Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Data management matters
Cerys
Willoughby
 * and
Jeremy Graham
Frey
* and
Jeremy Graham
Frey

School of Chemistry, University of Southampton, Southampton, UK. E-mail: cerys.willoughby@soton.ac.uk
First published on 3rd March 2022
Abstract
There are a number of issues that inhibit the replication and reproduction of research, and make it hard to utilise existing scientific data to make new discoveries. These include poor data management, competing standards, a lack of consideration of the usability of data, and a disconnect between the publication of science and the data and methods behind it. In this paper, we examine the benefits of good data management for not only ensuring that data are well organised, easy to find, and preserved for the future, but also for facilitating reproducibility and new discoveries in science. We consider the importance of documenting data and making them usable by both humans and machines, and consider the development of tools to support these processes in the future.
Introduction
Progression in science is achieved by building upon the discoveries of others. In order to do so, the scientific community must be able to scrutinize and validate the research and results of others. Fundamental to our ability to verify these results is access to the data and the methods used to generate it. In an ideal world, it would be possible to take the methods used and other resources related to an experiment and replicate the results. Often this is impractical, but at the very least the data from research should be accessible for scrutiny to ensure that results are reasonable, and no mistakes have been made in the calculations.The scientific journal article is the primary mechanism for sharing new research, but these articles rarely contain the full data from any specific experiment or study, and frequently do not contain enough information on the methods to enable the research to be adequately replicated. Historically it has been rare for any additional data or materials beyond the tables in the journal articles to be made available. Readers could request the data from a study, but in the majority of cases, the request would be refused, or the data would not be provided in a format suitable for interrogation or reuse.1
Many journals have introduced requirements for data to be provided in the form of supplementary information linked to the journal article. Supplementary information is typically provided in PDF format with data in the form of tables or images making it difficult to extract the data directly. Even when the full data are provided, there is still no guarantee that the data will be in a format that is usable or useful for others. Supplementary information in the form of PDFs are not machine readable, and a user typically must manually determine whether a particular article has any supplementary information because there is often a disconnect between the identifier of the article and identifier of the supplementary information (if there is one).
Licensing is also rarely declared on supplementary information, so it is often not clear if the data are available for reuse.2 Even if the full final data are provided in an appropriate format, the data will have gone through a variety of processes using different tools or scripts before analysis, and it is still relatively rare for such tools, code or models to be included with the supplementary information, meaning that it can be difficult to replicate the results even with the raw data.
There are a variety of reasons why researchers may be reluctant or indeed unable to share their research. Sharing may be restricted due to intellectual property concerns or privacy issues with sensitive data. However, other reasons often relate to poor data management making it difficult to make data available in suitable formats, or a general lack of awareness about what and how to share. It can be very time consuming to retrospectively attempt to organise data, convert data into a more appropriate format, and to add adequate descriptions to make them useful to a different audience. Other issues include the myriad of competing standards for both scientific data and the metadata that describes it.3
Although we tend to think of data as structured numerical information, scientific data can be much more diverse, and even change structure over time.4 Within chemistry very large amounts of data are in the form of images or visualisations. There are also many options for locations to share data. Consideration needs to be given to ensure that the chosen location is appropriate so that it can be found by those who need to make use of it. The reality is that most scientific data are collected with little consideration of whether the data could be useful or usable by other researchers outside of the originators group.
Even if the data are destined never to be shared outside an individual research group, if they are not properly managed and taken care of, they can become impossible to use even by the person who originally captured or created it. This may be because after a few months, or even weeks after an experiment or project has ended, we may not be able to remember the full context and circumstances of the generation of the data, for example the settings or configurations that were used. Data files may not contain details of the units with which measurements were made, and abbreviated titles or labels may now be incomprehensible, rendering the contents of the file meaningless. Although it is possible to retroactively fix some of these problems, it is much harder to do than to include good documentation and use appropriate formats from the beginning.
There are other factors that may cause the loss or degradation of digital data such as physical media being lost, files being moved or accidentally overwritten on shared media, and due to physical loss through hard-drive failure or bit rot. Data can also be vulnerable to deliberate tampering, malicious destruction, or virus attack. Even if the data are adequately stored, backed-up and periodically copied onto new media, they may become unusable if separated from the original context of the data or become obsolete when the format becomes incompatible with the latest hardware and software.
The development of technologies for automating the execution of research and the capture of data in digital form has exacerbated these challenges by significantly increasing the amount of data that may be produced during research. Most data are now ‘born digital’, as laboratories become more automated and data are produced using digital instruments. Very large-scale experiments and high-throughput experiments (HTE) generate very large datasets or so-called ‘Big Data’.5,6 Although mostly commonly associated with large-scale experiments in disciplines such as astronomy and genetics, Big Data are also being generated and exploited within chemistry.7–10 These large complex datasets present numerous additional challenges in terms of storage, transport, access and processing. Data can also be generated by various sensors, for example, those used to monitor the environment of the experiment or field sensors for water quality.11,12
Data are also increasingly generated through computational methods and data-intensive science such as simulations, modelling, and workflows. Digital data produced through all these methods are often very complex and generated in a multitude of non-standard formats making them even more difficult to organise and share. The data are also typically separated from the context of the experiment, which is often still captured in the pages of paper notebooks, making it difficult to retrospectively manage.
In this paper, we will examine the benefits of good data management for facilitating or enabling reproducibility of science by encouraging the addition of context that links together the published research with data, methods and rich metadata. Achieving such aims requires commitment at the outset of projects to planning and design; documentation; preservation and protection, thus enabling discovery and reuse.
Other aspects that need attention are interoperability, usability by both humans and machines, the development of services that expedite the processes of recording and management comprehensively, and not only the reinforcement of stricter requirements by publishers and funders but also recognition for researchers who share data effectively.
Scientific data management
Good data management ensures that you can always find your data, that they are well documented and therefore usable when you do find them. It also means that they can be found and understood by others who may gain value from them regardless of whether they were created last week or decades ago. Science builds upon the research of others and that works at both small and large scales.A common problem for researchers is trying and struggling to understand and build upon experiments either published in the literature or even carried out within the same research group, especially when the full record of the experiment is not available and digital data are separated from the context of their creation. Good data management helps by ensuring that all of the information about an experiment is available and well documented, and that links are maintained between the data and the conditions and methods under which it they were generated.
Researchers collect all kinds of data in a variety of forms; these unprocessed ‘raw’ data are processed, analysed and converted into new data to create new understanding and meaning. Data management covers a range of processes relating to the creation, organisation, and protection of the raw and processed data, including data architecture, data modelling and design, data processing and analysis, storage, security, governance, interoperability and integration, data quality and metadata. Data management not only helps to make sure that the data are effectively utilised and stored during use, but also helps to simplify the process of preparing the data to be shared. Although the term data now usually refers to digital data, some of the principles are relevant to physical artefacts such as print-outs, maps, photographs, notes, physical models, and samples.
Data governance encompasses the processes of data management, but also includes processes to ensure quality and consistency of the data and managing accessibility and usability of the data.
The term ‘digital curation’ describes processes for data management over the whole lifecycle of the data, from planning through to preservation, sharing and reuse. These processes also add value to the data in the form of additional context that can assist in the discovery and usability of the data. The processes are typically focused on data at the end of the lifecycle, when the data has been collected and analysed, and is ready to be published or put into storage.
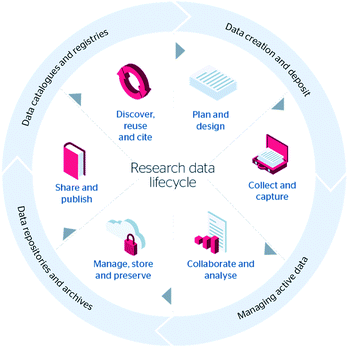
Fig. 1 shows one example of the various stages that research data go through during their lifecycle from planning to preservation and sharing, and ultimately to reuse. This diagram is from the JISC Research Management Toolkit,13 but there are many such lifecycle models of varying complexity. Typically, these diagrams contain a planning stage, a data collection stage, often represented as an iterative stage where the data is captured and described. There may be a collaborative stage where the data is shared and worked with by others but before final publication. Nearing the end of the life cycle the data is preserved and published, where it may then be discovered and reused by others. What the JISC Research Data Lifecycle diagram does not show is the importance of the role of metadata throughout the lifecycle, and in particular how it is used to prepare the data for the later stages of the lifecycle where it will be preserved and shared. The Digital Curation Centre (DCC) have created a Curation Lifecycle Model14 that describes in more detail the actions and responsibilities associated with curating and preserving data.
Metadata provides a simplified representation and description of the larger and more complex body of data and can be understood by both humans and machines. It can be used to not only help describe data and enable them to be discovered, but also to help organise them and create provenance trails that describe the processing and changes that the data have undergone during their lifecycle. Metadata are also fundamental for being able to make use of the data once they have been shared. Metadata are often generated automatically by the instruments and software that capture, generate and transform the data, but ‘user-defined metadata’ can also be created manually to add additional and more meaningful context about the data.
Effective data management requires understanding of a range of topics about data in general but also some specialist knowledge about data formats and use in the appropriate disciplinary domains, including how data are currently collected, processed, stored and utilised within the specific research organisation. An organisation may also be subject to legislative requirements or requirements imposed by funders related to data protection, information governance, copyright and IP and licensing.
Plan and design
It is often the case that the need for better organisation and management of data are not recognised until a problem occurs during a project or when trying to locate and make sense of data after the end of a project. For data to be effectively managed it is necessary to consider how the data will be managed long before any data has been collected. One formal and effective way to do this is the preparation of a Data Management Plan (DMP). The creation of DMPs as part of projects is increasingly being required by funders and institutions, especially for publicly funded research. The DMP describes not only how the data will be taken care of during the project, but also how the data will be prepared for sharing and preservation once the project is complete, or during the project if it is long running.The DMP describes how the data will be collected during the project, how it will be managed, standards that will be used, how the data will be kept safe during and after the project including issues such as data security and backups, and where and how it will be shared. The DMP also contains information about concerns such as sensitivity of the data and related ethical issues, how quality will be ensured, how long the data can or must be retained, and whether it will be subject to any particular licensing conditions. The DMP may also contain information about existing data sources that could be utilised as a part of the project to avoid unnecessary duplication of effort.
The DMP can also detail any tools that will be used or need to be developed in order to work with and manage the data. The plan should also consider the costs and resources required to manage the data, including storage, backup and security protection. The requirements for managing the data during and after the project are likely to be different. Long-term storage and preservation of data involves its own processes and are frequently handed over to a third-party service provider. Consideration needs to be given to the longevity and security of such services and who has ultimate responsibility for the data. Thought also needs to be given about who will be able to access the data, the conditions of use for that data, and how the data will be supported over their lifetime if it is published and shared.
A variety of organisations including national data services, universities and funders provide templates, case studies, tools, and other guidance to assist research teams in creating DMPs for their research projects and understanding the processes involved in data management. The DCC provides links to guidance and examples of DMPs for a variety of disciplines and funders in the UK.15 There are also wizard-style tools to help build tailored DMPs, for example, DMPOnline16 from the DCC is popular in the UK, whilst the US equivalent is DMPTool.17
Although most DMPs are produced in the form of word-processed documents, there has been discussion about the need to have machine actionable DMPs, also known as active or dynamic DMPs, to ensure that the DMP itself is available as a source of information about the research, is kept up to date during a project, and can be used for compliance checking.18 The Research Data Alliance (RDA) has a working group for the development of a common standard for machine-actionable data management plans.19
Data design typically refers to designing the appropriate structures for information in a database, although it is also an important concept within software engineering, and for data that are generated through software the design of data structures to store and manipulate the data is very important. Data can be inputted manually into a database, but for data collection at scale there is likely to be a machine interface to an instrument or sensor to automatically collect and store the data within a database of some kind. In order to design appropriate data structures and database tables it is necessary to have an understanding of the types and sizes of information that are being generated. The data types also determine the kind of database that may be appropriate, for example, for standard applications relational databases are most commonly used, but non-relational NoSQL databases are popular for Big Data because they are significantly more scalable. Resource Description Frame (RDF) triple stores are suitable for holding data describing relationships between objects but historically their scalability has been an issue.20 In addition to allowing greater amounts of data to be stored, NoSQL databases are also very flexible allowing different data structures for different data objects and enabling changes in the way that data are captured and manipulated over time.21 MongoDB, one of the most popular NoSQL databases, represents data in a form very similar to Python dictionaries enabling data to be easily added and extracted using Python code. As a relatively easy language to learn, this lowers the barriers for data management for smaller research groups.
Choices need to be made about how metadata will be captured during the project, and any standards that may be used. There are numerous metadata standards for different disciplines, with many groups choosing to create their own by extending existing standards. The Digital Curation Centre and Research Data Alliance maintain a list of standards, related tools and use cases by discipline.22,23 FAIRsharing.org maintains a list of over 1500 standards in addition to repositories, knowledgebases and policies.24 Decisions also need to be made about how the metadata will be stored, together with the data or separate from it. Metadata can be used to describe the contents of data files and if the metadata are included in the file, the data file is self-describing. For example, CSV data files or spreadsheets may contain a header row with a label to describe each field contained in the rows beneath and no other information is required to understand the meaning of the data. An alternative to self-describing files is to have separate files containing metadata that describe the contents of the data file. The advantage with using a separate metadata file is that more sophisticated descriptions can be provided, for example, linking to standard vocabularies and units of measurement. Also, the same metadata file can be used to describe multiple data files.
When the metadata are held in separate files, it is important that the metadata and data do not become separated, because the data become meaningless without it. A typical way to ensure the metadata and data remain associated is to include a link to the appropriate metadata file or files within the data file. Where many metadata files are related to a data file, an alternative is to create a structural metadata file that describes the relationship between the data files and the metadata files.
Documentation
Documentation of the data is incredibly important both to the owner of the experiment as a reminder of what was done and to help compare results from different activities, and for any other researcher trying to make use of the data. Documentation provides the context that explains not just what the values in the data mean, but also how the data was created. Without good documentation, the data are harder to understand, it is more difficult to assess the quality of the data, and the data are more likely to be misinterpreted or misused. As previously mentioned, it is important that the data and the documentation can be linked together. The documentation must also be high quality, meaning that it must be accurate, complete, and record the important observations, any problems that were encountered and changes that were made in order to adapt. The documentation must provide all the information that explains the data but also enables the experiment or study to be replicated. It is much harder to remember this important information after the event, and therefore the documentation needs to be captured whilst the experiment is being completed.The primary way for most scientists to capture documentation for their research is through paper or electronic notebooks, although some disciplines and in some industrial settings, such as sample testing laboratories, forms may be used to capture the relevant information. For paper notebooks, the content and structure of the documentation is largely determined by personal preferences, previous training and experience of the researcher, but this is also influenced by institutional and disciplinary norms. As an example, notebooks produced by synthetic chemists are usually very formally structured and consistent, whereas the notebooks from many physical chemists include a minimum of structure, often limited to only dates and occasional titles for experiments. Formal structures are sometimes dictated, and these are often the norms within Electronic Laboratory Notebooks (ELNs) where templates may be used to provide consistency or guidance as to what information needs to be recorded. There are pros and cons to providing a strict structure for researchers to use to record and document their research. The requested information is effectively and consistently recorded, but some information that would have been recorded with a less formal structure can sometimes be lost.25 In either case, without adequate documentation, a full understanding of the data cannot be gained.
Metadata are also an important part of the documentation and can be used to describe the documentation in both paper and electronic notebooks. For example, metadata may describe IDs for the notebook, author's name, dates, project funders and project IDs, table of contents containing links to individual pages, short descriptions of experiments, and other information such as experiment IDs, dates, sample IDs, chemicals used, instruments and so on. Although the use of a table of contents within notebooks is relatively rare now, for a small amount of effort they can significantly improve the ease with which a particular experiment or result can be found. Within electronic notebooks, metadata may be automatically captured, and users may also be able to define their own metadata properties and values. Such systems enable information to be found through search and filtering by making use of such metadata values. For example, a search could fetch a list of all experiments using a specified chemical compound.
Other documentation that may need to be created with data include descriptions of existing data, literature sources, instruments, sensors, code, configurations, software, workflows, models and other artefacts that are used as part of the design or execution of an experiment.26 Descriptive metadata can also be produced for these items, for example metadata describing literature sources, instrument configurations and prerequisite versions for software or code that are needed to process and analyse the data.
Organising the data
Regardless of the scale of data collection, starting with a plan for where and how to organise the data files and related resources is important to ensure the data can easily be retrieved and make it less likely that data will be inadvertently overwritten. If the data are stored outside of a database, then an effective directory structure and naming conventions are necessary. Hierarchies are not the only way to organise information, for example some systems may make use of some form of tagging or labelling to organise information, but for most operating systems and cloud-based storage, from a user-perspective, a system of files and folders is most common. High-throughput experiments can generate hundreds of data files for a single experiment, and it makes sense to store those files in their own directory, whereas a small-scale experiment may only require one folder for the whole project. Names for directories may be based on experiment IDs, dates, or topic based. A common strategy for individual file names is to include important IDs within the name of the file in order to facilitate search at a later stage, for example sample IDs, experiment IDs and date.It is important to ensure that raw data are not overwritten by transformations or processing, so either data at different stages of the experiment should be stored in different directories or given unique file names to create different versions of the data. It can be useful to include information about the transformation in the name, for example ‘cleaned’ or ‘analysed’. Dinneen and Julien provide an in-depth review of file management behaviours and issues.27
The data needs to be linked in some way to the documentation describing it. Although it is quite common in paper notebooks for data to be printed out and attached to the notebook, this is not an ideal solution. The data can easily become detached and lost, and the data are no longer in a format that it can be further processed or shared. Where an instrument produces data files in a digital format, an alternative is to list the files generated, together with their location, within the notebook. If the location of the files is later changed, then the list needs to be updated. Many researchers create a similar system when they capture their notes using tools such as word processors or generic electronic notebooks. The advantage with the digital note-taking tool is that a hyperlink can be created, so the files can be located or opened directly from the tool. The links would still need to be manually updated if the location of the files is changed. In an electronic laboratory notebook, it may be possible to attach the data files directly to the experiment record enabling the data and documentation to be stored together. This method is effective for small numbers of files, but most ELN systems are not designed for handling the numbers of files generated by large throughput experiments. An alternative could be to create links to the directories containing the data files or attaching compressed ‘zip files’ of the data.
Structural metadata can be produced and stored together with the data to describe how the data resources are related to each, for example describing the directory structures and how data files relate to one another within a data set. Technical metadata can describe the format, compression and encoding of the files, along with details of the software, version and settings used to produce it.
Storage and protection
Decisions need to be made about where the data are stored during the duration of the project. When data are automatically output by instruments, it may be possible to configure the instrument to put the data in a particular location, and to configure the directory structure and file names to use. Often instruments are connected to a single computer and deposit the data files on the one device. Moving the files from this device for processing and storage is often manual, by copying to a shared drive or using a USB key, but an alternative is to make use of scripts or other software to detect when new files are created and automatically move them to a shared drive or to upload them to an ELN. Consideration needs to be given to how to manage data from multiple users of the same instrument and for different experiments to ensure that data are assigned to the correct researcher and stored in the correct manner.Storing data on an individual's hard-drive or on portable media such as CDs or USB drives is not very safe. Such media can be lost, stolen, or become damaged or corrupted, and they are typically not subject to any form of backup process. Manual backups can be made, for example, copying the data to multiple locations, but the risk with this is that data in one location may be updated or altered, and it can become difficult to know which location holds the original and the most recent data files, especially if multiple users have access to it. Institutional shared drives are likely to have regular backups performed and the capability to restore data if there is some kind of failure or it is accidentally deleted or overwritten. A popular option is to store data in the cloud through services such as Microsoft OneDrive,28 Google Drive,29 and Dropbox.30
Simple operating system-based file management or storing documents on cloud storage is not scalable for environments where vast amounts of files and data are being produced or where many users may need to access the data. For organisations the use of specialist data management and curation platforms such as Cordra, iRODS or the National Institute of Standards and Technology (NIST)'s Configurable Data Curation System are more appropriate, particularly where there is a requirement for collaboration and interoperability.31–33 Commercial options are also available for storage of code and data for managing Big Data and to support applications such as high-performance computing, data analytics and metadata intensive applications such as Google's Filestore, Amazon's FSx, and Microsoft's Azure storage range.34–36
Whatever storage option is chosen, it is important to ensure that backups are actually being taken and that restore is possible; organisations often do not find out their backups are failing until they try to restore lost files. File versioning, as previously mentioned, is one important way to ensure that files are not accidently overwritten. Versioning enables readers to see what changes have been made from one stage to the next. Some tools, including some ELNs, automatically include versioning, but many do not. For tools that do not support versioning it is necessary to save files with a new name or version number at significant points during editing or processing.
Sensitive or valuable data may need additional protection to keep them from being accessed, copied, manipulated or damaged by unauthorised users. Data may also be subject to additional regulations or requirements; for example, data containing personally identifiable information may need to be anonymized. Where the data are stored has an impact on the security of the data; data stored on portable devices can easily be lost or stolen and data stored in the cloud or on institutional shared drives needs to be protected by methods to authorise and authenticate only those users with permission to view and modify the data. ELNs also frequently have sharing capabilities with or without authorisation and authentication. Such tools enable collaboration within a local group or even between global teams depending on the location of the data and accessibility of systems through firewalls.
Preserve and publish
When the data have been collected, analysed, and the findings determined, decisions need to be made about what data needs to be preserved, what can be disposed of, what data should be shared, and how the data will be managed for the remainder of their lifecycle. It is also important to consider what documentation and other resources need to be preserved alongside the data. For example, not only will the data need to be kept up to date as part of preservation, but also the code used to generate them may need to be updated or migrated to ensure that the data are still compatible with new operating systems and prerequisite software. For preservation, some kind of long-term storage is required. For data recorded in a paper notebook long-term storage may be a locked filing cabinet, but for digital data the most common solution is to deposit the data in a database or repository. Paper notebooks and other paper-based documents can be stored more effectively for long-term preservation through digitisation by scanning the documents. In order to make the captured content searchable, Optical Character Recognition (OCR), or more recently Intelligent Character Recognition (ICR) software can be used to extract handwritten text and numerical content from the images or directly during scanning.37There are many repositories and databases for data that range from the very generic that take any kind of dataset from any discipline to those that are very specific and require data to be provided in standard format. Examples of generic or generalist repositories include Dryad, Figshare, Hardvard Dataverse, Open Science Framework, Science Data Bank, and Zenodo.38–43 There are numerous subject specific repositories of relevance to chemistry. For example, Materials Cloud for computational materials science, which also includes code resources and lectures as well as curated datasets; and the NOMAD repository data provides open access to materials data in both raw form and in a machine-processable form in standard formats.44,45 PubChem allows users to upload chemical structures, names, links, spectra and associated bioassay test results, toxicity and other data, and the Crystallography Open Database allows users to upload, validate and deposit CIF files.46,47 Some databases provide services beyond search and storage, for example ioChem-DB provides tools for data creation, curation, analysis and publication of computational chemistry data.48
Data are not necessarily safer in a repository or long-term storage if the problems of media degradation and digital obsolescence are not effectively managed. Even if the repository itself is secure and backed-up the files may need to periodically be copied onto new media, and to be migrated to newer software versions or even transformed into a new standard format to ensure ongoing compatibility. The repository may be a local institutional database where the responsibility for some preservation actions such as keeping file formats up to date and data migration fall to the owner of the data. In some third-party repositories, the job of performing preservation tasks is the responsibility of specialist data curation experts or data stewards. In addition to ensuring that the data are in an appropriate format for preservation, ideally using community accepted standards, documentation and metadata describing the data sets and associated resources are likely to need to be prepared. This is a less onerous task if documentation and metadata describing the data, and how it was generated and processed, have been prepared during the project as the data was created.
A key aspect of preservation of data for sharing is that the data and associated resources including the metadata are supplied with persistent identifiers. However, there is currently a lack of persistent identifiers for data, datasets and metadata meaning that links between publications and their data or metadata can get broken if these resources change location over their lifecycles.3 Persistent identifiers, such as Uniform Resource Locators (URLs) for websites, Digital Object Identifiers (DOIs) for literature and data, and ORCID IDs for authors, are typically created using a third-party identifier registration service. For example, crossref49 for scholarly information, DataCite for Digital Object Identifiers for research data including scientific datasets,50 and ORCID for ORCID IDs for researchers.51
The metadata describing the data may be stored with the data or in a separate database where the database is registered. In this way, even data that are not shared in an externally accessible database can still be discovered by interested members of the community. The metadata should also include information about appropriate licenses for the data, so it is clear in what circumstances the data can be accessed and used, and how to access them. Administrative metadata describes the provenance of the data, intellectual property restrictions, licenses, and access rights. During preservation, additional metadata are created including both administrative and technical metadata describing the storage and operating environment of the data, and updates to new versions or standards. Additional metadata may be captured over time, for example, the number of times the dataset has been accessed or downloaded.
Discovery and reuse
Finally, the data lifecycle comes full circle as the metadata associated with the data enables researchers to discover, access, and license data that are relevant for their projects. Some researchers are entirely reliant on the data that are produced by others, and therefore discovery and reuse are of primary importance, but most researchers make use of the work of others in their experiment designs or to compare research. There are a variety of subject specific repositories and databases that can be searched for data of interest, but there are also data registry services and directories that aggregate metadata from different service providers. Examples include OpenDOAR that allows users to search through thousands of repositories based on location, software and types of material52 and re3data.org is a global registry of research repositories across academic disciplines.53 Google also provides a search engine specifically for searching for datasets.54When data are downloaded from a repository for reuse, the metadata and documentation associated with the data can be kept and updated to indicate the changes made to the data and the provenance path from the origin of the data to the new use and transformation.
Making data usable by machines
A set of principles, called the FAIR guiding principles, were published in 2015 to provide guidance on how to ensure data are effectively managed and made reusable for the benefit of future science.55 FAIR stands for Findable, Accessible, Interoperable, and Reusable and builds upon good data management and data curation behaviour, but with a focus on how data can be made discoverable, meaningful and usable by machines. This focus on interoperability is to enable machines to do the work of finding and assessing whether datasets could be useful for particular scientific research, leaving the researchers more time to focus on the science. The FAIR principles do not themselves provide specific technologies or standards to follow, but they do provide a number of examples of technology implementations that follow the guidelines.56 For chemical information, there is a drive towards the ‘FAIRification’ of existing services, to maximise the value of services that are already being used, and work that has already been done to increase sharing of scientific datasets.57 There is, however, a concern that the lack of prescribed standards and technologies may lead to continuing development of software and infrastructures that are not interoperable.58FAIR advocates for the use of persistent identifiers to enable data and associated documentation and metadata to be found; the use of rich metadata to describe the data so they can be found, understood and then be reused. In order for data to be findable by machines they need to be shared in repositories that machines can search. The Materials Project is an open-access database of materials and properties where each entry is assigned a DOI and a variety of different metadata to make the data discoverable. The content includes citations and links to methods, data, and calculations for each property and the contents of the database are accessible through various dataset search engines.59
Even if the data are not in a repository that can be accessed by the community, metadata describing them should be. The metadata should provide provenance information that enables the quality of the data to be assessed. The metadata used to describe the data must also provide access to licenses. A license selector tool can be used to determine what kind of license is appropriate for the data.60
Currently, even data that are stored in repositories may not be accessible to machines to read and access because of an inappropriate structure or metadata, and therefore require that human intervention or case-by-case scripts need to be created in order to extract the data.61 Competing standards for research data and metadata exacerbate the problem by requiring different software to discover, access, extract and process the data.3
For data and associated resources to be understood by machines they need to make use of standard open protocols and standard file formats; proprietary formats are inaccessible and unusable as they can only be accessed and read by software that has been specifically developed to work with them. The use of proprietary formats also increases the likelihood of digital obsolescence of the data if the software that created them ceases to be supported by the developer and no longer works on newer hardware or operating systems. Ideally community supported standards should be used; those that are already supported by the community have a much better chance of on-going support in the long term and also reduces the amount of duplication required to code machines that are capable of discovering, accessing, extracting and utilising the data. A challenge of scientific data is that each discipline and even sub-disciplines uses their own terminology. Often the same terms have different meanings in different disciplines, and this can cause problems when attempting to integrate or reuse datasets across interdisciplinary projects. schema.org is useful resource for searching and viewing community developed schemas and vocabularies for use on the internet.62
FAIR advocates for the use of standard vocabularies that are linked to the data. As an example, the International Union of Pure and Applied Chemistry (IUPAC) provides a standard vocabulary for chemistry in both printed (IUPAC Gold Book/Compendium of Chemical Terminology) and digital forms including an API.63,64 IUPAC are also developing new standards with FAIR in mind, for example, a standard for the FAIR data management of spectrographic data.65 For chemistry, another challenge for machine-readability is the myriad ways in which chemical information can be represented digitally along with a lack of an adopted standard format for molecular structure information with complete coverage and consistency.66 Groups such as the IUPAC's Data Interest Group/Chemistry, the Research Data Alliance's Chemistry Research Data Interest Group, and the Royal Society of Chemistry's Chemical Information and Computer Applications Group are working on developing standards and tools relating to FAIR and machine-readability of data.67–69
Making data usable by humans
With the current focus on machine-readability, it is also important to consider what elements of the data, associated documentation and metadata need to be understandable by humans as well as the machines. There is inevitably going to be a compromise in getting the balance right. There is no value to human-readability at the expense of machine-readability to ensure the data can be found and accessed, but the data must ultimately be usable by the researchers who need to use them. There are a variety of ways in which human-readability of data can be improved. Data files and metadata formats can become extremely complex and difficult to read. Labels included in such files should be meaningful, so it is clear what the data contained actually means and includes key information such as units of measurement.Machines of course can provide the solution through the development of tools that can read the file formats and present the information contained within in an easy-to-understand manner without the underlying complexity of the internal mark-up being exposed to the user. Documentation that describes the content of the data or metadata files and their structure can also facilitate a better understanding for the users. With the increasing reliance on scripts, workflows and other code to collect, process and analyse our data it becomes critically important that these code assets are properly documented. This includes not just documentation about the intended purpose of the tools, but also ensuring that the code itself is readable, with both readable and sensible variable and function names, most importantly detailed and comprehensible commenting within the code to explain how it works. In this way, a researcher can determine whether the code actually does what the originator intended and that the resulting outputs are correct, in the same way that a calculation within a paper would be assessed for accuracy and validity.
Beyond the data
As has been mentioned numerous times, it is not sufficient to share the datasets alone, more context is required in order to be able to verify and validate the results of research. This means that other researchers should be able to access the associated documentation along with the data, and other resources used in generating it, including any code, scripts and workflows used along with the raw data. Currently it is rare for these resources to be provided with the data.70There are a variety of websites and repositories that are utilised for the sharing of resources beyond the data. For example, websites for sharing protocols and methods including OpenWetWare,71 protocols.io,72 and Protocol Online,73 and also for workflows, such as myExperiment74 and AiiDAlab,75 and for models such as BioModels,76 Chembench77 and OCHEM.78 Several websites also facilitate the storage and sharing of code such as GitHub,79 Gitlab,80 SourceForge,81 and Bitbucket.82
For researchers using workflows as part of their research processes, a recommended practice is the creation of ‘Research Objects’ (RO) that aggregate the workflows and data together with other related resources and metadata so that all the appropriate context and documentation is available for others wanting to reuse the workflow.83,84 The ROs contain everything that is needed to replicate the results from the workflow including the scripts and raw data, and also contain rich metadata to describe the contents and structure of the RO itself. Similar aggregated bundles of materials could be supplied as packages for other data types to include additional context, code, and rich metadata. Such bundles may be generated manually, or tools can be used to automate the process. Science Capsule is an example of one such tool that automates the capture of end-to-end workflow processes including the data, scripts, and contextual metadata.85
Other kinds of information may be appropriate to include together with the data for preservation purposes and to provide the complete context for the data. For example, it could be argued that the DMP should be included along with the project's application for funding and biographies of the personnel who worked on the project. Willoughby provides a more in depth look at what constitutes the scientific record, and what additional materials we ought to consider preserving alongside the data from our research and why.86
For those researchers who spend more of their time working with code, Jupyter notebooks facilitate the sharing of methods and code for the more computationally focused disciplines. Jupyter notebooks (formally IPython notebooks) allow the user to create detailed commentary alongside their code, to attach their data, and enables the results of the code to be effectively viewed within the same document.87 Jupyter notebooks can also be shared so that others can manipulate the code or the data and replicate the results of the notebook author. For example, links to Jupyter notebooks are widely used in fields such as astrophysics and astronomy as an alternative or in addition to supplementary information in publications.88 Jupyter notebooks are also widely used as a teaching tool. The Jupyter notebooks are often shared through code repositories, but also through online services such as the Jupyterhub and Google's Colaboratory so that the author and viewers of a notebook do not need to install and configure a local copy of the software.89,90
Future of scientific data management
With the introduction of the FAIR guidelines and increasing requirements for the use of DMPs, data management has become a hot topic within science. In particular, the focus has been on the use of the Semantic Web to provide new ways to represent and model data, and increasingly how Big Data can be leveraged for scientific discovery through the use of machine-learning and artificial-intelligence technologies.91–94It is clear that there is a need for the establishment of research data management services that provide capabilities for planning research such as data management plans, data collection strategies including ethics and informed consent, and support for workflow design. Services and tools are required for supporting data collection and analysis, code design and sharing, collaboration, and version management. Tools are also required for creating documentation and metadata, organising and storing data. For preservation and long-term storage, tools are required for data storage, archiving, migrating, managing preservation metadata, generating persistent identifiers, and preservation services. The providers of such services need to go beyond just the production of the service and consider how they will provide support to the users of their services, for example providing training and support, as well as publicity to ensure that potential users are aware of the existence and benefits of the system.
At the moment development of such services is piecemeal with a lot of work being done by individual institutions or in relation to individual repositories, but a more joined up solution is required to progress the selection of appropriate community standards to make adoption of FAIR principles more likely. A current focus for infrastructure projects for data management is on the use of modular open-source components that can be developed and shared by different disciplines. Together with the development of user friendly and functional community standards, the use of such components enables the creation of loosely coupled infrastructures that can be more easily repurposed for different purposes and data types. Electronic laboratory notebooks have the potential to be a valuable resource for data management and in the preparation of data for sharing as they already provide a number of useful benefits. The data and the documentation of the experiment can be stored in the same place, depending upon the storage capabilities of the ELN, but even if the data cannot be stored directly, it is easier to create links to the data, and many ELNs enable data to be opened in the correct application from within the ELN. ELNs are also capable of generating useful metadata automatically and often enabling users to create their own user-defined metadata, thus making it easier to generate metadata at the time of creation of the record and data.95 ELNs can also become valuable tools for collaboration by becoming central repositories for storing, sharing, and discussing the research.12 ELNs also have the potential to benefit sharing of data with the wider scientific community, particularly for projects in Open Science where sharing methods and results as quickly and as widely as possible is often a primary goal. Open Notebook Science aims to share the whole research process as it is carried out, enabling rapid peer review and community problem solving. ELNs that are commonly used for open notebook science include LabTrove96 and OpenWetWare Labs,97 although many scientists engaged in open notebook science have created their own websites and blogs to share their research, examples can be found on openlabnotebooks.org.98 The fact that users are creating their own websites for the purpose suggests that there are more opportunities in this space to provide value to both individual researchers and institutions.
One of the challenges with ELNs is getting the data and the associated context and documentation out of them. A lot of ELNs are using proprietary formats and data export and migration is inconsistent, and not necessarily into appropriate machine-readable formats, or even in human-readable formats. Given the value of ELNs in the workflow of scientists more work is needed to ensure that the information captured within is high quality but can also be associated with data sets and exported for publication, sharing and long-term preservation.
Laboratory Information Systems (LIMS) already play an important role within data management for those laboratories where they are deployed, for automated processes, the LIMS can be responsible for the end-to-end management of samples and analysis results. Samples and appropriate metadata are typically registered with the LIMS in a standard format at the beginning of the process, stored within the LIMS and the results reported at the end of the process. Tracking of the data enables provenance information to be automatically captured. LIMS also facilitate the automated collection of data through interfaces to instruments, software and sensors, which enables large quantities of data to be captured and managed automatically. LIMS also have the potential to be connected to external repositories for long-term storage of data. The higher functionality of LIMS for data management and the ability for data stewards to curate the data within the system makes them a good candidate to produce data that are born FAIR.99,100
The Materials Experiment and Analysis Database (MEAD) provides an example of how a good data management framework and experimental science can be combined.101 MEAD contains raw data and metadata from millions of materials synthesis and characterisation experiments which have been distilled into property and performance metrics. The focus of the system is on managing metadata and relationships between the data, connecting the experiment, instruments, analysis and properties together. Instrument data is automatically ingested and linked together with experimental and processing methods, characterisations, associations and analyses. Users of the database can explore the data and download it complete with full provenance information.
Conclusions
Effective data management should be a goal for all scientific projects to ensure that time and money is not wasted on producing data that later become difficult to find or understand, or worse incomplete, lost, or incomprehensible. Good data management also makes the task of preparing data to be shared for the benefit of the whole scientific community much less onerous. Both education and appropriate tools are required to facilitate these processes. To reduce the burden on researchers there needs to be a commitment to usability in the development of community standards, tools and infrastructures for managing data and associated resources throughout the lifecycle.Data availability
As this is a [Review/Perspective article], no primary research results, data, software or code have been included.Author contributions
CW: conceptualization, investigation, writing; JGF: conceptualization, funding acquisition, supervision, review.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Artificial and Augmented Intelligence for Automated Scientific Discovery, EP/S000356/1, funding from Digital Economy IT as a Utility Network+ UKRI/EPSRC EP/K003569/1, PLATFORM: end-to-end pipeline for chemical information: from the laboratory to literature and back again, EP/C008863/1, structure–property mapping: combination chemistry & the grid (CombiChem), GR/R67729/01. We thank Colin Bird for his valuable review and feedback. We also thank the reviewers for their valuable feedback in helping to improve the manuscript.References
- V. Stodden, J. Seiler and Z. Ma, Proc. Natl. Acad. Sci., 2018, 115(11), 2584 CrossRef CAS PubMed.
- H. Rzepa, A. Mclean and M. Harvey, Chem. Int., 2016, 38(3–4), 24 CAS.
- R. Johnson, T. Parsons, A. Chiarelli and J. Kaye, Jisc Research Data Assessment Support - Findings of the 2016 data assessment framework (DAF) surveys, 2016 Search PubMed.
- J. Frey, D. De Roure, M. Schraefel, H. Mills, H. Fu and S. Peppe, in Proceedings of First International Workshop on Hypermedia and the Semantic Web, ed. D. Millard, 2003 Search PubMed.
- M. Shevlin, ACS Med. Chem. Lett., 2017, 8(6), 601 CrossRef CAS PubMed.
- V. Marx, Nature, 2013, 498, 255 CrossRef CAS PubMed.
- S. Lusher, R. McGuire, R. vanSchaik, C. Nicholson and J. deVlieg, Drug Discovery Today, 2014, 19(7), 859 CrossRef CAS PubMed.
- H. Zhu, J. Zhang, M. Kim, A. Boison, A. Sedykh and K. Moran, Chem. Res. Toxicol., 2014, 27, 1643 Search PubMed.
- B. Erickson, Chem. Eng. News, 2013, 91(7), 40 CrossRef.
- R. Mullin, Chem. Eng. News, 2013, 91(42), 19 CrossRef.
- N. Knight, S. Kanza, D. Cruickshank, W. Brocklesby and J. Frey, IEEE Internet Things J., 2020, 7(9), 8631 Search PubMed.
- K. Badiola, C. Bird and W. Brocklesby, et al. , Chem. Sci., 2015, 3, 1614 RSC.
- https://www.jisc.ac.uk/guides/rdm-toolkit, accessed February 2022.
- https://www.dcc.ac.uk/guidance/curation-lifecycle-model, accessed February 2022.
- https://www.dcc.ac.uk/resources/data-management-plans/guidance-examples, accessed November 2021.
- https://dmponline.dcc.ac.uk, accessed November 2021.
- https://dmptool.org, accessed November 2021.
- S. Simms, S. Jones, D. Mietchen and T. Miksa, Research Ideas and Outcomes, 2017, 3, e13086 CrossRef.
- RDA DMP Common Standard for Machine-actionable Data Management Plans|RDA (rd-alliance.org), accessed February 2022.
- R. Punroose, A. Crainiceanu and D. Rapp, Cloud-I '12: Proceedings of the 1st International Workshop on Cloud Intelligence, 2012, vol. 4, pp. 1–8 Search PubMed.
- G. Harrison, Next Generation Databases: NoSQL, NewSQL, and Big Data, Apress, Berkeley, 2015 Search PubMed.
- http://rd-alliance.github.io/metadata-directory/, accessed November 2021.
- https://www.dcc.ac.uk/guidance/standards/metadata, accessed November 2021.
- S. Sansone, P. McQuilton and P. Rocca-Serra, et al. , Nat. Biotechnol., 2019, 37, 358 CrossRef CAS PubMed.
- C. Willoughby, T. Logothetis and J. Frey, J. Chem. Inf., 2016, 8, 9 Search PubMed.
- C. Willoughby and J. Frey, International Journal of Digital Curation, 2017, 12(1), 72 CrossRef.
- J. Dinneen and C. Julien, J. Assoc. Inf. Sci. Technol., 2020, 71, E1 CrossRef.
- https://www.microsoft.com/en-us/microsoft-365/onedrive/online-cloud-storage, accessed February 2022.
- https://www.google.com/drive/, accessed February 2022.
- https://www.dropbox.com/, accessed February 2022.
- https://www.cordra.org/cordra.html, accessed February 2022.
- https://irods.org/, accessed February 2020.
- https://www.nist.gov/itl/ssd/information-systems-group/configurable-data-curation-system-cdcs, accessed February 2022.
- https://cloud.google.com/filestore, accessed February 2022.
- https://aws.amazon.com/fsx/?nc=sn%26loc=0,accessed February 2022.
- https://azure.microsoft.com/en-us/product-categories/storage/, accessed February 2022.
- J. Memon, M. Sami, R. A. Khan and M. Uddin, IEEE Access, 2020, 8, 142642 Search PubMed.
- https://datadryad.org/stash/, accessed February 2022.
- https://figshare.com/, accessed February 2022.
- https://dataverse.harvard.edu/, accessed February 2022.
- https://osf.io/, accessed February 2022.
- https://www.scidb.cn/en, accessed February 2022.
- https://zenodo.org/, accessed February 2022.
- https://www.materialscloud.org/home, accessed 2022.
- https://nomad-lab.eu/, accessed February 2022.
- https://pubchem.ncbi.nlm.nih.gov/, accessed February 2022.
- https://www.crystallography.net/cod/index.php, accessed February 2022.
- https://www.iochem-bd.org/, accessed February 2022.
- http://crossref.org, accessed November 2020.
- https://datacite.org, accessed November 2020.
- https://orcid.org, accessed November 2020.
- https://v2.sherpa.ac.uk/opendoar/, accessed February 2022.
- https://www.re3data.org/, accessed February 2022.
- https://datasetsearch.research.google.com/, accessed February 2022.
- M. Wilkinson, M. Dumontier and I. Aalbersberg, et al. , Sci. Data, 2016, 3, 160018 CrossRef PubMed.
- B. Mons, C. Neylon, J. Velterop, M. Dumontier, L. Da Silva Santos and M. Wilkinson, Inf. Serv. Use, 2017, 37(1), 49 Search PubMed.
- S. Coles, J. Frey, E. Willighagen and S. Chalk, Data Intelligence, 2020, 2(1–2), 131 CrossRef.
- A. Jacobsen, R. de Miranda Azevedo and N. Juty, et al. , Data Intelligence, 2020, 2(1–2), 10–29 CrossRef.
- https://next-gen.materialsproject.org/, accessed February 2022.
- https://ufal.github.io/public-license-selector/, accessed November 2021.
- M. Harvey, N. Mason and A. McLean, et al. , J. Cheminf., 2015, 7, 53 Search PubMed.
- https://schema.org/, accessed February 2022.
- https://goldbook.iupac.org, accessed November 2021.
- https://goldbook.iupac.org/pages/api, accessed November 2021.
- https://iupac.org/project/2019-031-1-024, accessed February 2022.
- L. McEwen, Chem. Int., 2017, 39(2), 6–9 CAS.
- https://iupac.org/digchem-a-vision-for-chemical-data-standards/, accessed February 2022.
- https://www.rd-alliance.org/groups/chemistry-research-data-interest-group.html, accessed February 2022.
- http://www.rsccicag.org/, accessed February 2022.
- C. Hutton, T. Wagener, J. Freer, D. Han, C. Duffy and B. Arheimer, Water Resour. Res., 2016, 52(10), 7548 CrossRef.
- https://openwetware.org/wiki/Protocols, accessed November 2021.
- https://www.protocols.io/protocols, accessed November 2021.
- http://www.protocol-online.org, accessed November 2021.
- D. De Roure, C. Goble and R. Stevens, Future Gener. Comput. Syst., 2009, 25(5), 561 CrossRef.
- A. Yakutovich, K. Eimre and O. Schütt, et al. , Comput. Mater. Sci., 2021, 188, 110165 CrossRef CAS.
- R. Malik-Sheriff, M. Glont and T. Nguyen, et al. , Nucleic Acids Res., 2020, 48(D1), D407 CAS.
- S. Capuzzi, I. Kim, W. Lam, T. Thornton, E. Muratov, D. Pozefsky and A. Tropsha, J. Chem. Inf. Model., 2017, 57(2), 105 CrossRef CAS PubMed.
- I. Sushko, S. Novotarskyi and R. Körner, et al. , J. Comput.-Aided Mol. Des., 2011, 25, 533 CrossRef CAS PubMed.
- https://github.com/, accessed November 2021.
- https://about.gitlab.com/, accessed November 2021.
- https://sourceforge.net/, accessed November 2021.
- https://bitbucket.org/product/, accessed February 2022.
- S. Bechhofer, D. De Roure, M. Gamble, C. Goble and I. Buchan, Nat. Preced., 2010 DOI:10.1038/npre.2010.4626.1.
- http://wiki.myexperiment.org/index.php/Research_Objects, accessed November 2021.
- D. Ghoshal, L. Bianchi, A. Essiari, M. Beach, D. Paine and L. Ramakrishnan, J. Open Source Softw., 2021, 6(62), 2484 CrossRef.
- C. Willoughby, Recording Science in the Digital Era: From Paper to Electronic Notebooks and Other Digital Tools, The Royal Society of Chemistry, 2019, p. 340 Search PubMed.
- https://jupyter.org/, accessed February 2022.
- M. Wofford, B. Boscoe, C. Borgman, I. Pasquetto and M. Golshan, Comput. Sci. Eng., 2019, 22(1), 5 Search PubMed.
- https://jupyter.org/hub, accessed February 2022.
- http://colab.research.google.com, accessed February 2022.
- J. Frey and C. Bird, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2013, 3(5), 465 CAS.
- A. Menon, N. Krdzavac and M. Kraft, Curr. Opin. Chem. Eng., 2019, 26, 33 CrossRef.
- M. Picklum and M. Beetz, Comput. Mater. Sci., 2019, 163, 50 CrossRef CAS.
- F. Strieth-Kalthoff, F. Sandfort and M. Segler, Chem. Soc. Rev., 2020, 49, 6154 RSC.
- J. Frey, S. Coles, C. Bird and C. Willoughby, International Journal of Digital Curation, 2015, 10(2), 1 CrossRef.
- A. Milsted, J. Hale and C. Neylon, PLoS One, 2013, 8(7), e67460 CrossRef CAS PubMed.
- https://openwetware.org/wiki/Labs, accessed November 2021.
- https://openlabnotebooks.org/, accessed November 2021.
- G. Mayer, W. Muller, K. Schork and J. Uszkoreit, et al. , Briefings Bioinf., 2021, 22(5), bbab010 CrossRef PubMed.
- M. Ghaffar, D. Schüler, P. König, D. Arend, A. Junker, U. Scholz and M. Lange, J. Integr. Bioinform., 2019, 16(4), 20190060 Search PubMed.
- E. Soedarmadji, H. Stein, S. Suram, D. Guevarra and J. Gregoire, npj Comput. Mater., 2019, 5, 79 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |