Open Access Article
Open Access ArticleMolecular recognition of enzymes and modulation of enzymatic activity by nanoparticle conformational sensors†
Kaiqian
Chen
and
Yan
Zhao
 *
*
Department of Chemistry, Iowa State University, Ames, Iowa 50011-3111, USA. E-mail: zhaoy@iastate.edu; Fax: +1-515-294-0105; Tel: +1-515-294-5845
First published on 7th January 2022
Abstract
Regulation of enzyme activity is key to dynamic processes in biology but is difficult to achieve with synthetic systems. We here report molecularly imprinted nanoparticles with strong binding for the N- and C-terminal peptides on lysozyme. Binding affinity for the enzyme correlated with conformational flexibility of the peptides in the protein structure. Significantly, binding at the C-terminus of lysozyme enhanced the performance of the enzyme at elevated temperatures and that at the N-terminus lowered the enzyme activity. These nanoparticles, when clicked onto magnetic nanoparticles, could also be used to fish out the protein of interest from a mixture in a single step.
Enzyme activities are frequently regulated through their binding with molecular partners inside a cell. Such regulation allows an enzyme to operate in different states, depending on molecular cues in the environment. Controlling enzyme activity by synthetic binding partners is difficult.1–3 The most popular method is probably a controlled inhibition, in which inhibitors are reversibly added to the enzyme.4–6 Reversible control of the accessibility to the enzyme is another interesting strategy although the enzyme activity is not directly regulated.7–9 For certain enzymes, controlled organization could also be used to influence enzyme activity.10,11
In this work, we report the binding of a model enzyme (lysozyme) at the C- or N-terminus by a protein-sized water-soluble nanoparticle. The binding was found to influence the conformation of the enzyme and, more importantly, impact the catalytic activity either in a positive or negative manner under challenging reaction conditions.
Our model enzyme is hen egg-white lysozyme, which catalyzes the hydrolysis of peptidoglycan.12 To create a material to bind the enzyme with high affinity in water, we employed the method of epitope imprinting, in which a linear peptide sequence of the protein is used as the template to prepare molecularly imprinted polymers (MIPs).13,14 Many successful examples of epitope imprinting have been reported; the materials, nonetheless, are generally used to capture the proteins from a mixture.13,15–23
In our case, we performed the imprinting in cross-linked micelles, to obtain protein-sized polymeric nanoparticles.24 As shown in Scheme 1, the preparation starts with spontaneous micellization of surfactant 1 and simultaneous inclusion of a peptide template within the micelle, together with divinyl benzene (DVB) and a photoinitiator 2,2-dimethoxy-2-phenylacetophenone (DMPA). Surface-cross-linking with diazide 2via the Cu(I)-catalyzed click reaction, followed by core-cross-linking by UV-induced radical polymerization completes the molecular imprinting process. The cross-linked micelles are generally decorated with a sugar-derived monoazide 3 by the click reaction, which facilitates the purification of the molecularly imprinted nanoparticles (MINPs) and removal of the templates by simple precipitation and solvent washing (ESI†).
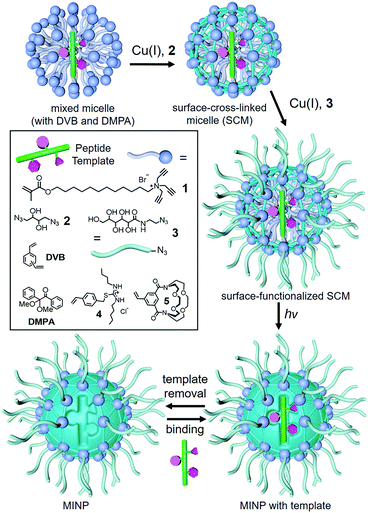 | ||
| Scheme 1 Preparation of peptide-binding MINP through molecular imprinting of a cross-linked micelle. | ||
The most obvious epitopes for the enzyme are the N- and C-terminal peptides. Micellar imprinting allowed us to quickly prepare ten water-soluble receptors for the last 5–9 residues on the N- and C-termini of lysozyme, respectively. Our intension was to use the MINPs to “scan” both tails to identify the receptors with the strongest abilities to modulate the enzyme activity.
Fig. 1a shows the binding of the N-terminal peptides and lysozyme by the corresponding MINPs. N-P5 (i.e., KVFGR) through N-P9 (KVFGRCELA) refer to the N-terminal peptides with the last 5–9 amino acid (AA) residues, all terminated with a carboxamide (CONH2) group. Our data shows that an increase of the chain length in the peptide template brought a steady increase in the binding free energy (−ΔG) by the MINP hosts, ranging from 7.10 to 8.10 kcal mol−1 (red line with data labels of filled squares). This trend is reasonable because a longer peptide template will create an imprinted site with a larger number of interactions with the template.
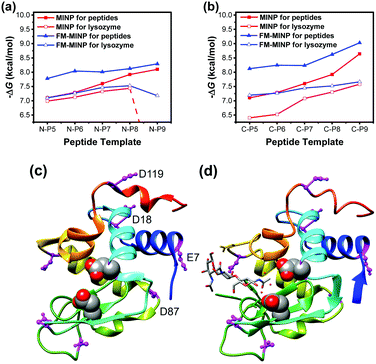 | ||
| Fig. 1 (a and b) Binding free energies for the peptides and lysozyme by the MINPs prepared with the corresponding (a) N-terminal and (b) C-terminal peptides as templates. The binding energies were determined by ITC (ESI†). (c and d) Crystal structure of lysozyme: PDB ID: 1DPX (c) and 1HEW (d). Molecular graphics was created using UCSF Chimera. The peptide chain is colored from blue (N-terminus) through the rainbow spectrum to red (C-terminus). The two acidic residues in the active site (Glu35 and Asp52) are highlighted with sphere models. Acidic residues are colored in magenta. | ||
The binding energies for the enzyme were generally lower (red line with data labels of empty squares) than those for the peptides. The overall weakening of the binding for the enzyme could be caused by steric repulsion from the two large binding partners. In addition, as shown in Fig. 1c, the (blue colored) N-terminal residues beyond the KVF are involved in an α-helix in the protein. Conformations of these peptides during micellar imprinting, however, are unlikely to be the same as those in the protein. Thus, MINP binding of these peptides in the protein would conflict with the α-helix formation of these peptides and indeed should be weaker than those for the peptide templates for which the MINPs were designed. The latter explanation is consistent with an extremely weak (nondetectable) binding of lysozyme by MINP(N-P9), i.e., the MINP prepared with the N-terminal peptide with 9 residues. Whereas the binding energy continued to increase for the peptides with an increase of chain length, that for the full protein displayed a precipitous drop at the end when the conflict between MINP binding and the α-helix formation was expected to be the strongest.
Functional monomers (FMs) 4 and 5 can hydrogen-bond with carboxylic acid and amino group, respectively. The micelle-stabilized hydrogen bonds can help the imprinting and binding of peptides significantly.25 Indeed, inclusion of these FMs in the imprinting strengthened the binding of the resultant FM-MINPs for their corresponding peptide templates (Fig. 1a, blue line with data labels of filled triangles). The binding for lysozyme (blue line with data labels of empty triangles) was lower probably for the same reasons discussed above, as the drop in the binding energy was most prominent at N-P9, the same as with the unfunctionalized MINPs.
Interestingly, conflicts between binding and secondary structure formation did not seem to exist for the C-terminal peptides (Fig. 1b). Even though binding was weaker for the enzyme than for the peptides, whether by the parent MINPs or FM-MINPs, the two series of binding data followed a similar trend, displaying an increase in binding energy with an increase in the chain length of the peptide template. Thus, the C-terminal peptide did not seem to have a strong preference for a particular conformation in the protein, at least not to the point to interfere with the MINP binding.
Our binding data suggests that the N- and C-terminal peptides of lysozyme have different conformational plasticity and the N-terminus is more resistant to conformational change. The conclusion is supported by crystallographic data of the enzyme. The lysozyme structure in Fig. 1c (PDB ID: 1DPX)26 shows a more ordered helix at the N-terminus than the C-terminus. Although crystal packing may influence the structure of a peptide, the better helical formation at the N-terminus is consistent with our binding data. Fig. 1d shows another lysozyme structure (PDB ID: 1HEW), obtained with a tri-N-acetylchitotriose inhibitor in the active site.27 This structure also shows a less ordered C-terminus. More importantly, the very last three N-terminal amino acids (KVF) are involved in a β-sheet with AA38–40 (FNT), which is very close to the active site. The two acidic residues (Glu35 and Asp52) in the active site are highlighted with sphere models in the structure.
Protein conformation can be monitored by CD spectroscopy.28 The most prominent band of lysozyme was at 208 nm, followed by a shoulder at 222 nm (Fig. 2a), consistent with a large number of α-helices in the protein (Fig. 1c and d). Addition of either 2 equivalents of MINP(N-P9) or MINP(C-P9) was found to reduce the intensity of these bands, indicating some of these helical structures were lost upon binding.
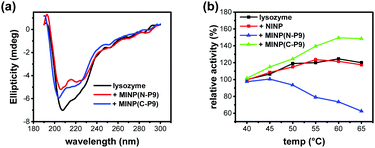 | ||
| Fig. 2 (a) CD spectra of lysozyme (black) and lysozyme in the presence of 3.0 equiv. MINP(N-P9) (red) or MINP(C-P9) (blue). [lysozyme] = 0.5 μM. (b) Relative activity of lysozyme as a function of temperature under different conditions, determined with glycol chitin as the substrate at pH 4.5 (ESI†). | ||
The results so far are exciting, because MINP binding with the N- and C-terminal peptides of lysozyme seem to correlate with the stability of the secondary structures formed by these peptides in the enzyme. Since the N-terminal peptide might be used to stabilize the active site structure, MINP binding at that location should lower the enzyme activity.
The activity of lysozyme can be analyzed by a widely used assay that employs glycol chitin as the substrate and potassium ferricyanide as the color reagent.29 Lysozyme has a remarkable thermal stability with a melting point ∼72 °C at pH 5.12 As shown in Fig. 2b, the enzyme was able to maintain its activity very well at elevated temperatures and was most active at 60 °C (black curve). Nonimprinted nanoparticles (NINPs) at a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 NINP/lysozyme ratio showed negligible effects on the enzyme (red curve). NINPs were prepared following the same micellar imprinting procedure, without any templates. Thus, the cross-linked cationic micelles had no abilities to modulate the enzyme activity. In contrast, MINP(N-P9), the MINP imprinted against the N-terminal peptide, displayed a negative effect on the enzyme (blue curve) while MINP(C-P9), the MINP imprinted against the C-terminal peptide, exhibited a positive effect (green curve).
1 NINP/lysozyme ratio showed negligible effects on the enzyme (red curve). NINPs were prepared following the same micellar imprinting procedure, without any templates. Thus, the cross-linked cationic micelles had no abilities to modulate the enzyme activity. In contrast, MINP(N-P9), the MINP imprinted against the N-terminal peptide, displayed a negative effect on the enzyme (blue curve) while MINP(C-P9), the MINP imprinted against the C-terminal peptide, exhibited a positive effect (green curve).
The negative effect of MINP(N-P9) is in line with the ITC binding and CD data. Since this particular MINP interferes with the secondary structure formation close to the active site, a negative effect is not surprising. It is interesting to see that both MINPs influence the enzyme activity of lysozyme minimally at 40 °C. Most likely, the extraordinary stability of lysozyme (with 4 disulfide bonds) allows it not only to operate at elevated temperatures but also to resist MINPs in their conformation-modifying binding.
If the negative effect of MINP(N-P9) could be inferred from its binding location, the positive, “protective” effect of MINP(C-P9) was completely unexpected. Lysozyme has 7 acidic residues (magenta-colored in Fig. 1c and d), in addition to Glu35 and Asp52 in the active site. Four of them—Glu7, ASP18, ASP87, and ASP119—reside on the same side of the protein, with the longest distance (2.8 nm) between ASP87 and ASP119. It is possible that MINP(N-P9) could not only bind at the designed position but, due to its multi-cationic nature, electrostatically interact with these acidic residues and thus “intramolecularly cross-link” the protein noncovalently. The side chains of ASP and Glu have a pKa of 4–4.5 and the enzymatic assay was performed at pH 4.5. However, the acidity/basicity of AA side chains can change dramatically depending on their local environment.30 Positive charges, which are abundant on our cationic MINPs, tend to promote the deprotonation of acids so that favorable electrostatic interactions can occur. One might be also concerned with electrostatic repulsion between MINP and many positively charged lysines on lysozyme. Lysine, however, has a remarkable ability to lower its pKa near positive charges31 or in a hydrophobic microenvironment32 and thus avoid such repulsion.
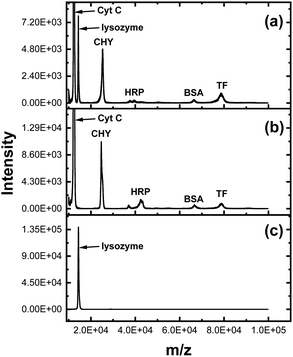
Surface ligands are generally clicked onto the (alkynyl-containing) SCM (i.e., surface-cross-linked micelle) shown in Scheme 1 in our MINP synthesis. However, the SCM could be clicked onto readily prepared azide-functionalized magnetic nanoparticles (MNPs).33 Since our MINP could bind lysozyme with a high affinity, the resultant MINP(C-P9)-MNP composite allowed us to isolate the enzyme from a six-component protein mixture. The lysozyme has distinctively difference N- and C-terminal sequences from the other proteins (Table S3, ESI†). To make the isolation more challenging, the concentrations of other proteins were 10 times higher than that of lysozyme. Fig. 3a shows the MALDI mass spectrum of the protein mixture before extraction. Peaks for cytochrome C (Cyt C, MW 12K Da) and lysozyme (MW 14K Da) nearly overlapped because of their similar molecular weight. Yet, incubation of the protein mixture with MINP(C-P9)-MNP) could completely remove lysozyme from the mixture (Fig. 3b) and the protein released from the material was pure lysozyme (Fig. 3c). A control experiment showed that the nonimprinted proteins were not extracted in the absence of lysozyme (Fig. S14, ESI†).
In summary, MINP binding can be used to up- or down-regulate an enzyme's activity and the effect gets stronger as the enzyme moves away from its most stable state. Facile synthesis of MINP makes it straightforward to “scan” multiple sequences to identify the most sensitive, whether to protect or to partly inhibit the enzyme. Although it is possible to study enzyme dynamics at different time scales, it is difficult to correlate the information with catalysis. Instead, our method not only allows one to identify the more and less conformational rigid sequences on the enzyme but readily identify those more influential in catalysis.
Protein purification is a lengthy process involving multiple techniques.34 Affinity tags are frequently included in recombinant proteins to aid purification but often require extra steps to be cleaved.35 In contrast, MINP-MNPs imprinted against the N- or C-terminal peptides have pre-determined binding selectivity for the natural, untagged proteins. Their stability at elevated temperature,36,37 in organic solvent,36 and at extreme pH38 are also attractive features for an affinity support.
We thank NSF (DMR-2002659) for financial support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- S. van Dun, C. Ottmann, L.-G. Milroy and L. Brunsveld, J. Am. Chem. Soc., 2017, 139, 13960–13968 CrossRef CAS PubMed.
- A. Ali, G. A. Bullen, B. Cross, T. R. Dafforn, H. A. Little, J. Manchester, A. F. A. Peacock and J. H. R. Tucker, Chem. Commun., 2019, 55, 5627–5630 RSC.
- T. Tian, Y. Y. Song, J. Q. Wang, B. S. Fu, Z. Y. He, X. Q. Xu, A. L. Li, X. Zhou, S. R. Wang and X. Zhou, J. Am. Chem. Soc., 2016, 138, 955–961 CrossRef CAS PubMed.
- J. H. Harvey and D. Trauner, ChemBioChem, 2008, 9, 191–193 CrossRef CAS PubMed.
- Y. M. Kim, J. A. Phillips, H. P. Liu, H. Z. Kang and W. H. Tan, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 6489–6494 CrossRef CAS PubMed.
- S. Ghosh and L. Isaacs, J. Am. Chem. Soc., 2010, 132, 4445–4454 CrossRef CAS PubMed.
- Y. Liang, S. Song, H. Yao and N. Hu, Electrochim. Acta, 2011, 56, 5166–5173 CrossRef CAS.
- O. Parlak, M. Ashaduzzaman, S. B. Kollipara, A. Tiwari and A. P. F. Turner, ACS Appl. Mater. Interfaces, 2015, 7, 23837–23847 CrossRef CAS PubMed.
- S. T. Xie, L. P. Qiu, L. Cui, H. L. Liu, Y. Sun, H. Liang, D. Ding, L. He, H. X. Liu, J. N. Zhang, Z. Chen, X. B. Zhang and W. H. Tan, Chem, 2017, 3, 1021–1035 CAS.
- D. T. Dang, H. D. Nguyen, M. Merkx and L. Brunsveld, Angew. Chem., Int. Ed., 2013, 52, 2915–2919 CrossRef CAS PubMed.
- E. M. Estirado, B. J. H. M. Rosier, T. F. A. de Greef and L. Brunsveld, Chem. Commun., 2020, 56, 5747–5750 RSC.
- S. Venkataramani, J. Truntzer and D. R. Coleman, J. Pharm. BioAllied Sci., 2013, 5, 148–153 CrossRef PubMed.
- A. Rachkov and N. Minoura, Biochim. Biophys. Acta, Protein Struct. Mol. Enzymol., 2001, 1544, 255–266 CrossRef CAS.
- S. P. B. Teixeira, R. L. Reis, N. A. Peppas, M. E. Gomes and R. M. A. Domingues, Sci. Adv., 2021, 7, eabi9884 CrossRef CAS PubMed.
- H. Nishino, C. S. Huang and K. J. Shea, Angew. Chem., Int. Ed., 2006, 45, 2392–2396 CrossRef CAS PubMed.
- A. M. Bossi, P. S. Sharma, L. Montana, G. Zoccatelli, O. Laub and R. Levi, Anal. Chem., 2012, 84, 4036–4041 CrossRef CAS PubMed.
- Y. Zhang, C. Deng, S. Liu, J. Wu, Z. Chen, C. Li and W. Lu, Angew. Chem., Int. Ed., 2015, 54, 5157–5160 CrossRef CAS PubMed.
- S. Li, K. Yang, N. Deng, Y. Min, L. Liu, L. Zhang and Y. Zhang, ACS Appl. Mater. Interfaces, 2016, 8, 5747–5751 CrossRef CAS PubMed.
- S. Schwark, W. Sun, J. Stute, D. Lutkemeyer, M. Ulbricht and B. Sellergren, RSC Adv., 2016, 6, 53162–53169 RSC.
- G. Pan, S. Shinde, S. Y. Yeung, M. Jakštaitė, Q. Li, A. G. Wingren and B. Sellergren, Angew. Chem., Int. Ed., 2017, 56, 15959–15963 CrossRef CAS PubMed.
- Y.-P. Qin, C. Jia, X.-W. He, W.-Y. Li and Y.-K. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 9060–9068 CrossRef CAS PubMed.
- R. Xing, Y. Ma, Y. Wang, Y. Wen and Z. Liu, Chem. Sci., 2019, 10, 1831–1835 RSC.
- K. G. Yang, S. W. Li, L. K. Liu, Y. W. Chen, W. Zhou, J. Q. Pei, Z. Liang, L. H. Zhang and Y. K. Zhang, Adv. Mater., 2019, 31, 1902048 CrossRef CAS PubMed.
- J. K. Awino, R. W. Gunasekara and Y. Zhao, J. Am. Chem. Soc., 2017, 139, 2188–2191 CrossRef CAS PubMed.
- S. Fa and Y. Zhao, Chem. Mater., 2019, 31, 4889–4896 CrossRef CAS PubMed.
- M. S. Weiss, G. J. Palm and R. Hilgenfeld, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2000, 56, 952–958 CrossRef CAS PubMed.
- J. C. Cheetham, P. J. Artymiuk and D. C. Phillips, J. Mol. Biol., 1992, 224, 613–628 CrossRef CAS PubMed.
- N. J. Greenfield, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- T. Imoto and K. Yagishita, Agric. Biol. Chem., 1971, 35, 1154–1156 CrossRef CAS.
- F. H. Westheimer, Tetrahedron, 1995, 51, 3–20 CrossRef CAS.
- J. D. Henao, Y.-W. Suh, J.-K. Lee, M. C. Kung and H. H. Kung, J. Am. Chem. Soc., 2008, 130, 16142–16143 CrossRef CAS PubMed.
- D. Matulis and V. A. Bloomfield, Biophys. Chem., 2001, 93, 37–51 CrossRef CAS PubMed.
- X. Liu, Z. Ma, J. Xing and H. Liu, J. Magn. Magn. Mater., 2004, 270, 1–6 CrossRef CAS.
- R. K. Scopes, Protein Purification: Principles and Practice, Springer-Verlag, New York, 3rd edn, 1994 Search PubMed.
- D. W. Wood, Curr. Opin. Struct. Biol., 2014, 26, 54–61 CrossRef CAS PubMed.
- X. Xing and Y. Zhao, Org. Biomol. Chem., 2018, 16, 2855–2859 RSC.
- J. K. Awino and Y. Zhao, J. Am. Chem. Soc., 2013, 135, 12552–12555 CrossRef CAS PubMed.
- X. Xing and Y. Zhao, New J. Chem., 2018, 42, 9377–9380 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, ITC titration curves, and spectroscopic data. See DOI: 10.1039/d1cc05699a |
| This journal is © The Royal Society of Chemistry 2022 |