobial, anticancer, immunogenic, ability to deliver other molecules, and signal peptides (Table 1). Here, we comprehensively review these deep generative models for peptide generation. Several reviews have described the use of deep generative models for proteins.32,33 Our paper, instead, focuses exclusively on peptide design. We should also point out that the focus of deep generative models on peptide design inevitably leads to a neglect of some generative models including flow-based34 and energy-based models.35 We refer the reader to a recent review33 for further details. Specifically, the rest of the paper is organized as follows: Before delving into specific generative algorithms, we first describe several commonly used datasets and feature representations of peptides. We then provide an overview of three popular deep generative model frameworks used in peptide generation: neural language models (NLMs), variational autoencoders (VAEs), and generative adversarial networks (GANs) (Fig. 1). The basic concepts of these models, as well as those of their variants, will be introduced and their applications in peptide generation will be reviewed. Finally, we conclude by discussing outstanding challenges and future directions in this exciting field. Although this review is primarily aimed at readers with a basic understanding of machine learning and deep neural networks, our goal is to reach a broader readership. Therefore, a glossary is provided (Table 2) to cover definitions of key machine learning terms that are not strictly defined in main text.
| Method | Feature Representation | Application | Citation | Year |
|---|---|---|---|---|
| NML | One-hot | AMP generation | Müller et al.56 | 2018 |
| NLM | Character sequence | AMP generation | Nagarajan et al.52 | 2018 |
| NLM | Character sequence | ACP generation | Grisoni et al.46 | 2018 |
| NLM | Learned representation using one-hot | Signal peptide generation | Wu et al.55 | 2020 |
| NLM | Learned representation using structural and evolutionary data | AMP generation | Caceres-Delpiano et al.54 | 2020 |
| NLM | One-hot | AMP generation | Wang et al.41 | 2021 |
| NLM | Character sequence | CPP generation | Tran et al.53 | 2021 |
| NLM | One-hot | AMP generation | Capecchi et al.42 | 2021 |
| NLM | Fingerprint, one-hot | PMO delivery peptide generation | Schissel et al.57 | 2021 |
| VAE | Learned representation using character sequence | AMP generation | Das et al.38 | 2018 |
| VAE | Learned representation using one-hot | AMP generation | Dean et al.44 | 2020 |
| VAE | Learned representation using character sequence | AMP generation | Das et al.45 | 2021 |
| GAN | Character sequence | AMP generation | Tucs et al.50 | 2020 |
| GAN | Character sequence/PDB structure | ACP generation | Rossetto et al.48 | 2020 |
| GAN | Learned representation using character sequence | AMP generation | Ferrell et al.39 | 2020 |
| GAN | Character sequence | AMP generation | Oort et al.40 | 2021 |
| GAN | Sequence of amino acid property vectors | Immunogenic peptide generation | Li et al.51 | 2021 |
| GAN | Character sequence | AMP generation | Surana et al.49 | 2021 |
| Term | Meaning |
|---|---|
| Active learning | Algorithms to query data samples from a dataset for labelling and model training so that the trained model has maximum performance gain |
| Backpropagation | An algorithm that trains neural networks. Gradients of the loss with respect to parameters in a neural net are first calculated by the chain rule. Then, gradient descent is performed to optimize the parameters |
| Bayesian optimization | A sequential search algorithm to optimize computationally expensive black-box functions |
| Bidirectional encoder representations from transformers | Transformer-based language model for feature/representation learning |
| Convolutional neural network | A class of neural networks that uses a series of convolution operations and nonlinear transformations to process structured inputs (e.g., images and sequences) and make predictions |
| Data augmentation | Supplementing a dataset with modified copies of the data or with synthetic data, often used to prevent overfitting and improve prediction performance |
| Decoder | A neural network converts compressed signals/features (usually represented as low-dimensional vectors) to raw signals |
| Embedding | A mapping from a high-dimensional input to a low-dimensional vector |
| Encoder | A neural network converts raw inputs/signals to compressed signals/features (usually represented as low-dimensional vectors) |
| Gradient | In machine learning, it usually refers to the partial derivative of loss with respect to machine learning model (e.g., neural network) parameters |
| Gradient descent | An optimization algorithm to minimize a differentiable function by iteratively moving in the opposite direction of the gradient of the function |
| Label | The answer of what machine learning models aim to predict |
| Loss | A measure between label and machine learning model prediction, indicating how far the prediction is from the corresponding label |
| Low-shot learning | Machine learning approaches that train effective models using a small number of training samples. Also known as few-shot learning |
| Multitask learning | Machine learning approaches that train models to solve multiple tasks simultaneously |
| Neural network | A model inspired by the brain that uses a series of nonlinear transformations to process inputs and make predictions |
| Objective | A value (e.g., loss) that machine learning models aim to minimize/maximize |
| One-hot encoding | A vector representation of categorical data, wherein all values are 0 except the singular 1 |
| Recurrent neural network | A class of neural networks that are suitable for modelling time series and sequential data by using the output from the last time step as input to the current time step |
| Training | A process to optimize parameters of machine learning models so that objective(s) are minimized/maximized |
| Training data/dataset/set | Data used to train machine learning models |
| Transfer learning | Machine learning approaches that transfer information/knowledge from one task to the other in order to improve prediction performance of models |
| Transformer | A neural network architecture based on attention for sequence-to-sequence learning |
Datasets
While there are publicly available datasets for protein informatics with labeled protein activity, their size is limited, particularly compared with the extensive datasets available in NLP and computer vision. The scarcity of data in this case is primarily due to the cost associated with running wet-lab experiments. For example, WordNet has over 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 well-organized words, and ImageNet has over 14 million labeled images for object recognition.36,37 On the contrary, as shown in Table 3, the size of databases with labeled data used by many of the studies covered in this review are in the order of hundreds or thousands. Further, the actual size of the datasets used in these studies is usually much smaller, after filtering for how activity was measured, specific targets, peptide length, presence or absence of non-canonical residues, unique sequences, synthesis difficulty, and other properties that help build a high-quality dataset. For properties such as antimicrobial activity, labeled negative data are often even more scarce than positive. To address the negative data scarcity problem and improve the generalizability of these models, many studies39–42 have resorted to using unlabeled proteins from databases like UniProt43 and DBAASP73 or randomly generated sequences42 as negative examples. The validity of this method is supported by Wang et al.41 To best address this issue, however, the field relies on experimentalists collecting and sharing data on peptides negative data on peptides. As an alternative to assuming unlabeled data is negative, unsupervised learning can take advantage of unlabeled data to learn feature representations. For example, Das et al.45 trained a general peptide generation model based on peptides from UniProt, whereas Grisoni et al.46 and Capecchi et al.42 incorporated transfer learning to be able to utilize more data.
000 well-organized words, and ImageNet has over 14 million labeled images for object recognition.36,37 On the contrary, as shown in Table 3, the size of databases with labeled data used by many of the studies covered in this review are in the order of hundreds or thousands. Further, the actual size of the datasets used in these studies is usually much smaller, after filtering for how activity was measured, specific targets, peptide length, presence or absence of non-canonical residues, unique sequences, synthesis difficulty, and other properties that help build a high-quality dataset. For properties such as antimicrobial activity, labeled negative data are often even more scarce than positive. To address the negative data scarcity problem and improve the generalizability of these models, many studies39–42 have resorted to using unlabeled proteins from databases like UniProt43 and DBAASP73 or randomly generated sequences42 as negative examples. The validity of this method is supported by Wang et al.41 To best address this issue, however, the field relies on experimentalists collecting and sharing data on peptides negative data on peptides. As an alternative to assuming unlabeled data is negative, unsupervised learning can take advantage of unlabeled data to learn feature representations. For example, Das et al.45 trained a general peptide generation model based on peptides from UniProt, whereas Grisoni et al.46 and Capecchi et al.42 incorporated transfer learning to be able to utilize more data.
| Name | Citation | Labels | Data size | Application |
|---|---|---|---|---|
| Uniprot | The UniProt Consortium43 | Sparse labels | 190 million sequences | Wu et al.55 |
| Das et al.38 | ||||
| Capecchi et al.42 | ||||
| Das et al.45 | ||||
| Oort et al.40 | ||||
| CPPsite 2.0 | Agrawal et al.116 | Cell-penetrating peptides | 1850 peptides,1,150 used in application | Schissel et al.57 |
| Pfam | Bateman et al.117 | Sparse labels | 47 million sequences, 21 million used in application | Caceres-Delpiano et al.54 |
| DBAASP | Pirtskhalava et al.73 | Antimicrobial, toxicity, anticancer and hemolytic activity | >15, 700 peptides | Wang et al.41 |
| Tran et al.53 | ||||
| Capecchi et al.42 | ||||
| Das et al.45 | ||||
| Tucs et al.50 | ||||
| Ferrell et al.39 | ||||
| Oort et al.40 | ||||
| ToxinPred's dataset | Gupta et al.118 | Toxicity | 1805 toxic peptides | Das et al.45 |
| AVPdb | Qureshi et al.119 | Antiviral | 2683 peptides | Oort et al.,40 |
| Surana et al.49 | ||||
| LAMP | Zhao et al.120 | Antimicrobial activity | 5547 peptides | Tran et al.53 |
| Tucs et al.50 | ||||
| THPdb | Usmani et al.121 | FDA approved therapeutic peptides | 239 peptides | Rossetto et al.48 |
| CAMP | Thomas et al.122 | Antimicrobial activity | 3782 peptides | Wang et al.41 |
| Tran et al.53 | ||||
| Tucs et al.50 | ||||
| Surana et al.49 | ||||
| DRAMP | Kang et al.123 | Antimicrobial activity | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 899 peptides 899 peptides |
Wang et al.41 |
| Surana et al.49 | ||||
| YADAMP | Piotto et al.124 | Antimicrobial activity | 2133 peptides | Nagarajan et al.52 |
| Wang et al.41 | ||||
| DADP | Novkovi'c et al.125 | Broad defence activity | 2571 peptides | Müller et al.56 |
| ADP | Wang et al.126 | Antimicrobial activity | 3273 peptides | Müller et al.56 |
| Tran et al.53 | ||||
| Dean et al.44 | ||||
| Tucs et al.50 | ||||
| DBAMP | Jhong et al.127 | Antimicrobial activity | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 389 peptides 389 peptides |
Surana et al.49 |
| IEDB | Fleri et al.128 | Immune epitope | > 1 million peptides, 8971 used in application | Li et al.51 |
Feature representations
A critical component of any machine learning task is the feature selection and input representation. An extra constraint imposed on the representation schemes used in peptide generative tasks, which does not apply broadly to all peptide property prediction tasks, is the need for a mapping of the input representation back to the peptide sequence. For example, global properties and amino acid composition have been extensively used to represent peptides in property prediction tasks,47 but an inverse mapping of a peptide represented by global properties back to a sequence currently does not exist. However, some peptide generation studies have used representations that cannot be mapped directly to a unique sequence in property predictors to filter out those peptides generated that possess undesired properties. For instance, Rossetto et al.48 used a 4D tensor representing structural information to calculate a reward function for the generated peptide, and Surana et al.49 used physicochemical properties, amino acid composition, and structural information to analyze the generated peptides. Schemes commonly used to represent peptides in the generation task itself include direct sequence representation41,42,45,46,49–53 and learned embeddings.38,39,44,45,54,55A natural way to encode peptides is through their primary structure (i.e., amino acid sequence). A peptide of length L can be represented by a string of characters or integers of length L,39,45,46,49,50 or a L × n matrix such that each amino acid has a unique n-dimensional vector. The n-dimensional vector may represent either experimentally51 or computationally derived properties,45 or be a one-hot encoding.41,42,44,56 In a one-hot encoding, the ith amino acid from an alphabet of size n is represented by a vector containing n − 1 0s and a 1 at the index i. Some studies use amino acid alphabets that expand beyond the standard 20 amino acids to represent non-canonical amino acids, markers for modified peptide terminals (e.g., an acetylated N-terminal), and padding (to allow encoding peptides of different lengths). A one-hot encoding does not retain information about amino acids, such as the similarity between leucine and isoleucine; however, gains in model performance when using more complex representations may be limited.51 Schissel et al.57 represented amino acids as topological fingerprints. These fingerprints contain bits to represent the presence of substructures and hence retain structural information about each residue. The authors found that such an encoding scheme led to models with lower accuracies, but with an enhanced generalizability to peptides with labels outside the range of the training data. ElAbd et al.58 showed that deep learning models can learn the amino representation scheme, whether it is a one-hot encoding or even a random vector, provided that the random vector has sufficiently high dimensionality. A substantial flaw in one-hot encodings, however, is their high dimensionality.58 Instead of using single amino acid residues as the smallest unit within a sequence, a k-mer can be considered the fundamental unit. A k-mer is a short amino acid sequence of length k, such as a 3-mer. These k-mers can be represented with a one-hot encoding. Depending on the choice of k, the dimensionality of this representation can explode, because there are many possible k-length amino acid combinations. However, using an embedding can reduce the dimensionality by orders of magnitude and ameliorate this problem.59
In addition to sequence representation, studies have focused on learning a latent representation from a deep neural network, such as an autoencoder.38,44,45 This latent representation can be used solely for the purpose of sequence generation,44 or additionally in property prediction models.45 These studies all used variational autoencoders, which can learn both a feature encoding and a distribution of the inputs to then generate peptides retaining properties present within the training set. However, a classical autoencoder (i.e., deterministic autoencoder) – which learns only feature encoding and its mapping to and from the sequence – could be used for the purpose of representation learning alone. Further, latent spaces leveraged to represent peptides can be learned from other model architectures, such as a GAN60 or an attention network like Bidirectional Encoder Representations from Transformers (BERT).22
Deep generative models
In this section, we delineate the basic concepts underlying several deep learning-based generative models (Fig. 1) and summarize their applications for peptide generation (Table 1).Neural language models (NLMs)
In NLP, given a sequence of words x = (w1, w2, …, wn), language models estimate the probability distribution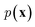 over it. How
over it. How 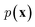 is factorized can reflect the generation process of a sentence. Here, a popular choice (but not the only one) to specify the generation process of a word sequence is based on the chain rule of probability:
is factorized can reflect the generation process of a sentence. Here, a popular choice (but not the only one) to specify the generation process of a word sequence is based on the chain rule of probability: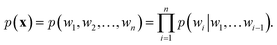 | (1) |
The models adopted in the generation process of eqn (1) are also known autoregressive models. That is, the next word wi is generated by taking all its previous words into account. In the context of deep learning, NLMs utilize neural networks to model the conditional probabilities of eqn (1). Here, we introduce two frameworks that are widely used in NLMs: recurrent neural networks (RNNs)61 and attention models.14,62 Note that it is also possible to use other network architecture like convolutional neural network as an autoregressive model.63
Specifically, RNNs such as long short-term memory (LSTMs)64 and gated recurrent unit (GRU)65 are commonly used to build autoregressive models as they model sequential data by storing the historical information (i.e., memory) into their hidden states. In the context of language modeling, RNNs are typically trained to predict the next word wi+1 given the current input word wi and the hidden state hi−1 that stores the information from word w1 to wi−1:
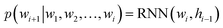 | (2) |
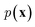 of a sequence of words in the training data. To generate novel sequences, a desired/random start word as well as a zero/random hidden state are first provided to RNNs to generate the distribution of the next word. Then, the same process is repeated by sampling a word from the predicted distribution and inputting it to the RNNs until some termination criterion is met (e.g., an “end” word is generated or a maximum generation length is reached).
of a sequence of words in the training data. To generate novel sequences, a desired/random start word as well as a zero/random hidden state are first provided to RNNs to generate the distribution of the next word. Then, the same process is repeated by sampling a word from the predicted distribution and inputting it to the RNNs until some termination criterion is met (e.g., an “end” word is generated or a maximum generation length is reached).
While RNNs implicitly model the long-range interactions of inputs by assuming the hidden states always retain all previous input information (eqn (2)), attention models utilize neural networks to directly model such interactions. Generally, given a feature sequence (x1, x2, …, xn), an attention model can be formulated as:
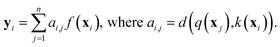 | (3) |
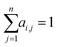 or treated as a parameter to sample a binary value (i.e., 0 or 1)66 as its replacement in eqn (3) (also known as hard attention). Unlike RNNs, attention models do not utilize the sequential information of input data. To address this issue, positional information like positional embedding14 can be incorporated into input features. To model the generation of a sequence, eqn (3) can be modified as:
or treated as a parameter to sample a binary value (i.e., 0 or 1)66 as its replacement in eqn (3) (also known as hard attention). Unlike RNNs, attention models do not utilize the sequential information of input data. To address this issue, positional information like positional embedding14 can be incorporated into input features. To model the generation of a sequence, eqn (3) can be modified as: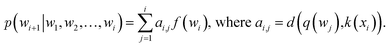 | (4) |
The above NLMs can be further extended to perform sequence to sequence (seq2seq) modeling,71 in which we aim to learn a mapping from a source sequence (x1, x2, …, xm) to a target sequence (y1, y2, …, yn). For instance, Winter et al.72 trained a RNN-based seq2seq method to obtain expressive latent features for compounds by translating the compounds' IUPAC representations to SMILES representations.
For peptide design and discovery, by treating peptides as sequences of amino acids, NLMs have been widely used to generate antimicrobial,41,42,52,54,56 anticancer,46 cell-penetrating,53 phosphorodiamidate morpholino oligomer (PMO) delivery,57 and signal55 peptides. To generate antimicrobial peptides (AMPs), previous work41,42,52,56 focused on using RNNs to model AMPs acquired from public datasets. Then, property filters like AMP predictors (i.e., trained classifiers or public prediction servers) were used to remove undesired peptide sequences generated by RNNs. In addition to these approaches, Capecchi et al.42 utilized a transfer learning strategy to fine tune the RNN model trained on DBAASP73 to two smaller non-hemolytic AMP datasets in order to address the scarcity of non-hemolytic AMP data. Taking a different approach, Caceres-Delpiano et al.54 adopted a multitask training strategy to simultaneously perform language modeling, secondary structure, contact map, and structure similarity prediction. After model training, novel peptide sequences were generated by mutating the target sequence and after meeting certain structure similarity and energy criteria based on the trained multitask model. Through wet-lab validation, Nagarajan,52 Capecchi et al.,42 and Caceres-Delpiano et al.54 managed to identify two, eight, and two novel AMPs, respectively. Grisoni46 also used transfer learning to fine tune the RNNs trained on 10K α-helical cationic amphipathic peptide sequences to 26 known anticancer peptides (ACPs). The authors further experimentally validated the anticancer activity of the ten generated peptides. Although both leveraged RNNs to generate cell-penetrating peptides (CPPs), Tran et al.53 used molecular dynamics simulations to prioritize generated CPPs for downstream validation, whereas Schissel et al.57 further utilized a deep learning-based PMO delivery predictor as well as a genetic algorithm to optimize the generated CPPs to PMO delivery peptides and demonstrated their safety and efficacy in animals. Moreover, Wu et al.55 formulated the signal peptide (SP) generation challenge as a machine translation problem (i.e., seq2seq), in which the transformer14 model was used to translate a mature protein with the SP sequence removed to the corresponding SP sequence. Furthermore, the authors validated the generated SPs by demonstrating their industrial-level enzyme secretion activity.
Variational autoencoders (VAEs)
VAEs are similar to classic autoencoders in terms of their model architecture, which typically consist of an encoder and a decoder neural network. However, they differ substantially in mathematical formulation. Merging probabilistic graphical models and deep learning, VAEs23,24 assume the data is generated from certain latent variables and provide a systematic approach to model data generation (i.e., decoding process) and infer the latent variables (i.e., encoding process) based on deep neural networks. On the one hand, the decoder neural network d(·) explicitly specifies how the observed data x can be generated given the latent variables z (i.e., x = d(z)) as well as the corresponding likelihood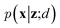 . On the other hand, given the observed data x, the encoder neural network e(·) is responsible for estimating posterior distribution
. On the other hand, given the observed data x, the encoder neural network e(·) is responsible for estimating posterior distribution 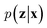 via variational posterior q(z|x;e), where z = e(x). Here, the introduction of low-dimensional latent variables subjected to certain prior distribution
via variational posterior q(z|x;e), where z = e(x). Here, the introduction of low-dimensional latent variables subjected to certain prior distribution 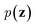 not only makes it easier to model high-dimensional data (e.g., images, text and biological sequences), but also allows users to draw samples
not only makes it easier to model high-dimensional data (e.g., images, text and biological sequences), but also allows users to draw samples 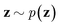 from the prior distribution and obtain novel data through decoding.
from the prior distribution and obtain novel data through decoding.
To train VAEs, we want to maximize the marginal likelihood with respect to the parameters of e(·) and d(·):
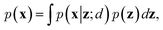 | (5) |
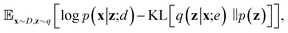 | (6) |
Since the introduction of VAEs, a number of variants were proposed to further enhance their modeling abilities. For instance, hierarchical VAEs introduced multiple layers of latent variables to create richer prior distributions.74,75 To incorporate supervised information into a VAE framework, conditional VAEs (CVAEs)76,77 introduced label variable y into the processes of data generation 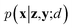 and posterior inference q(z|x,y;e). Instead of randomly sampling the latent variables from prior distribution, CVAEs explicitly specify the label variable during decoding and generate samples (i.e., conditional sampling) with the desired properties.78 Moreover, methods like maximum mean discrepancy VAE (MMD-VAE)79 and β-VAE80 replaced the original KL divergence term in eqn (6) with alternatives to encourage the models to learn more meaningful latent variables (i.e., features). Today, VAEs have been widely used in various types of tasks in computer vision,81,82 NLP,78,83 recommendation systems,84 and bioinformatics.17,30,85
and posterior inference q(z|x,y;e). Instead of randomly sampling the latent variables from prior distribution, CVAEs explicitly specify the label variable during decoding and generate samples (i.e., conditional sampling) with the desired properties.78 Moreover, methods like maximum mean discrepancy VAE (MMD-VAE)79 and β-VAE80 replaced the original KL divergence term in eqn (6) with alternatives to encourage the models to learn more meaningful latent variables (i.e., features). Today, VAEs have been widely used in various types of tasks in computer vision,81,82 NLP,78,83 recommendation systems,84 and bioinformatics.17,30,85
In the context of peptide generation, VAEs have mainly been used to discover AMPs.38,44,45 Dean et al.44 directly trained a VAE to model 6K peptides derived from an AMP database. Das et al.,38,45 on the other hand, utilized a larger unlabeled peptide dataset obtained from Uniprot to train the VAEs. Specifically, PepCVAE38 is a semi-supervised learning framework based on CVAE to jointly model 15K labeled AMPs and non-AMPs as well as 1.7M unlabeled peptide sequences from UniProt. By explicitly disentangling the AMP/non-AMP property from other latent features, PepCVAE was able to control the generation of peptides with desired AMP/non-AMP properties. Although trained on similar data, the VAE framework described in Das et al.45 improved the peptide generation process by replacing KL divergence loss with MMD loss to get more meaningful latent representations of peptides. To sample peptides with desired properties (e.g., AMPs and low toxicity), the authors first fitted a Gaussian mixture density estimator and linear property predictors on latent variables of labeled peptide data. Then, they developed a rejection sampling to sample desired latent variables from the density estimator with probability derived from the property predictors and obtained peptides by passing the sampled latent variables to the decoder of the VAE. Instead of sampling from prior distribution, the authors argued that the learnt distribution of latent variables q(z|x;d) could be quite different from the prior distribution and therefore a separate density estimator was required. Das et al. also showed that the combination of their VAE framework with molecular dynamics simulations and wet-lab experimentation yielded two novel and safe AMPs within 48 days. These studies demonstrate the potential of VAEs in peptide drug discovery and development.
Generative adversarial networks (GANs)
Inspired by game theory, GANs25 assume a two-player zero-sum game scenario, in which a discriminator d aims to distinguish fake and real data while a generator g tries to generate fake data as realistic as possible in order to fool the discriminator. As this competition between generator g and discriminator d proceeds, an equilibrium occurs where the fake examples generated by g are indistinguishable from real ones and d can only take a random guess as to whether a given example is real or not. In such an equilibrium, no further improvement can be made for g and d, and consequently we consider that the generator g captures the distribution of real data. In practice, both generator g and discriminator d are usually implemented as deep neural networks. For generator neural network g(·), it typically takes a random vector z drawn from a noise distribution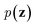 (e.g., standard Gaussian distribution) as input and outputs the generated data xfake = d(z). For classic discriminator neural network d(·)→(0,1) (i.e., a classifier), it receives both real samples xreal drawn from training dataset
(e.g., standard Gaussian distribution) as input and outputs the generated data xfake = d(z). For classic discriminator neural network d(·)→(0,1) (i.e., a classifier), it receives both real samples xreal drawn from training dataset 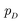 as well as fake samples xfake and classifies them to be real (i.e., d(xreal)→1) or not (i.e., d(xfake)→0), respectively. The above competition process between generator and discriminator can be mathematically formulated as a minimax loss function to optimize the learnable parameters of g(·) and d(·):
as well as fake samples xfake and classifies them to be real (i.e., d(xreal)→1) or not (i.e., d(xfake)→0), respectively. The above competition process between generator and discriminator can be mathematically formulated as a minimax loss function to optimize the learnable parameters of g(·) and d(·):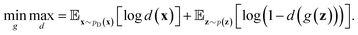 | (7) |
While the previously described NLMs and VAEs are explicit density models that specify the form of 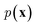 and optimize the models to maximize
and optimize the models to maximize 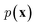 or its lower bound, GANs use implicit density models. From the model architecture and loss function shown in eqn (7), we can see that GANs do not fit and estimate the data distribution
or its lower bound, GANs use implicit density models. From the model architecture and loss function shown in eqn (7), we can see that GANs do not fit and estimate the data distribution 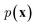 directly. However, the original GANs paper25 showed that training GANs implicitly minimized the Jensen-Shannon divergence (JSD) between generated data distribution
directly. However, the original GANs paper25 showed that training GANs implicitly minimized the Jensen-Shannon divergence (JSD) between generated data distribution 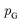 and real data distribution
and real data distribution 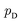 . Similar to VAEs, GANs can be extended to incorporate supervised information. By enabling generator x = g(z,y) and discriminator d(x,y)→(0,1) to take label information y into account, conditional GANs (CGANs)86,87 demonstrated their ability to generate data possessing desired properties. Without modifying the distribution of training data, CGANs and CVAEs encourages the models to draw samples with desired properties by introducing label information during sampling. FB-GAN,88 on the other hand, encourages the models to capture the sample distribution with desired properties by shifting the distribution of training data during model training. Specifically, an external property predictor is introduced to iteratively replace the data point with predicted undesired property with generated data point with predicted desired property.
. Similar to VAEs, GANs can be extended to incorporate supervised information. By enabling generator x = g(z,y) and discriminator d(x,y)→(0,1) to take label information y into account, conditional GANs (CGANs)86,87 demonstrated their ability to generate data possessing desired properties. Without modifying the distribution of training data, CGANs and CVAEs encourages the models to draw samples with desired properties by introducing label information during sampling. FB-GAN,88 on the other hand, encourages the models to capture the sample distribution with desired properties by shifting the distribution of training data during model training. Specifically, an external property predictor is introduced to iteratively replace the data point with predicted undesired property with generated data point with predicted desired property.
Although GANs have received great attention since they came out in 2014, problems like gradients vanishing89 and difficulty in converging to equilibrium90 make GANs notoriously difficult and unstable to train. To address these issues, a number of loss functions were proposed to replace the JSD.91–96 Among these variants, we discuss a widely adopted framework called Wasserstein GANs (WGANs).95 In WGANs, instead of being a binary classifier, the discriminator d(·) is used to measure the Wasserstein distance. Specifically, the Wasserstein distance (also known as earth mover's distance) of two distributions 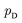 and
and 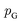 can be mathematically written as:
can be mathematically written as:
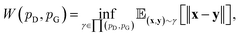 | (8) |
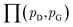 is the set of all joint distributions γ(x,y) whose marginal distributions are
is the set of all joint distributions γ(x,y) whose marginal distributions are 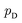 and
and 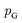 , respectively. Intuitively, Wasserstein distance measures the cost of transforming one distribution into another. This intractable distance
, respectively. Intuitively, Wasserstein distance measures the cost of transforming one distribution into another. This intractable distance 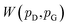 can be approximated by:
can be approximated by: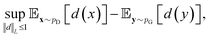 | (9) |
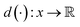 belongs to 1-Lipschitz functions. In the context of WGANs, d(·) is the discriminator and we can write the objective of WGANs as:
belongs to 1-Lipschitz functions. In the context of WGANs, d(·) is the discriminator and we can write the objective of WGANs as: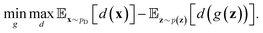 | (10) |
Note that the original discriminator of GANs is a binary classifier. On the other hand, in WGANs, the sigmoid function in the last layer of the discriminator should be removed in order to approximate the Wasserstein distance. In addition, the discriminator d(·) should satisfy the Lipschitz constraint. To this end, several approaches were proposed including weight clipping,95 gradient penalty,96 and spectral normalization.97 We refer the reader to these original papers for technical details. The smoother gradients provided by the loss function of WGANs can greatly improve the training process. Therefore, WGANs are widely used in various kinds of data generation tasks.88,98,99
Regarding peptide generation, several attempts have been made to utilize GANs and their variants to generate antimicrobial,39,40,49,50 anticancer,48 and immunogenic51 peptides. For example, upon training on 16K AMPs and 5K non-AMPs acquired from several datasets, PepGAN50 designed a mixed loss function to utilize AMP label information so that the discriminator was encouraged to distinguish real AMP sequences. Through experimental validation, the authors further showed that PepGAN was able to generate an AMP twice as strong as the conventional antibiotic ampicillin. In addition, AMPGAN39 and the follow-up model AMPGAN v240 adopted a bidirectional CGAN framework to generate peptides with desired targets, mechanisms, and minimum inhibitory concentration (MIC) values. By introducing an encoder neural network to map peptide sequence to latent representations, the learnt latent space can be more structural and consequently leads to improved data modeling. Trained on around 6K–8K AMPs and 500K non-AMPs, AMP-GAN was able to generate three experimental validated AMPs. Its improved version, AMP-GAN v2, exhibited better training stability and generated a higher percentage of peptide antibiotic candidates, as predicted by several machine learning-based AMP classifiers. PandoraGAN49 utilized around 100–400 active antiviral peptides to generate novel peptides targeting viruses. To address the data scarcity problem, a novel training framework proposed in LeakGAN100 was adopted to provide richer signals and guide the model training during the intermediate process of peptide generation. Moreover, DeepImmuno-GAN51 used a WGAN with gradient penalty to generate immunogenic peptides that can bind to HLA-A*0201. GANDALF48 generated peptides that target cancer-related proteins such as PD-1, PDL-1, and CTLA-4. Specifically, GANDALF decomposed the peptide generation into sequence and structure generation steps and used two GANs to model these two generation processes. Despite the fact that GANs have been used to generate peptides with different properties, most still need wet-lab validation to support their effectiveness.
Discussion
In this review, we summarize available deep generative models for peptides. Since peptide therapeutics have numerous applications, including in infectious diseases and cancer, the models described here represent foundational frameworks for the design of novel drugs. Recent advancements in software and hardware have allowed deep generative models to be trained at great speed and have enabled the generation of novel synthetic peptides displaying the desired properties. Such advances hold promise to accelerate peptide drug development by saving time, reducing cost, and increasing the likelihood of success. Indeed, the combination of generative models with deep neural networks has already generated promising peptides with therapeutic potential.Despite these initial advances, several challenges still need to be addressed. For example, there is currently no single deep generative model framework that consistently yields better results compared to other deep generative models. As a result, selecting a suitable model from various deep generative frameworks can be difficult given a peptide dataset of interest. In addition, there is a lack of benchmarking datasets and metrics in peptide generation evaluation that further hinders model comparison and selection. In the area of molecular generation, benchmarking platforms like GuacaMol101 and MOSES102 have been developed to systematically evaluate the quality of generated data by using various metrics like novelty, uniqueness, validity, and Fréchet ChemNet Distance.103 There is an urgent need to establish similar platforms for benchmarking peptide generation models.
Apart from benchmarking platforms for generative model assessment, we discuss several future directions that can possibly lead to better peptide generation systems. (1) Better peptide property predictors/filters. To prioritize generated peptides for wet-lab validation, a number of generative methods require property predictors to filter and counterselect for undesired peptides. Consequently, the quality of property predictors can significantly influence the outcome of any peptide design project. Recent advances in multitask learning,104 transfer learning,105 and low-shot learning106,107 may be adopted to better utilize labeled peptide data and improve property predictors. In addition, by quantifying uncertainty of prediction (e.g., peptide property predictions), active learning108 can select generated peptides with high uncertainty.109 In return, the experimental results of selected samples can be used as feedback to refine generative and property predictor models by efficiently expanding training data space and reducing model uncertainty. Similar to the case of generative model assessment, there is a lack of studies in ML-based peptide property prediction efforts aimed at systematically studying datasets, features, and model selection. Differences in dataset construction (i.e., positive data selection and negative data generation) and a lack of comprehensive feature selection prevent direct comparison among various models.110 Similar to MoleculeNet111 in molecular ML, a peptide property prediction benchmark incorporating commonly used peptide datasets, peptide features (e.g., physicochemical descriptors, one-hot encoding and learned representations) and ML models (e.g., linear models, SVM, tree-based models and neural networks) can greatly help researchers to standardize the process of model evaluation and selection. (2) Further optimization on generated peptides. By combining deep generative models with optimization/searching methods like genetic algorithms, Bayesian optimization and etc.,112–115 generated samples can be further optimized to acquire improved properties and functions. Schissel et al.57 studied this notion to generate peptides using a deep generative model in combination with a genetic algorithm. (3) Incorporation of peptide structure information into deep generative models. While peptide structures can provide mechanistic information to better guide the model to generate peptides with desired functions, the majority of deep generative models (except Caceres-Delpiano et al.54 and Rossetto et al.48) in peptide design have only used sequence information. We hypothesize that the dynamic and flexible nature of peptide structures make them difficult to be inputted into deep generative models. For example, a single PDB structure of a peptide may be too static to capture sufficient information for computational modelling. Instead of using a single PDB structure, generative models that take a set of peptide structure conformers or a trajectory of structure changes (computed by molecular dynamics) as input may lead to more optimal peptide representation learning and subsequent peptide generation. With our increasing ability to generate peptide structural and functional data, coupled with advancements in deep learning, we anticipate that deep generative models will play a major role in drug discovery in years to come.
Data availability
As this is a Review article, no primary research results, data, software or code have been included.Author contributions
Conceptualization: FW, DKH, CFN. Funding acquisition: CFN. Investigation: FW, DKH. Supervision: CFN. Visualization: FW. Writing – original draft: FW, DKH. Writing – review & editing: FW, DKH, CFN.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Cesar de la Fuente-Nunez holds a Presidential Professorship at the University of Pennsylvania, is a recipient of the Langer Prize by the AIChE Foundation and acknowledges funding from the Institute for Diabetes, Obesity, and Metabolism, the Penn Mental Health AIDS Research Center of the University of Pennsylvania, the Nemirovsky Prize, the Dean's Innovation Fund from the Perelman School of Medicine at the University of Pennsylvania, the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM138201, and the Defense Threat Reduction Agency (DTRA; HDTRA11810041 and HDTRA1-21-1-0014). Daphne Kontogiorgos-Heintz acknowledges funding from the Littlejohn Undergraduate Research Program from the School of Engineering, the Blair Research Fellowship from the Bioengineering department, and the Grant for Faculty Mentoring Undergraduate Research, all at the University of Pennsylvania.References
- K. Fosgerau and T. Hoffmann, Peptide therapeutics: current status and future directions, Drug Discovery Today, 2015, 20(1), 122–128 CrossRef CAS PubMed.
- M. C. Morris, J. Depollier, J. Mery, F. Heitz and G. Divita, A peptide carrier for the delivery of biologically active proteins into mammalian cells, Nat. Biotechnol., 2001, 19(12), 1173–1176 CrossRef CAS PubMed.
- A. Henninot, J. C. Collins and J. M. Nuss, The current state of peptide drug discovery: back to the future?, J. Med. Chem., 2018, 61(4), 1382–1414 CrossRef CAS PubMed.
- M. Muttenthaler, G. F. King, D. J. Adams and P. F. Alewood, Trends in peptide drug discovery, Nat. Rev. Drug Discovery, 2021, 20(4), 309–325 CrossRef CAS PubMed.
- M. D. Torres, S. Sothiselvam, T. K. Lu and C. de la Fuente-Nunez, Peptide design principles for antimicrobial applications, J. Mol. Biol., 2019, 431(18), 3547–3567 CrossRef CAS PubMed.
- M. Magana, M. Pushpanathan, A. L. Santos, L. Leanse, M. Fernandez and A. Ioannidis, et al., The value of antimicrobial peptides in the age of resistance, Lancet Infect. Dis., 2020, 20(9), e216–e30 CrossRef CAS PubMed.
- M. D. Torres, M. C. Melo, O. Crescenzi, E. Notomista and C. de la Fuente-Nunez, Mining for encrypted peptide antibiotics in the human proteome, Nature Biomedical Engineering, 2022, 6(1), 67–75 CrossRef PubMed.
- S. Basith, B. Manavalan, T. Hwan Shin and G. Lee, Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening, Med. Res. Rev., 2020, 40(4), 1276–1314 CrossRef CAS PubMed.
- M. C. Melo, J. R. Maasch and C. de la Fuente-Nunez, Accelerating antibiotic discovery through artificial intelligence, Communications Biology, 2021, 4(1), 1–13 CrossRef PubMed.
- C. de la Fuente-Nunez, Toward autonomous antibiotic discovery, Msystems, 2019, 4(3), e00151–19 CrossRef CAS PubMed.
- M. D. T. Torres and C. de la Fuente-Nunez, Toward computer-made artificial antibiotics, Curr. Opin. Microbiol., 2019, 51, 30–38 CrossRef CAS PubMed.
- M. D. T. Torres and C. de la Fuente-Nunez, Reprogramming biological peptides to combat infectious diseases, Chem. Commun., 2019, 55(100), 15020–15032 RSC.
- A. Krizhevsky, I. Sutskever and G. E. Hinton, Imagenet classification with deep convolutional neural networks, Adv. Neural Inf. Process. Syst., 2012, 25, 1097–1105 Search PubMed.
- Advances in Neural Information Processing Systems; ed. Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A. N., et al., Attention is all you need, 2017 Search PubMed.
- D. Silver, J. Schrittwieser, K. Simonyan, I. Antonoglou, A. Huang and A. Guez, et al., Mastering the game of go without human knowledge, Nature, 2017, 550(7676), 354–359 CrossRef CAS PubMed.
- B. Alipanahi, A. Delong, M. T. Weirauch and B. J. Frey, Predicting the sequence specificities of DNA-and RNA-binding proteins by deep learning, Nat. Biotechnol., 2015, 33(8), 831–838 CrossRef CAS PubMed.
- R. Lopez, J. Regier, M. B. Cole, M. I. Jordan and N. Yosef, Deep generative modeling for single-cell transcriptomics, Nat. Methods, 2018, 15(12), 1053–1058 CrossRef CAS PubMed.
- E. C. Alley, G. Khimulya, S. Biswas, M. AlQuraishi and G. M. Church, Unified rational protein engineering with sequence-based deep representation learning, Nat. Methods, 2019, 16(12), 1315–1322 CrossRef CAS PubMed.
- S. Li, F. Wan, H. Shu, T. Jiang, D. Zhao and J. Zeng, MONN: A Multi-objective Neural Network for Predicting Compound-Protein Interactions and Affinities, Cell Systems, 2020, 10(4), 308–322.e11 CrossRef CAS.
- B. C. Csáji, Approximation with Artificial Neural Networks, MSc thesis, Faculty of Sciences, Etvs Lornd University, Hungary, 2001, vol. 24, ch. 48, p. 7.
- G. E. Hinton and R. R. Salakhutdinov, Reducing the dimensionality of data with neural networks, Science, 2006, 313(5786), 504–507 CrossRef CAS PubMed.
- Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019, ed. Devlin J., Chang M., Wei, Lee K. and Toutanova K., NAACL-HLT 2019, Minneapolis, MN, USA, June 2-7, 2019, vol. 1 (Long and Short Papers); 2019 Search PubMed.
- Auto-Encoding Variational Bayes. 2nd International Conference on Learning Representations, ICLR 2014, Banff, AB, Canada, April 14-16, 2014,ed. Kingma D. P. and Welling M., Conference Track Proceedings; 2014 Search PubMed.
- Stochastic backpropagation and approximate inference in deep generative models, ed. Rezende D. J., Mohamed S. and Wierstra D., International Conference on Machine Learning; 2014 Search PubMed.
- I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, Advances in Neural Information Processing Systems, ed. Z. Ghahramani, M. Welling, C. Cortes, N. Lawrence, K. Q. Weinberger, Curran Associates, Inc., Generative Adversarial Nets, https://proceedings.neurips.cc/paper/2014/file/5ca3e9b122f61f8f06494c97b1afccf3-Paper.pdf, 2014, vol. 27 Search PubMed.
- C. Louizos, U. Shalit, J. Mooij, D. Sontag, R. Zemel and M. Welling. Causal effect inference with deep latent-variable models. Proceedings of the 31st International Conference on Neural Information Processing Systems, Curran Associates Inc.; Long Beach, California, USA: 2017. pp. 6449–59 Search PubMed.
- Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks. 4th International Conference on Learning Representations, ICLR 2016, ed. Radford A., Metz L. and Chintala S., Conference Track Proceedings; San Juan, Puerto Rico, May 2-4, 2016 Search PubMed.
- Pixel recurrent neural networks, ed. Van Oord A., Kalchbrenner N. and Kavukcuoglu K., International Conference on Machine Learning; 2016 Search PubMed.
- Syntax-Directed Variational Autoencoder for Structured Data. 6th International Conference on Learning Representations, ICLR 2018, Vancouver, BC, Canada, April 30 - May 3, 2018, ed. Dai H., Tian Y., Dai B., Skiena S. and Song L., Conference Track Proceedings; 2018 Search PubMed.
- R. Gómez-Bombarelli, J. N. Wei, D. Duvenaud, J. M. Hernández-Lobato, B. Sánchez-Lengeling and D. Sheberla, et al., Automatic chemical design using a data-driven continuous representation of molecules, ACS Cent. Sci., 2018, 4(2), 268–276 CrossRef PubMed.
- X. Chen, C. Li, M. T. Bernards, Y. Shi, Q. Shao, Y. He. Sequence-based peptide identification, generation, and property prediction with deep learning: a review. Molecular Systems Design & Engineering. 2021 Search PubMed.
- Z. Wu, K. E. Johnston, F. H. Arnold and K. K. Yang, Protein sequence design with deep generative models, Curr. Opin. Chem. Biol., 2021, 65, 18–27 CrossRef CAS PubMed.
- A. Strokach and P. M. Kim, Deep generative modeling for protein design, Curr. Opin. Struct. Biol., 2022, 72, 226–236 CrossRef CAS PubMed.
- Variational inference with normalizing flows. International Conference on Machine Learning; Rezende D. and Mohamed S., 2015 Search PubMed.
- Y. LeCun, S. Chopra, R. Hadsell, M. Ranzato and F. Huang, Energy-Based Models, Predicting structured data, 2006, 191–246 Search PubMed.
- G. A. Miller, WordNet: a lexical database for English, Commun. ACM, 1995, 38(11), 39–41 CrossRef.
- Imagenet: A large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition; ed. Deng J., Dong W., Socher R., Li L-J., Li K., and Fei-Fei L.: Ieee, 2009 Search PubMed.
- P. Das, K. Wadhawan, O. Chang, T. Sercu, C. N. dos Santos, M. Riemer, et al., PepCVAE: Semi-Supervised Targeted Design of Antimicrobial Peptide Sequences. CoRR. 2018;abs/1810.07743.
- J. B. Ferrell, J. M. Remington, C. M. Van Oort, M. Sharafi, R. Aboushousha and Y. Janssen-Heininger, et al., A Generative Approach toward Precision Antimicrobial Peptide Design, bioRxiv, 2021 DOI:10.1101/2020.10.02.324087.
- C. M. Van Oort, J. B. Ferrell, J. M. Remington, S. Wshah and J. Li, AMPGAN v2: Machine Learning-Guided Design of Antimicrobial Peptides, J. Chem. Inf. Model., 2021, 61(5), 2198–2207 CrossRef CAS PubMed.
- C. Wang, S. Garlick and M. Zloh, Deep Learning for Novel Antimicrobial Peptide Design, Biomolecules, 2021, 11(3), 471 CrossRef CAS PubMed.
- A. Capecchi, X. Cai, H. Personne, T. Kohler, C. van Delden, J.-L. Reymond. Machine Learning Designs Non-Hemolytic Antimicrobial Peptides. Chemical Science. 2021 Search PubMed.
- U. Consortium, UniProt: a worldwide hub of protein knowledge, Nucleic Acids Res., 2019, 47(D1), D506–D15 CrossRef PubMed.
- S. N. Dean and S. A. Walper, Variational autoencoder for generation of antimicrobial peptides, ACS Omega, 2020, 5(33), 20746–20754 CrossRef CAS PubMed.
- P. Das, T. Sercu, K. Wadhawan, I. Padhi, S. Gehrmann and F. Cipcigan, et al., Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations, Nature Biomedical Engineering, 2021, 5(6), 613–623 CrossRef CAS PubMed.
- F. Grisoni, C. S. Neuhaus, G. Gabernet, A. T. Müller, J. A. Hiss and G. Schneider, Designing anticancer peptides by constructive machine learning, ChemMedChem, 2018, 13(13), 1300–1302 CrossRef CAS PubMed.
- S. Spänig and D. Heider, Encodings and models for antimicrobial peptide classification for multi-resistant pathogens, BioData Mining, 2019, 12(1), 1–29 CrossRef PubMed.
- GANDALF: Peptide Generation for Drug Design using Sequential and Structural Generative Adversarial Networks. Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics; ed. Rossetto A., and Zhou W., 2020 Search PubMed.
- S. Surana, P. Arora, D. Singh, D. Sahasrabuddhe and J. Valadi, PandoraGAN: Generating antiviral peptides using Generative Adversarial Network, bioRxiv, 2021 Search PubMed.
- A. Tucs, D. P. Tran, A. Yumoto, Y. Ito, T. Uzawa and K. Tsuda, Generating ampicillin-level antimicrobial peptides with activity-aware generative adversarial networks, ACS Omega, 2020, 5(36), 22847–22851 CrossRef CAS PubMed.
- G. Li, B. Iyer, V. B. S. Prasath, Y. Ni and N. Salomonis, DeepImmuno: deep learning-empowered prediction and generation of immunogenic peptides for T-cell immunity, Briefings Bioinf., 2021, 22(6), bbab160 CrossRef PubMed.
- D. Nagarajan, T. Nagarajan, N. Roy, O. Kulkarni, S. Ravichandran and M. Mishra, et al., Computational antimicrobial peptide design and evaluation against multidrug-resistant clinical isolates of bacteria, J. Biol. Chem., 2018, 293(10), 3492–3509 CrossRef CAS PubMed.
- D. P. Tran, S. Tada, A. Yumoto, A. Kitao, Y. Ito and T. Uzawa, et al., Using molecular dynamics simulations to prioritize and understand AI-generated cell penetrating peptides, Sci. Rep., 2021, 11(1), 1–9 CrossRef PubMed.
- J. Caceres-Delpiano, R. Ibañez, P. Alegre, C. Sanhueza, R. Paz and S. Correa, et al., Deep learning enables the design of functional de novo antimicrobial proteins, bioRxiv, 2020 DOI:10.1101/2020.08.26.266940.
- Z. Wu, K. K. Yang, M. J. Liszka, A. Lee, A. Batzilla and D. Wernick, et al., Signal peptides generated by attention-based neural networks, ACS Synth. Biol., 2020, 9(8), 2154–2161 CrossRef CAS PubMed.
- A. T. Müller, J. A. Hiss and G. Schneider, Recurrent neural network model for constructive peptide design, J. Chem. Inf. Model., 2018, 58(2), 472–479 CrossRef PubMed.
- C. K. Schissel, S. Mohapatra, J. M. Wolfe, C. M. Fadzen, K. Bellovoda and C.-L. Wu, et al., Deep learning to design nuclear-targeting abiotic miniproteins, Nat. Chem., 2021, 1–9 Search PubMed.
- H. ElAbd, Y. Bromberg, A. Hoarfrost, T. Lenz, A. Franke and M. Wendorff, Amino acid encoding for deep learning applications, BMC Bioinf., 2020, 21(1), 1–14 CrossRef PubMed.
- K. K. Yang, Z. Wu, C. N. Bedbrook and F. H. Arnold, Learned protein embeddings for machine learning, Bioinformatics, 2018, 34(15), 2642–2648 CrossRef CAS PubMed.
- Adversarial Feature Learning. 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, ed. Donahue J., Krähenbühl P. and Darrell T., Conference Track Proceedings; 2017 Search PubMed.
- Y. Bengio, R. Ducharme, P. Vincent and C. Janvin, A neural probabilistic language model, J. Mach. Learn. Res., 2003, 3, 1137–1155 Search PubMed.
- Neural Machine Translation by Jointly Learning to Align and Translate. 3rd International Conference on Learning Representations, ICLR 2015, May 7-9, 2015, ed. Bahdanau D., Cho K. and Bengio Y., Conference Track Proceedings, San Diego, CA, USA; 2015 Search PubMed.
- J.-E. Shin, A. J. Riesselman, A. W. Kollasch, C. McMahon, E. Simon and C. Sander, et al., Protein design and variant prediction using autoregressive generative models, Nat. Commun., 2021, 12(1), 1–11 CrossRef PubMed.
- S. Hochreiter and J. Schmidhuber, Long short-term memory, Neural Comput., 1997, 9(8), 1735–1780 CrossRef CAS PubMed.
- Learning Phrase Representations using RNN Encoder-Decoder for Statistical Machine Translation. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing, EMNLP 2014, October 25-29, 2014, Doha, Qatar, A meeting of SIGDAT, ed. Cho K., van Merrienboer B., Gulcehre C., Bahdanau D., Bougares F., Schwenk H., et al, a Special Interest Group of the ACL; 2014 Search PubMed.
- Rationalizing Neural Predictions. Empirical Methods in Natural Language Processing (EMNLP); ed. Lei T., Barzilay R. and Jaakkola T., 2016 Search PubMed.
- A. Rives, J. Meier, T. Sercu, S. Goyal, Z. Lin and J. Liu, et al., Biological structure and function emerge from scaling unsupervised learning to 250 million protein sequences, Proc. Natl. Acad. Sci. U.S.A., 2021, 118(15), e2016239118 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov and O. Ronneberger, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596(7873), 583–589 CrossRef CAS PubMed.
- A. Madani, B. McCann, N. Naik, N. S. Keskar, N. Anand, R. R. Eguchi, et al.Progen: Language modeling for protein generation. arXiv preprint arXiv:200403497. 2020 Search PubMed.
- MSA transformer. International Conference on Machine Learning; ed. Rao R. M., Liu J., Verkuil R., Meier J., Canny J., Abbeel P., et al.: PMLR, 2021 Search PubMed.
- Sequence to sequence learning with neural networks. Advances in Neural Information Processing Systems; ed. Sutskever I., Vinyals O. and Le Q. V., 2014 Search PubMed.
- R. Winter, F. Montanari, F. Noé and D.-A. Clevert, Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations, Chem. Sci., 2019, 10(6), 1692–1701 RSC.
- M. Pirtskhalava, A. A. Amstrong, M. Grigolava, M. Chubinidze, E. Alimbarashvili and B. Vishnepolsky, et al., DBAASP v3: Database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics, Nucleic Acids Res., 2021, 49(D1), D288–D97 CrossRef CAS PubMed.
- S. C. Kaae, T. Raiko, L. Maaløe, S. K. Sønderby and O. Winther, Ladder variational autoencoders, Adv. Neural Inf. Process. Syst., 2016, 29, 3738–3746 Search PubMed.
- NVAE: A Deep Hierarchical Variational Autoencoder. Advances in Neural Information Processing Systems 33: Annual Conference on Neural Information Processing Systems 2020, NeurIPS 2020, December 6-12, 2020, virtual; ed. Vahdat A., and Kautz J., 2020 Search PubMed.
- K. Sohn, H. Lee and X. Yan, Learning structured output representation using deep conditional generative models, Adv. Neural Inf. Process. Syst., 2015, 28, 3483–3491 Search PubMed.
- J. Lim, S. Ryu, J. W. Kim and W. Y. Kim, Molecular generative model based on conditional variational autoencoder for de novo molecular design, J. Cheminf., 2018, 10(1), 1–9 Search PubMed.
- Toward controlled generation of text. International Conference on Machine Learning, ed. Hu Z., Yang Z., Liang X., Salakhutdinov R., and Xing E. P., 2017 Search PubMed.
- InfoVAE: Balancing Learning and Inference in Variational Autoencoders. The Thirty-Third AAAI Conference on Artificial Intelligence, AAAI 2019, The Thirty-First Innovative Applications of Artificial Intelligence Conference, IAAI 2019, The Ninth AAAI Symposium on Educational Advances in Artificial Intelligence, EAAI 2019, Honolulu, Hawaii, USA, January 27 - February 1, 2019; ed. Zhao S., Song J., and Ermon S.: AAAI Press, 2019 Search PubMed.
- beta-VAE: Learning Basic Visual Concepts with a Constrained Variational Framework. 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, ed. Higgins I., Matthey L., Pal A., Burgess C., Glorot X., and Botvinick M., et al., Conference Track Proceedings; 2017: https://OpenReview.net Search PubMed.
- Y. Pu, Z. Gan, R. Henao, X. Yuan, C. Li and A. Stevens, et al., Variational autoencoder for deep learning of images, labels and captions, Adv. Neural Inf. Process. Syst., 2016, 29, 2352–2360 Search PubMed.
- Y. Fan, G. Wen, D. Li, S. Qiu, M. D. Levine and F. Xiao, Video anomaly detection and localization via gaussian mixture fully convolutional variational autoencoder, Comput. Vis. Image Understand., 2020, 195, 102920 CrossRef.
- Variational autoencoder for semi-supervised text classification. Thirty-First AAAI Conference on Artificial Intelligence; ed. Xu W., Sun H., Deng C., and Tan Y., 2017 Search PubMed.
- Variational autoencoders for collaborative filtering. Proceedings of the 2018 World Wide Web Conference; ed. Liang D., Krishnan R. G., Hoffman M. D., Jebara T., 2018 Search PubMed.
- Junction tree variational autoencoder for molecular graph generation. International Conference on Machine Learning; ed. Jin W., Barzilay R., and Jaakkola T., 2018 Search PubMed.
- M. Mirza, S. Osindero. Conditional Generative Adversarial Nets. CoRR. 2014; abs/1411.1784.
- Towards diverse and natural image descriptions via a conditional gan. Proceedings of the IEEE International Conference on Computer Vision; Dai B., Fidler S., Urtasun R., and Lin D., 2017 Search PubMed.
- A. Gupta and J. Zou, Feedback GAN for DNA optimizes protein functions, Nature Machine Intelligence, 2019, 1(2), 105–111 CrossRef.
- Towards Principled Methods for Training Generative Adversarial Networks. 5th International Conference on Learning Representations, ICLR 2017, Toulon, France, April 24-26, 2017, ed. Arjovsky M., Bottou L., Conference Track Proceedings; 2017: http://OpenReview.net Search PubMed.
- T. Salimans, I. Goodfellow, W. Zaremba, V. Cheung, A. Radford and X. Chen, Improved techniques for training gans, Adv. Neural Inf. Process. Syst., 2016, 29, 2234–2242 Search PubMed.
- f-gan: Training generative neural samplers using variational divergence minimization. Proceedings of the 30th International Conference on Neural Information Processing Systems; ed. Nowozin S., Cseke B. and Tomioka R., 2016 Search PubMed.
- Infogan: Interpretable representation learning by information maximizing generative adversarial nets. Proceedings of the 30th International Conference on Neural Information Processing Systems; ed. Chen X., Duan Y., Houthooft R., Schulman J., Sutskever I., Abbeel P., 2016 Search PubMed.
- Least squares generative adversarial networks. Proceedings of the IEEE International Conference on Computer Vision; ed. Mao X., Li Q., Xie H., Lau R. Y. K., Wang Z., and Paul Smolley S., 2017 Search PubMed.
- MMD GAN: Towards Deeper Understanding of Moment Matching Network. Advances in Neural Information Processing Systems 30: Annual Conference on Neural Information Processing Systems 2017, December 4-9, 2017, ed. Li C., Liang, Chang W., Cheng, Cheng Y., Yang Y., and Póczos B., Long Beach, CA, USA; 2017 Search PubMed.
- Wasserstein Generative Adversarial Networks. Proceedings of the 34th International Conference on Machine Learning; 2017 06--11 Aug: ed. Arjovsky M., Chintala S., and Bottou L., PMLR, 2017 Search PubMed.
- Improved Training of Wasserstein GANs. Advances in Neural Information Processing Systemsed. Gulrajani I., Ahmed F., Arjovsky M., Dumoulin V. and Courville A. C.: Curran Associates, Inc.; 2017 Search PubMed.
- Spectral Normalization for Generative Adversarial Networks. 6th International Conference on Learning Representations, ICLR 2018, Vancouver, BC, Canada, April 30 - May 3, 2018, ed. Miyato T., Kataoka T., Koyama M. and Yoshida Y., Conference Track Proceedings; 2018 Search PubMed.
- MolGAN: An implicit generative model for small molecular graphs. CoRR. ed. Cao N. D. and Kipf T., 2018;abs/1805.11973 Search PubMed.
- Stargan: Unified generative adversarial networks for multi-domain image-to-image translation. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; ed. Choi Y., Choi M., Kim M., Ha J-W., Kim S., and Choo J., 2018 Search PubMed.
- Long text generation via adversarial training with leaked information. Proceedings of the AAAI Conference on Artificial Intelligence; ed. Guo J., Lu S., Cai H., Zhang W., Yu Y. and Wang J., 2018 Search PubMed.
- N. Brown, M. Fiscato, M. H. S. Segler and A. C. Vaucher, GuacaMol: benchmarking models for de novo molecular design, J. Chem. Inf. Model., 2019, 59(3), 1096–1108 CrossRef CAS PubMed.
- D. Polykovskiy, A. Zhebrak, B. Sanchez-Lengeling, S. Golovanov, O. Tatanov and S. Belyaev, et al., Molecular sets (MOSES): a benchmarking platform for molecular generation models, Front. Pharmacol., 2020, 11, 1931 Search PubMed.
- K. Preuer, P. Renz, T. Unterthiner, S. Hochreiter and K. Gn, Frechet ChemNet distance: a metric for generative models for molecules in drug discovery, J. Chem. Inf. Model., 2018, 58(9), 1736–1741 CrossRef CAS PubMed.
- Y. Zhang and Q. Yang, An overview of multi-task learning, Natl. Sci. Rev., 2018, 5(1), 30–43 CrossRef.
- A survey on deep transfer learning. International Conference on Artificial Neural Networks; ed. Tan C., Sun F., Kong T., Zhang W., Yang C. and Liu C., 2018 Search PubMed.
- H. Altae-Tran, B. Ramsundar, A. S. Pappu and V. Pande, Low data drug discovery with one-shot learning, ACS Cent. Sci., 2017, 3(4), 283–293 CrossRef CAS PubMed.
- I. Olier, N. Sadawi, G. R. Bickerton, J. Vanschoren, C. Grosan and L. Soldatova, et al., Meta-QSAR: a large-scale application of meta-learning to drug design and discovery, Mach. Learn., 2018, 107(1), 285–311 CrossRef PubMed.
- Y. Zhang and others, Bayesian semi-supervised learning for uncertainty-calibrated prediction of molecular properties and active learning, Chem. Sci., 2019, 10(35), 8154–8163 RSC.
- B. Hie, B. D. Bryson and B. Berger, Leveraging uncertainty in machine learning accelerates biological discovery and design, Cell systems, 2020, 11(5), 461–477.e9 CrossRef CAS PubMed.
- J. Xu, F. Li, A. Leier, D. Xiang, H.-H. Shen and T. T. Marquez Lago, et al., Comprehensive assessment of machine learning-based methods for predicting antimicrobial peptides, Briefings Bioinf., 2021, 22(5), bbab083 CrossRef PubMed.
- Z. Wu, B. Ramsundar, E. N. Feinberg, J. Gomes, C. Geniesse and A. S. Pappu, et al., MoleculeNet: a benchmark for molecular machine learning, Chem. Sci., 2018, 9(2), 513–530 RSC.
- R.-R. Griffiths, J. Hernández-Lobato and Miguel, Constrained Bayesian optimization for automatic chemical design using variational autoencoders, Chem. Sci., 2020, 11(2), 577–586 RSC.
- A. Kumar, S. Levine. Model inversion networks for model-based optimization. arXiv preprint. 2019 Search PubMed.
- A. Chan Guo Wei, A. Madani, B. Krause and N. Naik, Deep Extrapolation for Attribute-Enhanced Generation, Advances in Neural Information Processing Systems, 2021, vol. 34 Search PubMed.
- W. F. Porto, L. Irazazabal, E. S. Alves, S. M. Ribeiro, C. O. Matos and Á. S. Pires, et al., In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design, Nat. Commun., 2018, 9(1), 1–12 CAS.
- P. Agrawal, S. Bhalla, S. S. Usmani, S. Singh, K. Chaudhary and G. P. S. Raghava, et al., CPPsite 2.0: a repository of experimentally validated cell-penetrating peptides, Nucleic Acids Res., 2016, 44(D1), D1098–D103 CrossRef CAS PubMed.
- A. Bateman, L. Coin, R. Durbin, R. D. Finn, V. Hollich and S. Griffiths-Jones, et al., The Pfam protein families database, Nucleic Acids Res., 2004, 32(suppl\\_1), D138–D41 CrossRef CAS PubMed.
- S. Gupta, P. Kapoor, K. Chaudhary, A. Gautam, R. Kumar and O. S. D. D. Consortium, et al., In Silico Approach for Predicting Toxicity of Peptides and Proteins, PloS One, 2013, 8(9), e73957 CrossRef CAS PubMed.
- A. Qureshi, N. Thakur, H. Tandon and M. Kumar, AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses, Nucleic Acids Res., 2014, 42(D1), D1147–D53 CrossRef CAS PubMed.
- X. Zhao, H. Wu, H. Lu, G. Li and Q. Huang, LAMP: A Database Linking Antimicrobial Peptides, PLoS One, 2013, 8(6), e66557 CrossRef CAS PubMed.
- S. S. Usmani, G. Bedi, J. S. Samuel, S. Singh, S. Kalra and P. Kumar, et al., THPdb: Database of FDA-approved peptide and protein therapeutics, PloS One, 2017, 12(7), e0181748 CrossRef PubMed.
- S. Thomas, S. Karnik, R. S. Barai, V. K. Jayaraman and S. Idicula-Thomas, CAMP: a useful resource for research on antimicrobial peptides, Nucleic Acids Res., 2010, 38(suppl\\_1), D774–D80 CrossRef CAS PubMed.
- X. Kang, F. Dong, C. Shi, S. Liu, J. Sun and J. Chen, et al., DRAMP 2.0, an updated data repository of antimicrobial peptides, Sci. Data, 2019, 6(1), 148 CrossRef PubMed.
- S. P. Piotto, L. Sessa, S. Concilio and P. Iannelli, YADAMP: yet another database of antimicrobial peptides, Int. J. Antimicrob. Agents, 2012, 39(4), 346–351 CrossRef CAS PubMed.
- M. Novković, J. Simunić, V. Bojović, A. Tossi and D. Juretić, DADP: the database of anuran defense peptides, Bioinformatics, 2012, 28(10), 1406–1407 CrossRef PubMed.
- G. Wang, X. Li and Z. Wang, APD3: the antimicrobial peptide database as a tool for research and education, Nucleic Acids Res., 2016, 44(Database issue), D1087–D93 CrossRef CAS PubMed.
- J.-H. Jhong, Y.-H. Chi, W.-C. Li, T.-H. Lin, K.-Y. Huang and T.-Y. Lee, dbAMP: an integrated resource for exploring antimicrobial peptides with functional activities and physicochemical properties on transcriptome and proteome data, Nucleic Acids Res., 2019, 47(D1), D285–D97 CrossRef CAS PubMed.
- W. Fleri, S. Paul, S. K. Dhanda, S. Mahajan, X. Xu and B. Peters, et al., The Immune Epitope Database and Analysis Resource in Epitope Discovery and Synthetic Vaccine Design, Front. Immunol., 2017, 8, 278 Search PubMed.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2022 |