 Open Access Article
Open Access ArticleTrehalose matrices for high temperature dynamic nuclear polarization enhanced solid state NMR†
Monu
Kaushik‡
a,
Hugo
Lingua‡
b,
Gabriele
Stevanato
c,
Margarita
Elokova
a,
Moreno
Lelli
 de,
Anne
Lesage
de,
Anne
Lesage
 *a and
Olivier
Ouari
*a and
Olivier
Ouari
 *b
*b
aCentre de RMN à Très Hauts Champs, Université de Lyon/CNRS/ENS Lyon/UCB Lyon 1, 5 Rue de la Doua, 69100 Villeurbanne, France. E-mail: anne.lesage@ens-lyon.fr
bAix Marseille Univ., CNRS, Institut de Chimie Radicalaire UMR 7273, 13013 Marseille, France. E-mail: olivier.ouari@univ-amu.fr
cInstitut des Sciences et Ingénierie Chimiques, Ecole Polytechnique Fédérale de Lausanne, Av. F.-A. Forel 2, CH-1015 Lausanne, Switzerland
dCenter of Magnetic Resonance (CERM), Department of Chemistry, University of Florence, Via della Lastruccia 3, 50019 Sesto Fiorentino
eConsorzio Interuniversitario Risonanze Magnetiche Metallo Proteine (CIRMMP), Via Luigi Sacconi 6, 50019 Sesto Fiorentino
First published on 21st April 2022
Abstract
Dynamic nuclear polarization (DNP) at cryogenic temperatures has proved to be a valuable technique to enhance the sensitivity of solid-state NMR spectroscopy. Over the years, sample formulations have been optimized for experiments at cryogenic temperatures. At 9.4 T, the best performing polarizing agents are dinitroxides such as AMUPol and TEKPol that lead to enhancement factors of around 250 at 100 K. However, the performance of these radicals plummets at higher temperatures. Here we introduce trehalose-based DNP polarizing matrices, suitable to embed biomolecular assemblies. Several formulation protocols are investigated, in combination with various polarizing agents, including a new biradical structure chemically tethered to a trehalose molecule. The DNP efficiency of these new polarizing media is screened as a function of the radical concentration, the hydration level of the matrix and the protein content. Sizeable enhancement factors are reported at 100 K and 9.4 T. More importantly, we show that the DNP performance of these new polarizing media outperform the conventionally used water/glycerol mixture at temperatures above 180 K. This study establishes trehalose matrices as a promising DNP medium for experiments at temperatures >150 K where conventional water-based formulations soften and are no longer viable, thus opening new avenues for DNP enhanced solid-state NMR spectroscopy at temperatures close to ambient temperature.
Introduction
Solid-state nuclear magnetic resonance (NMR) spectroscopy has developed as an attractive analytical technique to investigate with atomic-resolution the structure, dynamics and interactions of an extremely wide range of solid substrates, from biological assemblies, polymers, glasses, functional materials or pharmaceuticals to name but a few. Despite the versatility of solid-state NMR, its application remains challenging in many research fields due to its intrinsic low sensitivity, which often leads to prohibitively long acquisition times or precludes the analysis of mass-limited samples and of dilute substrates. These limitations have stimulated the development of high frequency high field Dynamic Nuclear Polarization (DNP).1 This approach enables sensitivity increase by 1 to 2 orders of magnitude in solid-state magic angle spinning (MAS) NMR, and has developed as a unique methodology to characterize a wide range of substrates that were previously inaccessible to this spectroscopy.2 This boost in sensitivity is produced by introducing suitable polarizing agents, such as organic free radicals or paramagnetic metal complexes in the sample and by applying a micro-wave irradiation to transfer the spin polarization of the intrinsically highly polarized unpaired electrons of the dopant to the neighbouring nuclear spins. The power and frequency of the microwave source, the choice of the polarizing agent, its concentration as well as the temperature of the experiment dictate the DNP mechanism invoked during the process.3 Today, the most efficient polarization transfer scheme for MAS DNP NMR experiments at 100 K is the cross-effect (CE) mechanism, which is particularly effective with organic biradicals. A series of dinitroxides and mixed narrow-/broad-EPR line biradicals of tailor-designed molecular structure are available today for efficient DNP at high magnetic fields (9.4 to 21 T).4The vast majority of DNP solid-state NMR experiments are performed at cryogenic temperatures (∼100 K). Cryogenic temperatures provide long enough electron and nuclear spin relaxation times, which ensures an efficient transfer of polarization and in turn high signal amplification factors. Yet, this temperature requirement often impairs the spectral resolution and hence limits the potential of DNP MAS NMR, notably for biological applications. In frozen samples, the spectral resolution of biomolecules is indeed affected by a significant inhomogeneous broadening of the NMR lines, which arises partly from the coexistence of a distribution of conformers trapped at low temperature.5 Several methods have been investigated to restore room temperature resolution, such as in situ temperature jump by sample melting with laser irradiation.6 This approach however necessitates a dedicated home-made instrumentation. In the 2010s, Oschkinat and co-workers suggested to perform protein DNP NMR experiments at temperatures of around 180 K instead of 100 K to improve spectral resolution.7 However the effective improvement in resolution comes at the expense of significantly lower sensitivity. Indeed, the efficiency of the polarization transfer decreases as the temperature increases and relatively poor enhancements are usually reported above 150–180 K.8 Combining sensitivity and resolution is thus one of the key challenges of modern DNP MAS NMR. While the recent advents of DNP instrumentation operating at very high magnetic field (up to 21.1 T) and fast MAS (up to 60 kHz) are milestones to reach this goal,9 the development of DNP sample formulations that would preserve high enhancement factors at elevated temperature is highly desirable.
The most common formulation consists in embedding the substrate of interest in a radical-containing medium known to form a glassy matrix at cryogenic temperatures. Mixtures of water and glycerol, or of water and DMSO10 as well as non-aqueous solvents such as TCE (1,1,2,2-tetrachloroethane)8a or OTP (ortho-terphenyl)11 are typically employed as DNP polarizing glassy matrices. Microcrystalline organic solids and hybrid or inorganic materials are usually prepared by incipient wetness impregnation, while biomolecules are simply dissolved or immersed in the radical-containing solution after precipitation by crystallization or sedimentation. Alternative polarizing media have been proposed such as acrylamide gels12 that can be used to disperse colloidal nanoparticles. It has also been shown that the solvent phase could be reduced in amount and this approach has been successfully applied to characterize lipids of liposomes, crystalline cellulose and membrane polypeptides.13 Finally, solid materials incorporating radicals covalently bond to their organic or inorganic framework have been proposed to polarize bulk frozen solutions.14 Despite all these developments, there is currently no DNP sample formulation compatible with biomolecular samples that allows experiments at temperatures above 180 K. Some of us recently demonstrated that enhancement factors as high as 65 could be obtained at 230 K and 18.8 T with the HyTEK2 radical in ortho-terphenyl (OTP), a high glass transition temperature (Tg) rigid matrix.15 While OTP is not a good solvent candidate for biomolecules, this result highlights that high Tg matrices potentially offer promising opportunities for DNP MAS NMR at non-cryogenic temperatures.
Here we introduce a new polarizing matrix composed of trehalose sugar. Trehalose is a natural cryo- and lyoprotectant found in lifeforms that survive extreme weather conditions thanks to its extensive network of hydrogen-bonds, its capacity to stabilize proteins in their biologically active state and its high melting point.16 At cryogenic temperatures, trehalose forms an amorphous matrix that has a glass transition temperature of about 380 K.17 Several formulation protocols have been proposed to uniformly disperse biomolecules in sugar glasses. The film formulation method involves spraying an aqueous solution of protein and sugar on a surface and letting the solvent evaporate.18 Another commonly-employed method consists in freeze-drying (or lyophilizing) the protein/sugar solution followed by a rehydration step in a controlled atmosphere.19 These sugar matrices preserve the structural integrity of embedded biomolecules and in addition enable longer electron relaxation time in nitroxide labels, which has been exploited to measure distances between spin-labelled protein segments by EPR spectroscopy at ambient temperature.20 Their unique immobilization capabilities critically depend on the propensity of sugars to create intermolecular hydrogen bond networks and long-range, homogeneous connectivity within the glass.20d,21 The presence of hydration water molecules forming a layer between the protein surface and the trehalose matrix remains however essential to maintain protein dynamics.19b,22 Of interest, it has been observed that the transition from aqueous solution to the dense and highly concentrated glassy state of trehalose maintains the stabilizing properties of the trehalose matrix, a key feature in bioadaptation processes such as desiccation stress.34
Trehalose and glucose have also been exploited for DNP MAS NMR, at 100 K in a reduced solvent content approach, where sugars in the presence of glycerol were proposed to act as a gluing agent between the polarizing agent and the biomolecule of interest,23 while retaining structural water molecules around the substrate and maintaining a uniform distribution of the polarizing agent during the drying process under vacuum.
In this work, we introduce a new approach to high temperature DNP MAS NMR of biomolecules using trehalose-based formulations. We hypothesized that the high glass transition temperature of trehalose can be exploited to preserve the efficiency of the DNP transfer mechanism at temperatures above 180 K. Specifically, we present several experimental protocols to formulate trehalose matrices in combination with various nitroxide-based radicals and for various hydration levels. We demonstrate that casein as a model protein can be incorporated in these sugar-based DNP media and investigate its temperature-dependence enhancement profile. We show that an enhancement factor of above 10 at 9.4 T is preserved for the protein resonances at around 230 K.
Results and discussion, experimental
Polarizing agents
Three free radicals, O-MbPyTol, AMUPol and PyT, were tested as polarizing agents in sugar matrices. Their structures are depicted in Fig. 1(a). They were selected after an initial screening by EPR where the propensity of these radicals to disperse homogeneously in the sugar matrix after the freeze drying process (see below) was checked (Fig. S3, ESI†).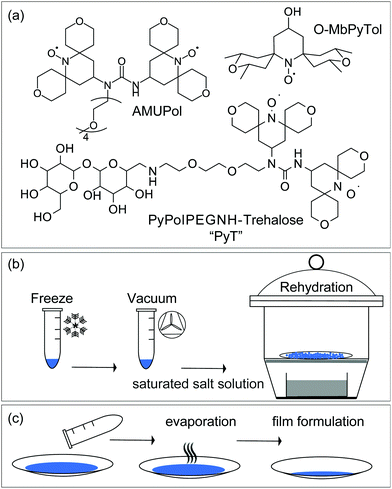 | ||
| Fig. 1 (a) Molecular structures of O-MbPyTol, AMUPol and PyT. Schematic presentation of (b) the freeze drying and rehydration method, and (c) the film formulation method, that were used for DNP sample formulation in sugar matrices. In (b), a precise control of the hydration level was achieved by using the isopiestic method, according to the protocol described by Malferrari et al.22a Two hydration levels were obtained using relative humidity levels (RH) of 11 and 74% in a gastight chamber as detailed in the ESI,† corresponding to H2O/sugar molar ratio of ∼0.5 and 2.5 respectively. | ||
O-MbPyTol24 is the ‘open’ form of a spirotetrahydropyrane mononitroxide and has been recently shown to yield enhancements of 119 ± 2 in DMSO/water mixture at 9.4 T and 100 K, the highest value reported so far for monoradicals under these conditions. AMUPol8b is currently the most efficient polarizing agent for CE DNP in aqueous solutions at intermediate magnetic fields, with enhancements as high as ∼250 at 9.4 T in frozen glycerol/water solutions. PyT (PyPolPEGNH-Trehalose) is a new biradical introduced in this study that corresponds to a PyPol moiety8b tethered to trehalose via an ethylene glycol chain. The synthesis of PyT is reported in the ESI,† while those of O-MbPyTol and AMUPOL have been described in previous publications.8b,24
Formulation methods
Each of these free radicals were incorporated at various concentrations in sugar matrices, following either the freeze-drying and rehydration process (Fig. 1b), or the film formulation method (Fig. 1c). The two protocols are detailed in the ESI.†DNP features of trehalose matrix prepared by freeze drying
For freeze dried samples, a precise control of the hydration level was achieved using an isopiestic method to avoid partial or complete crystallization of the sugar matrix. In detail, for rehydration after freeze drying, the samples were immediately exposed to a controlled relative humidity (RH) in a gastight chamber containing a saturated aqueous solution of salt (RH = 11% and 74% for LiCl and NaCl solutions, respectively, Fig. 1b).20a These hydration levels correspond to H2O/sugar molar ratios of 0.5 and 2.5, which is represents roughly 3 and 11 weight percent of water, respectively (see ref. 19b and TGA data in Table S5 and Fig. S8, ESI†). Two radical concentrations were investigated, corresponding to sugar/free radical molar ratios of 325 and 130, corresponding to respectively 0.3 and 0.8 mole percent, or 0.9 and 2 weight percent of free radical. The film formulation is more straightforward but less reproducible. A solution of trehalose and free radicals in water is exposed to air and let evaporate to get a film (Fig. 1c). The moisture content is therefore not controlled in the preparation protocol.EPR spectra of trehalose formulations are shown in Fig. S3 (ESI†). For the mononitroxide O-MbPyTol, the EPR signal exhibits a powder pattern characteristic of no or limited amount of aggregation. For PyT and AMUPOL, the EPR signals are more complex due to intramolecular spin exchange and dipolar magnetic interactions. However, for PyT, the pattern is very similar to the EPR spectrum recorded in glycerol/water solution at 193 K, where the radical is homogeneously distributed.
The DNP performance of the different samples was then evaluated at 9.4 T and 100 K. The DNP enhancement factors obtained for O-MbPyTol, AMUPol and PyT incorporated in trehalose matrices at 0.3 and 0.8 molar ratio are reported in Fig. 2a.
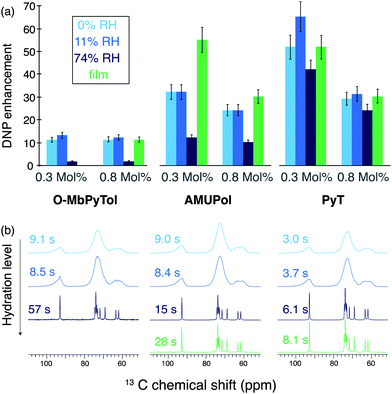 | ||
| Fig. 2 (a) DNP enhancement factors ε for 0.3 and 0.8 mole percent in free radical O-MbPyTol, AMUPol and PyT incorporated in trehalose matrices. Samples are prepared by freeze drying aqueous solutions (blue color) or by forming films (green color). Data are reported at different relative humidity (RH) levels for the freeze-drying method. The enhancements correspond to proton enhancements measured on the 13C resonances of trehalose after a cross-polarization step. (b) One-dimensional 13C CPMAS NMR spectra and proton polarization build-up times TB,on of the trehalose matrices (the color code is the same as in (a)). The spectra were recorded with a radical molar ratio of 0.3% in free radical. One-dimensional 13C CPMAS NMR spectra corresponding to 0.8 mole percent in radical are shown in the Fig. S4 (ESI†). Enhancements were measured from the peak intensity of the 13C resonance at 74 ppm of trehalose. The data were recorded at 9.4 T (263 GHz microwave frequency) with a 3.2 mm probe. A MAS frequency of 8.0 kHz and a sample temperature of 94 ± 2 K were maintained. The proton build-up times indicated next to the spectra were measured indirectly on carbon-13 spectra, by inserting a 1H saturation and recovery period before the CP step. | ||
For O-MbPyTol, AMUPol and PyT, the exposure of the freeze-dried matrix to moisture at a relative humidity (RH) level of 11% has hardly any effect on the enhancement factors with regard to direct measurements after freeze drying. This is consistent with the fact that the hydration level of the matrix is nearly unchanged (Table S5, ESI†). However, a higher moisture content (RH of 74%) leads to a deterioration of the DNP performance for O-MbPyTol and AMUPol biradicals. Such a drastic drop of the enhancement factor at high degrees of hydration is less marked for the PyT biradical.
Additional important information is obtained by analyzing the significant differences observed in proton polarization build-up times TB,on and in 13C spectral resolution depending on the radical and formulation protocol used. Indeed, the polarization build-up times are indicative of the radical distribution within the matrix, while spectral resolution provides clues about the crystalline versus amorphous nature of the polarizing medium.
Fig. 2(b) shows 13C cross-polarization (CP) spectra of the trehalose matrix recorded for the 0.3 mole% (mol%) radical concentration at different hydration level prepared with the freeze-drying method (in blue). The same trend is observed for all three polarizing agents. In the dehydrated matrices and in the samples exposed to a RH of 11%, the 13C NMR lines are relatively broad (∼5 ppm) indicating the presence of amorphous glassy matrix preserved after the freeze-drying process. In contrast, the resonances become considerably sharper (∼0.4 ppm) and 1H relaxation times notably increase for higher hydration level (RH of 74%).
These narrow resonances and long build-up times suggest the formation of crystalline domains or domains more ordered than those present at lower hydration levels. This observation agrees with the EPR studies carried out by Malferrari and co-workers,19b showing the presence of crystalline clusters upon rehydration of the trehalose matrix. Here we note that the two types of 13C spectra are in line with those reported previously in literature, for example by Lefort et al. on amorphous and crystalline forms of trehalose.25
Proton buildup times (TB,on) were measured for all types of formulations and are reported to the left-hand side of the spectra. Relatively long 1H TB,on were observed for O-MbPyTol and AMUPol at high hydration level for the freeze-dried samples (57 and 15 s, respectively), which is consistent with the formation of ordered domains of trehalose, from which these nitroxides are excluded. In these cases, the transfer of polarization from the radical to the trehalose molecules would mainly occur by spin-diffusion relayed mechanisms.26 This segregation explains the drastic drop observed in the DNP enhancement factors for O-MbPyTol and to a lesser extend for AMUPol at a RH of 74%. In contrast, in the rehydrated matrix that incorporates PyT, the polarization build-up time is only slightly elongated at high water content (6.1 s versus 3.7 s), which suggests that the radical is embedded within, rather than excluded from, ordered domains. This is most likely due to the specific molecular structure of PyT. The trehalose-like part of the molecule is indeed expected to facilitate its incorporation into those domains. This in turn leads to a more homogeneous distribution of PyT and explains why relatively high DNP enhancements are preserved at high rehydration levels, namely 42 for a concentration of 0.3 mol% in PyT versus 12 and 1.7 for AMUPol and O-MbPyTol respectively [dark blue bars in Fig. 2(a)], and 24 for a concentration of 0.8 mol% in PyT versus 10 and 1.5 for AMUPol and O-MbPyTol respectively [dark red bars in Fig. 2(a)]. Carbon-13 spectra and 1H TB,on of trehalose matrices containing a free radical concentration of 0.8 mol% are presented in Fig. S4 (ESI†). As expected at this higher radical concentration, shorter build-up times are measured for glassy formulations enabling a homogeneous distribution of the polarizing agent.
DNP enhancement factor (ε) is of direct importance in evaluating the DNP process, but the relative signal intensity per square root of time affords a better evaluation of the performance when comparing different polarizing formulations.27Table 1 summarizes both the enhancement factors ε as well as the values of ε/√TB,on for the different freeze-dried formulations.
| Radical | RH (%) | 0.3 mol% | 0.8 mol% | ||
|---|---|---|---|---|---|
| ε(CP) | ε/√TB,on | ε(CP) | ε/√TB,on | ||
| O-MbPyTol | 0 | 11 | 3.6 | 11 | 6.1 |
| AMUPol | 32 | 10.7 | 24 | 10.7 | |
| PyT | 52 | 30.0 | 29 | 32.4 | |
| O-MbPyTol | 11 | 13 | 4.5 | 12 | 7.2 |
| AMUPol | 32 | 11.1 | 24 | 10.5 | |
| PyT | 65 | 33.8 | 31 | 32.7 | |
| O-MbPyTol | 74 | 1.7 | 1.4 | 1.5 | 0.3 |
| AMUPol | 12 | 3.1 | 10 | 2.5 | |
| PyT | 42 | 17 | 24 | 14 | |
We note that PyT yields shorter build-up times than AMUPol at the two concentrations, which is also a benefit in terms of overall sensitivity enhancement. The relative values for the overall DNP sensitivity expressed as ε/√TB,on clearly reflect the superior performance of PyT over AMUPol in trehalose matrices.
Trehalose matrices were also prepared in the absence of any polarizing agent to validate that the presence of the biradical does not have any impact on the formation of either disordered or crystalline phases (Fig. S4, ESI†). A series of freeze-dried samples with different rehydration levels were prepared and the corresponding NMR spectra were measured at room temperature. The spectral resolution of 13C CPMAS spectra and proton T1 were found to be directly linked to RH levels, exactly in the same way as observed for doped formulations.
DNP features of trehalose matrix prepared by film formation
During this process, a solution of trehalose and free radicals in water is exposed to air and let evaporate (Fig. 1c). The moisture content is therefore not controlled in the preparation protocol. Nevertheless, physical attributes as well as NMR data suggest that the hydration level in the films is higher than that achieved for the freeze-dried samples upon rehydration at 11% RH. Indeed, the films have a wet and paste-like aspect, in contrast to the flaky powder consistency of the 0 and 11% RH level freeze-dried samples, which becomes increasingly granular at higher hydration levels.In terms of DNP performance, O-MbPyTol (only evaluated at a concentration of 0.8 mol%) yields a poor enhancement factor. Both AMUPol and PyT at 0.3 mol% are giving competitive enhancement factors in air-dried trehalose films (green bars in Fig. 2a). The 13C NMR spectra are all dominated by narrow lines that reflect the formation of ordered domains. At a concentration of 0.8 mol% (Fig. S4, ESI†), for O-MbPyTol and to a lesser extend for AMUPol, the presence of amorphous phases is also observed as broad resonances under the sharp ones. As was the case for the freeze-dried samples at a RH level of 74%, a much longer build-up time TB,on is measured for AMUPol than for PyT (28 s versus 8.1 s at a concentration of 0.3 mol%, and 58 s versus 12 s at a concentration of 0.8 mol%, see Fig. S4, ESI†). This suggests a homogeneous distribution of PyT in trehalose films. We note that for 0.8 mol% O-MbPyTol, a bi-exponential fit was applied to extract the TB,on. It provides two distinct values, indicating the joint formation of both amorphous and crystalline phases in the film.
In conclusion, substantial enhancement factors (ε13C = 65) and short polarization build-up times were measured by incorporating PyT in trehalose matrix either by freeze-drying the samples or by forming a film. This latter approach provides well-ordered phases, while glassy domains were obtained in the freeze-dried samples with no or low rehydration. Overall, these data suggest that PyT in trehalose matrices can be a viable alternative to the cryoprotecting glassy DNP media currently used for biomolecules (typically solution of 10 mM AMUPol in 60% glycerol-d8, 30% D2O, and 10% H2O, the so-called DNP juice). In the following, we report DNP experiments on a model protein embedded in PyT/sugar polarizing matrices using the freeze-drying formulation protocol.
Protein in polarizing trehalose matrix
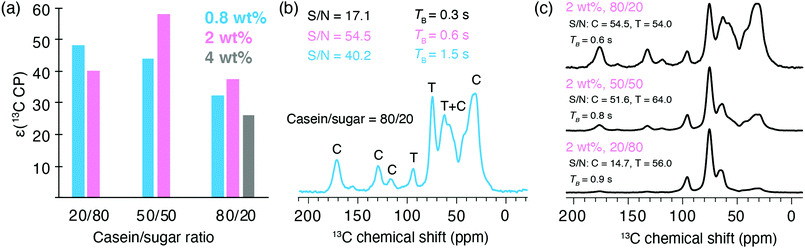
Casein (at natural abundance) from bovine milk was chosen as a model protein as it is readily available, stable, and easy-to-handle.The protein/sugar ratio in the trehalose glass was varied from 20/80 to 50/50 and 80/20 (w/w). A detailed description of sample formulation is given in the SI. Several free radical concentrations were screened around the 0.3 mole percent (0.9 weight percent) value previously optimized for the trehalose matrix. The protein/sugar formulations are expected to significantly differ from the pure trehalose matrix, notably at the highest concentration of the biomolecule. Free radical concentrations are indicated in the following, respectively, in weight percent (wt%) as there is a distribution of molar mass in the casein sample. The solutions were first freeze-dried using the aforementioned protocol (Fig. 1b) without any rehydration. Fig. 3a shows 13C enhancement factors measured on the protein bulk resonance at 30 ppm. Proton and carbon-13 enhancements measured on the trehalose resonances are also reported in Table S3 (ESI†). Significant enhancement factors are measured for the protein resonances in the formulations containing 20 and 50% (w/w) of protein, at a radical concentration of 0.8 and 2 wt%, with ε values between 40 and 58. These values are comparable to those reported for the pure trehalose matrix (Fig. 2a). Relatively lower DNP enhancements were obtained with a protein content of 80%, which might be due to protein aggregation due the low amount of trehalose. In this latter case, a radical concentration of 4 wt% was also evaluated but did not yield higher enhancement factors. Proton build-up times were measured for each formulation and signal-to-noise ratio per unit of time and unit of mass were calculated from one-dimensional spectra recorded with a recycle delay of 5 times TB,on (Table S3, ESI†). Fig. 3(b) shows the 13C spectrum corresponding to the 80/20 casein/sugar formulation doped with 0.8 wt% in PyT. The line broadening of the trehalose resonances clearly indicate that the protein is embedded in a glassy sugar matrix. The fact that similar enhancements are measured for the casein and trehalose signal also supports this hypothesis (Table S3, ESI†). The build-up times and signal-to-noise ratio are also indicated for the 80/20 protein/sugar ratio at the 3 different radical concentrations. As expected, the DNP build-up times are getting shorter with increasing radical concentrations. Despite a short TB, the formulation prepared with a concentration of 4 wt% in PyT leads to a drop in the overall sensitivity gain likely due to strong paramagnetic bleaching. The 2 wt% concentration in PyT radical leads to the highest S/N ratio. Fig. 3c shows the signal to noise ratio calculated on protein and trehalose regions of 13C spectra of samples formulated with 2 wt% in PyT and various casein/sugar ratio. The 80/20 and 50/50 ratio yield similar overall sensitivity on the protein signals.
In order to evaluate the effect of sample hydration, the freeze-dried formulation containing 50/50 casein/sugar and doped 2 wt% in PyT was exposed to relative humidity levels of 11% and 74% (Fig. S5, ESI†). In presence of embedded biomolecules, the trehalose matrix was previously reported to remain homogenous even at 74% RH.20a DNP Enhancement factors and DNP build-up times were recorded at 100 K for each sample and are reported in Fig. S5 (ESI†), with the corresponding carbon-13 spectra. Enhancements of 64 and 56 were measured on the rehydrated formulations for the 11% RH and 74% RH levels respectively. As was the case of the pure matrix, at a RH of 74%, the trehalose resonances are narrower than at lower hydration levels, which is indicative of the formation of ordered sugar domains around the protein, while the 1H enhancement is not affected by the increased hydration level. These experiments demonstrate that hydrated biomolecules can be successfully and homogeneously incorporated within DNP trehalose formulations at high protein to matrix ratio, and that enhancement factors as high as 64 can be achieved on the protein resonances.
DNP at elevated temperatures in polarizing trehalose matrix
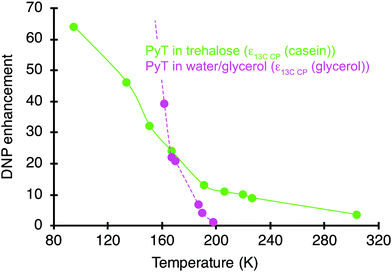
The DNP enhancements were monitored on the hydrated casein/sugar formulations as a function of temperature. The longitudinal relaxation time (T1) of 79Br in KBr was used to measure each temperature point precisely28 inside the rotor by packing a pellet of KBr at the bottom of the rotor. Fig. 4 shows 13C DNP enhancement factors measured on the casein resonances for the 11% rehydrated 50/50 casein/sugar formulation prepared with 2 wt% in PyT. A gradual decline in the DNP performance is clearly evident with still a sizable enhancement of 10 preserved up to a temperature of 220 K. It is the most efficient polarizing media found so far for water soluble samples at temperature above 200 K. At 304 K, an enhancement of ∼4 is recorded. Similar data were recorded for a RH of 74% and are shown in Fig. S6a (ESI†).Pure trehalose matrices have been reported to have glass transition temperature Tg of 379 K.17 Hydration content in sugar matrices is known to influence the glass transition temperature. The glass transition temperature is also possibly modified by the addition of polarizing agents and proteins. The ‘glass transition’ of proteins occurs at temperatures around 180 and 240 K and the freezing of the hydration shell is described to happen below 200 K.35 To get an insight into these temperatures, DSC measurements were conducted for several freeze-dried formulations, including on the pure and doped sugar matrix and the DNP formulations containing 50/50 casein/sugar and 2 wt% PyT at the 0, 11 and 74% RH levels. Measurements were performed from 110 to 373 K (Fig. S9, ESI†). Glass transition temperatures could be clearly seen at 318 and 291 K for the pure dry sugar matrix after freeze-drying and for DNP sample formulation with casein at a RH = 11%, respectively. The sugar matrix thus remains probably rigid in the temperature range on which the DNP NMR experiments were carried out (Fig. 4). However, as the temperature increases, the rigidity of the trehalose matrix is expected to diminish due to entropy-induced motions.
It is noteworthy that the spin polarization is transferred from the polarizing agents to the protein via the trehalose matrix in a freeze dried powder-like sample containing no bulk solvent molecules.
Despite a continuous decrease, sizable enhancements are still measured up to room temperature. This performance is compared with that of a pure solution of PyT dissolved in 60% glycerol-d8, 30% D2O, and 10% H2O (pink curve in Fig. 4). While a higher enhancement is achieved at 100 K in the aqueous matrix (ε = 210), a clear difference in the slopes of the two curves is evidenced. Notably, at around 200 K the glycerol–water mixture is no longer a working matrix for DNP, with no enhancement. The temperature dependence of the enhancement was also recorded for a standard DNP formulation of 12 mM AMUPol in a 60/30/10 glycerol-d8/D2O/H2O solution containing 0.2 M proline (Fig. S6b, ESI†) and follows the same trend. As the frozen glycerol–water soften around 180 K,29 it can no longer be used as an amorphous matrix for DNP polarization transfer above this temperature. These results are in line with data reported by Akbey et al. who obtained an enhancement of 11 at 180 K on the microcrystalline SH3 protein in a glycerol–water mixture and no enhancement above 200 K.7 The proposed formulations based on trehalose matrices thus represent a novel approach for biomolecular DNP at temperatures higher than 200 K. The effective sample volume is maximized by the removal of the solvent while the hydration shell around the protein is retained.
An enhancement factor of 10 is obtained for the casein signals in the trehalose matrix at 220 K. This temperature is above the glass transition temperature of many hydrated proteins30 and therefore, an improvement in spectral resolution can be expected. Such an improvement was however not detected in the carbon-13 spectra recorded here (Fig. S7, ESI†), as the size of the protein (∼25 kDa) does not allow to get resolved individual carbon-13 resonances in one-dimensional spectra.
Conclusions
In this work we investigated a new sample formulation for high temperature DNP MAS NMR that relies on the use of trehalose as matrix. Using PyT, a new radical specifically designed to homogeneously disperse into sugars, enhancements of around 50–60 are achieved at 100 K in freeze dried trehalose matrices. While at these cryogenic temperatures, these enhancement factors are lower than the value reported with AMUPol in frozen solutions of water and glycerol, we show that the trend is opposite at higher temperatures. The sensitivity gain achieved with PyT in sugar matrices becomes substantially higher at temperatures >180 K. These results are confirmed for a trehalose DNP formulation incorporating casein as a model protein, with a record enhancement factor of 10 at 220 K and 4 at 304 K on a biomolecular sample. Our results demonstrate the significant benefits of this formulation for the study of samples available in limited quantities and of unlabeled biomolecules at temperatures around 200 K, a regime where conventional water-based matrices soften and are no longer viable for efficient polarization transfer. We believe that this is the first time that sizable DNP enhancements are reported on a protein at a temperature above 200 K. This proof-of-principle sets a milestone for combining DNP enhancements and improved resolution in biomolecular solid-state NMR. We expect that the deuteration of the matrix, 2 – and 3D experiments, DNP at higher magnetic field and faster MAS rates are the next steps and thus open up the perspective of achieving higher resolution, high temperature DNP in biosolids, which is currently one of the main drawbacks of DNP ssNMR in biological applications.Experimental section
Free radicals of appropriate concentration are embedded in sugar matrix by preparing an aqueous solution and removing excess water by either of the methods explained below.Freeze dry and rehydration
Freeze dry method includes an initial step of lyophilization followed by an additional step of rehydration. Controlled amount of moisture is re-introduced into the samples by exposure to relative humidity (RH). The solution of trehalose and radical in water are lyophilized for >15 hours. The flaky white solid thus obtained is rehydrated in the next step that requires exposing the dried powder to controlled levels of humidity in a desiccator, schematic of the setup is shown in Fig. 1(b). A saturated salt solution of LiCl or NaCl in a hermetic environment equilibrates at 11% or 74% relative humidity respectively at ambient temperature of ∼25 °C. After a prolonged (>12 hours) exposure to humidity the water content in the samples correspond to 0.5 and 2.4 water per sugar molecule in trehalose.19bFilm formulation
An aqueous solution of sugar and radical is transferred to a watch glass for a large surface area and left under fume hood for approximately 3 hours to facilitate evaporation, schematic of the setup is shown in Fig. 1(c). The concentration of water is not controlled following this method.31DNP NMR experiments
The MAS DNP NMR experiments were performed on a Bruker Avance III 400 MHz wide bore spectrometer, equipped with a triple resonance 3.2 mm low-temperature DNP MAS probe. The samples were irradiated with continuous wave high-power microwaves at a frequency of 263 GHz, with a power stability better than ±1%. The 1H 1D spectra were recorded with a background suppression and phase cycled DEPTH pulse sequence using a proton 90° pulse of 2.5 μs.32 The 13C spectra were recorded with a 1H–13C cross-polarization (CP) pulse sequence, with a contact time of 1 ms. For protons a linear ramped CP was used to optimize the magnetization transfer efficiency. A proton radio frequency (RF) field of 70 kHz in the center of the ramp was applied, while the RF field on carbon-13 was adjusted for optimal sensitivity. SPINAL-64 proton decoupling33 was applied during acquisition at a radio-frequency field of 100 kHz. 1H buildup times were measured via saturation recovery pulse sequence either directly on 1H or by CP to 13C, each case being specified in the text wherever applicable. DNP enhancement factor was measured by scaling intensity of ‘microwave off’ spectrum to ‘microwave on’ spectrum that are both recorded with the same delay time and receiver gain. Additional details are provided in the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from Equipex contracts, ANR-15-CE29-0022-01, and ANR-17-CE29-0006-01 are gratefully acknowledged. M. K. gratefully acknowledges financial support from the Deutsche Forschungsgemeinschaft (KA 5221/1-1). M. L. gratefully acknowledges financial support from Fondazione CR Firenze and from Instruct R&D project (Appid 1514).Notes and references
- (a) Q. Z. Ni, E. Daviso, T. V. Can, E. Markhasin, S. K. Jawla, T. M. Swager, R. J. Temkin, J. Herzfeld and R. G. Griffin, Acc. Chem. Res., 2013, 46, 1933 CrossRef CAS PubMed; (b) R. G. Griffin, T. M. Swager and R. J. Temkin, J. Magn. Reson., 2019, 306, 128 CrossRef CAS PubMed.
- (a) P. Berruyer, L. Emsley and A. Lesage, eMagRes, 2018, 7, 93 CAS; (b) A. G. M. Rankin, J. Trébosc, F. Pourpoint, J.-P. Amoureux and O. Lafon, Solid State Nucl. Magn. Reson., 2019, 101, 116 CrossRef CAS PubMed; (c) A. L. Paioni, M. A. M. Renault and M. Baldus, eMagRes, 2020, 15, 51 Search PubMed; (d) K. Jaudzems, T. Polenova, G. Pintacuda, H. Oschkinat and A. Lesage, J. Struct. Biol., 2019, 206, 90 CrossRef CAS PubMed; (e) L. D. Fernando, W. C. Zhao, M. C. D. Widanage, F. Mentink-Vigier and T. Wang, eMagRes, 2020, 9, 251 CAS.
- K. Kundu, F. Mentink-Vigier, A. Feintuch and S. Vega, eMagRes, 2019, 8, 295 CAS.
- G. Casano, H. Karoui and O. Ouari, eMagRes, 2018, 7, 195 CAS.
- (a) A. H. Linden, W. T. Franks, U. Akbey, S. Lange, B. J. van Rossum and H. Oschkinat, J. Biomol. NMR, 2011, 51, 283 CrossRef CAS PubMed; (b) A. B. Siemer, K. Y. Huang and A. E. McDermott, PLoS One, 2012, 7, e47242 CrossRef CAS PubMed; (c) P. Fricke, D. Mance, V. Chevelkov, K. Giller, S. Becker, M. Baldus and A. Lange, J. Biomol. NMR, 2016, 65, 121 CrossRef CAS PubMed; (d) K. Jaudzems, A. Bertarello, S. R. Chaudhari, A. Pica, D. Cala-De Paepe, E. Barbet-Massin, A. J. Pell, I. Akopjana, S. Kotelovica and D. Gajan, et al. , Angew. Chem., Int. Ed., 2018, 57, 7458 CrossRef CAS PubMed.
- C. G. Joo, A. Casey, C. J. Turner and R. G. Griffin, J. Am. Chem. Soc., 2009, 131, 12 CrossRef CAS PubMed.
- Ü. Akbey, A. H. Linden and H. Oschkinat, Appl. Magn. Reson., 2012, 43, 81 CrossRef.
- (a) A. Zagdoun, G. Casano, O. Ouari, M. Schwarzwalder, A. J. Rossini, F. Aussenac, M. Yulikov, G. Jeschke, C. Coperet and A. Lesage, et al. , J. Am. Chem. Soc., 2013, 135, 12790 CrossRef CAS PubMed; (b) C. Sauvée, M. Rosay, G. Casano, F. Aussenac, R. T. Weber, O. Ouari and P. Tordo, Angew. Chem., Int. Ed., 2013, 52, 10858 CrossRef PubMed.
- P. Berruyer, S. Bjorgvinsdottir, A. Bertarello, G. Stevanato, Y. Rao, G. Karthikeyan, G. Casano, O. Ouari, M. Lelli and C. Reiter, et al. , J. Phys. Chem. Lett., 2020, 11, 8386 CrossRef CAS PubMed.
- A. B. Barnes, G. De Paëpe, P. C. A. van der Wel, K. N. Hu, C. G. Joo, V. S. Bajaj, M. L. Mak-Jurkauskas, J. R. Sirigiri, J. Herzfeld and R. J. Temkin, et al. , Appl. Magn. Reson., 2008, 34, 237 CrossRef CAS PubMed.
- (a) T.-C. Ong, M. L. Mak-Jurkauskas, J. J. Walish, V. K. Michaelis, B. Corzilius, A. A. Smith, A. M. Clausen, J. C. Cheetham, T. M. Swager and R. G. Griffin, J. Phys. Chem. B, 2013, 117, 3040 CrossRef CAS PubMed; (b) M. Lelli, S. R. Chaudhari, D. Gajan, G. Casano, A. J. Rossini, O. Ouari, P. Tordo, A. Lesage and L. Emsley, J. Am. Chem. Soc., 2015, 137, 14558 CrossRef CAS PubMed.
- J. Viger-Gravel, P. Berruyer, D. Gajan, J. M. Basset, A. Lesage, P. Tordo, O. Ouari and L. Emsley, Angew. Chem., Int. Ed., 2017, 56, 8726 CrossRef CAS PubMed.
- (a) C. Fernández-de-Alba, H. Takahashi, A. Richard, Y. Chenavier, L. Dubois, V. Maurel, D. Lee, S. Hediger and G. De Paëpe, Chem. – Eur. J., 2015, 21, 4512 CrossRef PubMed; (b) H. Takahashi, D. Lee, L. Dubois, M. Bardet, S. Hediger and G. De Paëpe, Angew. Chem., Int. Ed., 2012, 51, 11766 CrossRef CAS PubMed; (c) E. Salnikov, M. Rosay, S. Pawsey, O. Ouari, P. Tordo and B. Bechinger, J. Am. Chem. Soc., 2010, 132, 5940 CrossRef CAS PubMed; (d) E. S. Salnikov, S. Abel, G. N. Karthikeyan, H. Karoui, F. Aussenac, P. Tordo, B. Bechinger and O. Ouari, ChemPhysChem, 2017, 18, 2103 CrossRef CAS PubMed.
- (a) W. Cao, W. D. Wang, H.-S. Xu, I. V. Sergeyev, J. Struppe, X. Wang, F. Mentink-Vigier, Z. Gan, M.-X. Xiao and L.-Y. Wang, et al. , J. Am. Chem. Soc., 2018, 140, 6969 CrossRef CAS PubMed; (b) D. Gajan, M. Schwarzwälder, M. P. Conley, W. R. Grüning, A. J. Rossini, A. Zagdoun, M. Lelli, M. Yulikov, G. Jeschke and C. Sauvée, et al. , J. Am. Chem. Soc., 2013, 135, 15459 CrossRef CAS PubMed; (c) E. Besson, F. Ziarelli, E. Bloch, G. Gerbaud, S. Queyroy, S. Viel and S. Gastaldi, Chem. Commun., 2016, 52, 5531 RSC; (d) D. L. Silverio, H. A. van Kalkeren, T.-C. Ong, M. Baudin, M. Yulikov, L. Veyre, P. Berruyer, S. Chaudhari, D. Gajan and D. Baudouin, et al. , Helv. Chim. Acta, 2017, 100, e1700101 CrossRef; (e) W. R. Grüning, H. Bieringer, M. Schwarzwälder, D. Gajan, A. Bornet, B. Vuichoud, J. Milani, D. Baudouin, L. Veyre and A. Lesage, et al. , Helv. Chim. Acta, 2017, 100, e1600122 CrossRef.
- G. Menzildjian, A. Lund, M. Yulikov, D. Gajan, L. Niccoli, G. Karthikeyan, G. Casano, G. Jeschke, O. Ouari and M. Lelli, et al. , J. Phys. Chem. B, 2021, 125, 13329 CrossRef CAS PubMed.
- H. Tapia and D. E. Koshland, Curr. Biol., 2014, 24, 2758 CrossRef CAS PubMed.
- A. Simperler, A. Kornherr, R. Chopra, P. A. Bonnet, W. Jones, W. D. S. Motherwell and G. Zifferer, J. Phys. Chem. B, 2006, 110, 19678 CrossRef CAS PubMed.
- L. Cordone, G. Cottone, A. Cupane, A. Emanuele, S. Giuffrida and M. Levantino, Curr. Org. Chem., 2015, 19, 1684 CrossRef CAS.
- (a) L. Greenspan, Phys. Chem., 1977, 81, 89 CrossRef; (b) M. Malferrari, A. Nalepa, G. Venturoli, F. Francia, W. Lubitz, K. Mobius and A. Savitsky, Phys. Chem. Chem. Phys., 2014, 16, 9831 RSC.
- (a) M. Malferrari, A. Savitsky, M. D. Mamedov, G. E. Milanovsky, W. Lubitz, K. Mobius, A. Y. Semenov and G. Venturoli, Biochim. Biophys. Acta, Bioenerg., 2016, 1857, 1440 CrossRef CAS PubMed; (b) C. Olsson, H. Jansson and J. Swenson, J. Phys. Chem. B, 2016, 120, 4723 CrossRef CAS PubMed; (c) V. Meyer, M. A. Swanson, L. J. Clouston, P. J. Boratyński, R. A. Stein, H. S. McHaourab, A. Rajca, S. S. Eaton and G. R. Eaton, Biophys. J., 2015, 108, 1213 CrossRef CAS PubMed; (d) A. A. Kuzhelev, G. Y. Shevelev, O. A. Krumkacheva, V. M. Tormyshev, D. V. Pyshnyi, M. V. Fedin and E. G. Bagryanskaya, J. Phys. Chem. Lett., 2016, 7, 2544 CrossRef CAS PubMed.
- (a) C. Selva, M. Malferrari, R. Ballardini, A. Ventola, F. Francia and G. Venturoli, J. Pharm. Sci., 2013, 102, 649 CrossRef CAS PubMed; (b) D. Corradini, E. G. Strekalova, H. E. Stanley and P. Gallo, Sci. Rep., 2013, 3, 1218 CrossRef PubMed; (c) K. Mobius, A. Savitsky, M. Malferrari, F. Francia, M. D. Mamedov, A. Y. Semenov, W. Lubitz and G. Venturoli, Appl. Magn. Reson., 2020, 51, 773 CrossRef.
- (a) M. Malferrari, A. Savitsky, W. Lubitz, K. Möbius and G. Venturoli, J. Phys. Chem. Lett., 2016, 7, 4871 CrossRef CAS PubMed; (b) C. Hackel, T. Zinkevich, P. Belton, A. Achilles, D. Reichert and A. Krushelnitsky, Phys. Chem. Chem. Phys., 2012, 14, 2727 RSC.
- H. Takahashi, S. Hediger and G. De Paepe, Chem. Commun., 2013, 49, 9479 RSC.
- G. Stevanato, G. Casano, D. J. Kubicki, Y. Rao, L. Esteban Hofer, G. Menzildjian, H. Karoui, D. Siri, M. Cordova and M. Yulikov, et al. , J. Am. Chem. Soc., 2020, 142, 16587 CrossRef CAS PubMed.
- R. Lefort, P. Bordat, A. Cesaro and M. Descamps, J. Chem. Phys., 2007, 126, 014510 CrossRef PubMed.
- A. J. Rossini, A. Zagdoun, F. Hegner, M. Schwarzwälder, D. Gajan, C. Copéret, A. Lesage and L. Emsley, J. Am. Chem. Soc., 2012, 134, 16899 CrossRef CAS PubMed.
- (a) A. J. Rossini, A. Zagdoun, M. Lelli, D. Gajan, F. Rascon, M. Rosay, W. E. Maas, C. Coperet, A. Lesage and L. Emsley, Chem. Sci., 2012, 3, 108 RSC; (b) S. Hediger, D. Lee, F. Mentink-Vigier and G. De Paepe, Emagres, 2018, 7, 105 CAS.
- K. R. Thurber and R. Tycko, J. Magn. Reson., 2009, 196, 84 CrossRef CAS PubMed.
- A. Leavesley, C. B. Wilson, M. Sherwin and S. Han, Phys. Chem. Chem. Phys., 2018, 20, 9897 RSC.
- (a) K. L. Ngai, S. Capaccioli and N. Shinyashiki, J. Phys. Chem. B, 2008, 112, 3826–3832 CrossRef CAS PubMed; (b) S. Khodadadi, A. Malkovskiy, A. Kisliuk and A. P. Sokolov, Biochim. Biophys. Acta, Proteins Proteom., 2010, 1804, 15 CrossRef CAS PubMed.
- A. Nalepa, M. Malferrari, W. Lubitz, G. Venturoli, K. Möbius and A. Savitsky, Phys. Chem. Chem. Phys., 2017, 19, 28388 RSC.
- D. G. Cory and W. M. Ritchey, J. Magn. Reson., 1988, 80, 128 Search PubMed.
- B. M. Fung, A. K. Khitrin and K. Ermolaev, J. Magn. Reson., 2000, 142, 97 CrossRef CAS PubMed.
- H. Tapia, L. Young, D. Fox, C. R. Bertozzi and D. Koshland, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 6122 CrossRef CAS PubMed.
- (a) K. Kawai, T. Suzuki and M. Oguni, Biophys. J., 2006, 90, 3732–3738 CrossRef CAS PubMed; (b) A. B. Siemer, K.-Y. Huang and A. E. McDermott, PLoS One, 2012, 7, e47242 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cp00970f |
| ‡ These authors contributed equally to this work. |
| This journal is © the Owner Societies 2022 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
: