Open Access Article
Open Access ArticleHydrophobic cavity-directed azide-acetyllysine photochemistry for profiling non-histone interacting partners of bromodomain protein 1†
Jordan
Kuwik‡
a,
Shana
Wagner‡
a,
Babu
Sudhamalla‡
ab,
Ronald
Debiec
a and
Kabirul
Islam
 *a
*a
aDepartment of Chemistry, University of Pittsburgh, Pittsburgh, PA 15260, USA. E-mail: kai27@pitt.edu
bCurrent address: Department of Biological Sciences, Indian Institute of Science Education and Research-Kolkata, Mohanpur 741246, India
First published on 14th June 2022
Abstract
Bromodomain containing protein 1 (BRD1) plays critical roles in chromatin acetylation, gene transcription, erythropoiesis, and brain development. BRD1 is also implicated in several human conditions and is a therapeutic target for cancer. Although, the bromodomain is known to bind acetylated histones, how the function of BRD1 is regulated via non-histone acetylation is unexplored. To identify the non-histone acetylome of BRD1, we develop an R585AzF variant carrying photo responsive 4-azido phenylalanine (AzF) via amber suppressor mutagenesis. We demonstrate biochemical integrity of the AzF-containing analogue and its ability to crosslink non-histone interacting partners present in human cells. Subsequent proteomic experiments led to the identification of the novel BRD1 interactome representing diverse signaling pathways. As a proof-of-concept demonstration, we validated acetylated PDIA1 protein as a bona fide binding partner of BRD1. Our work suggests that BRD1 interacts with additional acetyllysine motifs, beyond those characterized in histone proteins.
Introduction
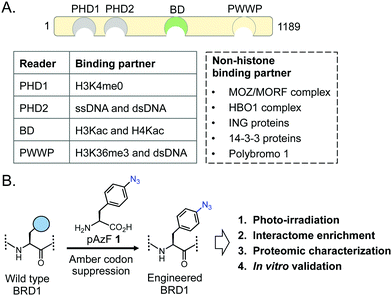
Chemical modifications on histones and DNA are critical for gene expression and regulation of cellular processes.1–3 Three classes of proteins, identified as writers, readers, and erasers, are involved in establishing, recognizing and removing these marks, respectively, thereby controlling gene transcription in a context-dependent manner.4,5 A majority of such chromatin-modifying proteins contain several interacting domains, which serve as recruiting platforms for transcriptional regulators, facilitating the formation of multiprotein complexes at specific loci in the genome. A systematic dissection of these transient but productive interactions between a chromatin modifier and its binding partners is key to the understanding of gene regulatory functions of such multi-protein complexes.Among histone modifications, lysine acetylation constitutes a dominant mechanism for gene activation. Transfer of an acetyl unit to the ε-amino group of lysine neutralizes its positive charge and attenuates the electrostatic interaction between histone and DNA; this contributes to an open chromatin conformation and gene expression.6,7 Acetyllysine (Kac) sites on histones are also recognized by bromodomain (BD) containing proteins that are essential for regulating gene expression in normal and disease physiology, including cancer and autoimmune disorders.8 The human genome encodes 46 distinct BDs, present in more than 60 proteins implicated in chromatin-dependent processes.9 Recently, several studies have reported that certain BDs, particularly the members of the bromo and extra-terminal (BET) family, can bind acetyllysine motifs present in non-histone proteins to control biological processes such as DNA repair, splicing and cellular metabolism.10,11 However, little is known about the proteome-wide binding partners of the non-BET human BDs (>40), and many of them are considered “orphans” due to such lack of functional characterization. Identifying the non-histone interactome of such poorly characterized BDs is essential for the molecular understanding of their functions in diverse cellular processes, ranging from gene transcription and splicing to metabolism.
Among bromodomain proteins, BRD1 is known to interact with several histone acetyltransferase complexes containing the MOZ/MORF and HBO1 transcriptional coactivators to affect gene expression (Fig. 1A).12,13 It has been implicated in erythropoiesis, brain development and several related human conditions such as susceptibility to schizophrenia and bipolar disorder.12,14–17 BRD1 belongs to the bromodomain and plant homeodomain (PHD) finger (BRPF) subfamily, which comprises three paralogs, BRPF1, BRPF2/BRD1 and BRPF3.18 BRPFs possess a multitude of interacting modules, including C2H2, PHD-linked fingers, and bromo and PWWP domains (Fig. 1A).18–22 Despite the domain similarity, each BRPF member is functionally distinct. Loss of either BRPF1 or BRPF2/BRD1 in mice leads to embryonic lethality at E9.5 and E15.5, respectively, due to different developmental defects; in contrast, loss of BRPF3 has no effect on mouse development without compensatory expression of BRPF1 or BRPF2, suggesting nonoverlapping functions of the members of the BRPF subfamily.23–25 BRPF1 is important for maintaining Hox gene expression by linking the MOZ/MORF complex to other subunits such as ING5 and hEAF6.26 On the other hand, BRPF2/BRD1 preferentially forms complexes with ING4 and HBO1 to acetylate H3K14 and promote erythroid differentiation via expression of key erythroid regulator gene GATA1.12 Thus, non-overlapping biological functions of the BRPF members are manifested by their ability to interact with distinct sets of non-histone transcriptional regulators through recognition of acetyllysine motifs present in these proteins by the BRPF bromodomains.
Recently, immunoprecipitation studies coupled with proteomics have revealed that BRD1 interacts with a range of cellular proteins other than histones.27 Given that a full-length BRD1 construct was employed in the study, it was challenging to assign the BRD1 bromodomain-specific interactome. Furthermore, given the weaker interaction between the bromodomain and acetyllysine (a typical dissociation constant is on the order of 10–100 μM), traditional immunoprecipitation methods may not capture the full range of bromodomain interacting partners. To advance our understanding of the role of BRD1 in diverse cellular processes, we set out to characterize its bromodomain-specific interactome network present in human cells.
Proteins carrying photoactivable amino acids have been developed to capture transient interactions.28–30 A particularly notable method includes introduction of a photo-crosslinkable amino acid (PCAA) using an evolved synthetase-tRNA pair (Fig. 1A).31–35 We recently developed a chemoproteomic platform called interaction based protein profiling (IBPP) to identify transient protein–protein interactions mediated by chromatin readers (Fig. 1B).36 In this approach, a wildtype bromodomain is engineered to carry a PCAA in the acetyllysine recognition cavity via amber suppressor mutagenesis; the mutant protein is amenable to binding and crosslinking with interacting partners upon photo-irradiation. The crosslinked proteins are then subjected to affinity enrichment and proteomic analysis. We have successfully employed the IBPP tool to identify novel non-histone binding partners of BRD4, a member of the BET family. Herein, we report our efforts to develop photo-crosslinkable BRD1, and biochemical characterization of the variant, along with proteomic characterization and validation of a novel non-histone interactome of BRD1.
Results and discussion
The bromodomain, a conserved motif with approximately 110 amino acids, carries a hydrophobic cavity or ‘aromatic cage’ that can recognize acetyllysine residues in interacting proteins (Fig. 2A).7 We have previously demonstrated that certain amino acids in the hydrophobic cavity of the bromodomain can be replaced with photo-crosslinkable para-azido phenylalanine (AzF) 1 without any significant change in the binding affinity and sequence specificity.36,37 For example, BRD4 variants carrying either W81AzF, L92AzF or M149AzF mutations in the first bromodomain (BD1) are capable of binding and crosslinking with acetylated histone H4 peptide and full-length protein. Sequence and structure alignments of the bromodomains reveal that R585 of BRD1 and W81 of BRD4 occupy a similar position in the hydrophobic pocket (Fig. 2A and B). We surmised that replacement of R585 with an aromatic residue such as AzF will likely result in W81-like orientation in BRD1 for interactome binding and crosslinking. To develop a photo-activable analogue of BRD1 for profiling its non-histone interactome, we generated the mutant R585AzF via amber suppressor mutagenesis; the protein was purified in homogeneity with excellent yield and confirmed by LC-MS analysis (Fig. 2C and Fig. S1, ESI†).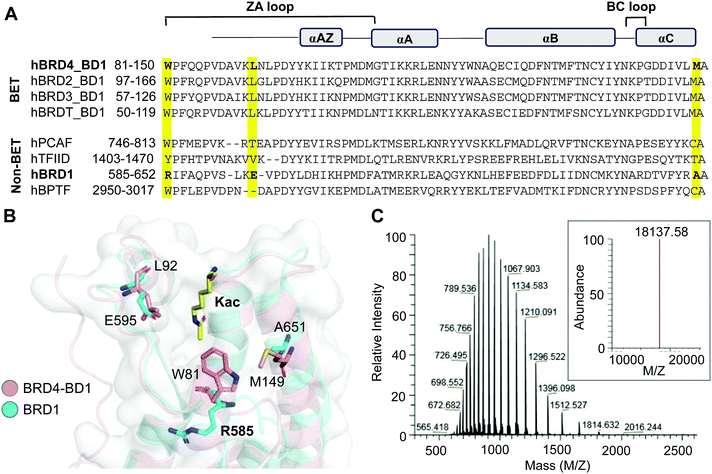 | ||
| Fig. 2 Engineering BRD1. (A) Sequence alignment of BET and representative non-BET bromodomains. W81, L92 and M149 of BRD4-BD1 and the corresponding residues in BRD1-BD are highlighted. (B) Superimposed structures of BRD4-BD1 (PDB 3UVW) and BRD1-BD (3RCW) showing the positions of selected residues. R585 of BRD1 was subjected to AzF incorporation for the development of photo-crosslinkable BRD1. (C) The LC-HRMS spectrum confirming the R585AzF variant. The inset is the average mass of the mutant. | ||
To examine the ability of R585AzF to bind acetylated protein segments, we synthesized peptide 2 corresponding to the first 21 amino acids of the histone H4 tail carrying acetyllysine groups at positions K5 and K8 (H4Kac2) (Fig. 3 and Fig. S2, ESI†). To calculate solution-phase dissociation constants (Kd) of the peptide from reader proteins, we performed isothermal titration calorimetric (ITC) measurements at 15 °C. The Kd for wild type BRD1-BD was measured to be 17.3 ± 3.6 μM, closely agreeing with reported values (Fig. 3A),38 while R585AzF showed a Kd value of 110 ± 35 μM (Fig. 3B). Although the mutation resulted in decreased binding of the peptide compared to the wild type protein, the Kd value is within the range for bromodomains to bind their cognate acetylated histones.7 Collectively, these results demonstrate that BRD1-BD can be successfully engineered to introduce an unnatural amino acid at the acetyllysine binding site without compromising its structural and functional integrity.
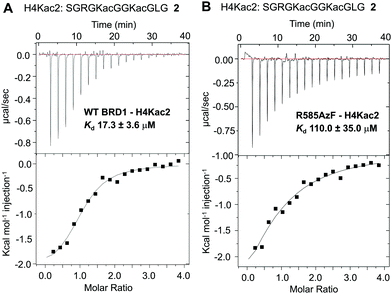 | ||
| Fig. 3 Isothermal calorimetric measurements of the dissociation constants of H4Kac2 peptide 2 from wild type BRD1 (A) and the R585AzF mutant (B). | ||
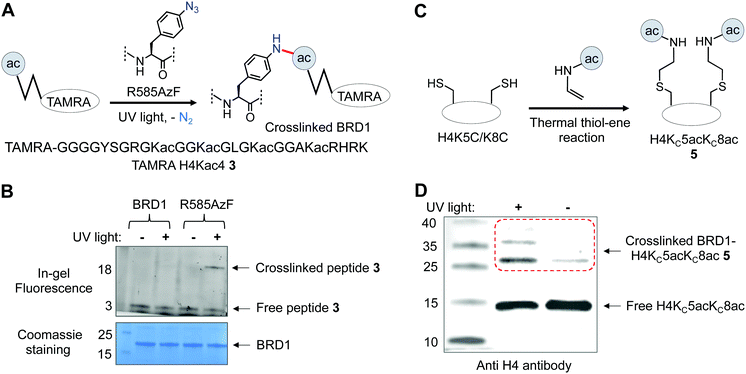
After confirming the binding efficiency of the BRD1 mutant, we sought to examine its ability to crosslink with known interacting partners. We synthesized tetramethyl rhodamine (TAMRA) attached H4Kac4 peptide 3 and subjected it to crosslinking with wild type and R585AzF, one at a time, using UV light at 365 nm; the extent of crosslinking was gauged by in-gel fluorescence (Fig. 4A). The BRD1 mutant carrying the AzF moiety underwent successful crosslinking with the peptide as evident from the appearance of a fluorescent band of higher molecular weight only when the sample was irradiated with 365 nm light (Fig. 4B and Fig. S3, ESI†). Wild type BRD1, on the other hand, failed to undergo any photo-crosslinking in spite of its strong affinity towards the peptide, confirming that the AzF group is indeed essential for crosslinking with the peptide (Fig. 4B). Furthermore, the mutant failed to undergo crosslinking with H4 peptide 4 lacking the acetylated lysine residues (Fig. S3, ESI†), demonstrating that the acetyllysine moiety is also key to binding and crosslinking with the engineered reader.
We next analyzed the crosslinking efficiency of R585AzF with full-length H4 in order to test the mutant's ability to interact with physiologically relevant substrates. Employing a semi-synthetic approach that combines recombinant protein expression and posttranslational chemical modification, we developed full-length H4 carrying acetylated thia-lysine (KCac) at positions 5 and 8 (H4KC5acKC8ac) 5 (Fig. 4C).39 The KCac motif has been shown to functionally mimic Kac in biochemical assays.39 We subjected the semi-synthetic thia-H4 to crosslinking with the BRD1 mutant in the presence of light (λmax = 365 nm). Protein bands were separated on SDS-PAGE gel and analyzed by immunoblotting using H4 antibody. The full-length thia-H4 indeed underwent UV-light-mediated bond formation with the BRD1 mutant as evident from the appearance of a higher molecular band on the gel (Fig. 4D and Fig. S3, ESI†); a control sample that was not exposed to UV light produced marginal crosslinking. Taken together, our results consolidate the potential of R585AzF to bind and crosslink with physiologically relevant histone H4.
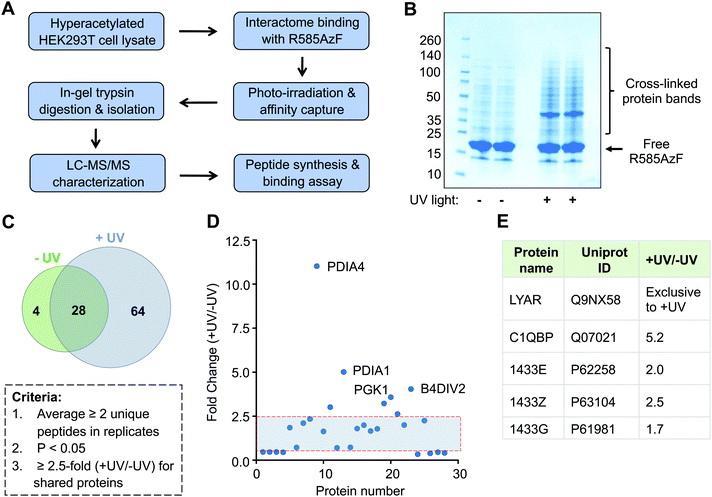
Successful crosslinking of R585AzF with semisynthetic H4 prompted us to undertake an unbiased proteomic study to characterize the non-histone interacting partners of BRD1 present in human cells (Fig. 5A). Toward this end, HEK293T cells were cultured in the presence of a broad-spectrum deacetylase inhibitor called suberanilohydroxamic acid (SAHA) to generate a hyperacetylated proteome. Cellular extracts were then incubated with R585AzF and irradiated at 365 nm for 30 minutes. The UV light-mediated crosslinked proteins were enriched by affinity purification using Ni-NTA beads as the mutant carries a 6xHis tag at the N-terminus. We observed several protein bands of higher molecular weight on polyacrylamide gel, indicating successful crosslinking of the engineered BRD1 with putative non-histone interacting partners (Fig. 5B). Importantly, no significant enrichment was observed in cell extracts that were not subjected to photo-irradiation at 365 nm. Furthermore, affinity pull down from cell extracts incubated with wildtype BRD1 or no bromodomain control did not lead to a significant enrichment (Fig. S4, ESI†). This result further suggests that the AzF group is indispensable for the crosslinking of the cellular interactome of BRD1.
Subsequently, we performed liquid chromatography-tandem mass spectrometry (LC-MS/MS) on trypsin-digested peptides of the enriched crosslinked proteins (Fig. 5A and Table S1, ESI†). To improve confidence in the proteomic analyses, two independent biological replicates were carried out for each treatment group (±UV). We applied several criteria in a sequential manner to analyze the proteomic data (Fig. 5C). First, exclusive unique peptides for proteins identified in both biological replicates for a given condition (±UV) were averaged. Proteins that were present in both the replicates and had a minimum of two unique peptides were selected. Next, proteins within a set with a p-value of ≤0.05, obtained from a t-test between UV-treated and non-treated samples, were chosen. Applying these criteria, we identified 64 proteins present exclusively in the UV-treated samples and only 4 in the non-treated samples (Fig. 5C and Table S1, ESI†). These results further confirm that light-induced covalent bond formation between two interacting proteins through the AzF moiety is key to enrich the transient interactome of BRD1. We also enlisted 28 proteins shared by both the treatment groups. We next implemented a 2.5-fold enrichment filter for the shared proteins, which led to 7 proteins significantly enriched in UV-treated samples compared to 3 in the non-treated group (Fig. 4D). Taken together, we identified a total of 71 proteins as a high-confidence non-histone interactome of BRD1, exclusively (64 proteins) or significantly (7 proteins) enriched by UV-mediated crosslinking (Fig. 5C, D and Table S1, ESI†).
Analysis of the proteomic results revealed that the newly identified BRD1 interactome represents diverse signaling pathways previously not recognized to be associated with the bromodomain protein (Table S1, ESI†). The interactome includes transcription elongation factor (TCEA1), ribonucleoproteins (hnRNPA/B, hnRNPD), translation initiation factor (EIF4B), metabolic enzymes (LDHA, ALDH2, and ECHS1), isomerases (PDIA1/3/4, FKBP3), proteasome subunits (PSMA4-5, PSMB3/6), signaling proteins (14-3-3), and enzymes involved in oxidative pathways (TXNRD1, CRYZL1, PRDX5, and ETFB). We identified several 14-3-3 regulatory proteins, including YWHAE, YWHAZ, and YWHAQ, which are consistent with the previous proteomic study that employed full-length BRD1.27 Other common proteins included subcomponent-binding protein C1QBP and cell growth-regulating protein LYAR, suggesting that the IBPP platform can identify both known and novel acetylated non-histone binding partners of the BRD1 bromodomain (Fig. 5E).
Several recent studies have shown that, in mammalian cells, a large number of proteins are acetylated at specific lysine residues.40 Biological significance of such acetylation events is poorly defined, primarily due to lack of knowledge about their respective reader modules. Our proteomic data revealed that subsets of these known acetylated proteins are binding partners of BRD1. To examine if the BRD1 bromodomain could bind the novel non-histone proteins identified through IBPP, we selected PDIA1 as this protein was significantly enriched in the UV-treated samples (Fig. 4D). Furthermore, PDIA1 is also a binding partner of BRD4-BD1 as demonstrated previously.36 To compare the binding efficiency of the two bromodomains, we synthesized a segment of PDIA1, AKacAAGKacLKacA 6, known to be acetylated in HEK293T cells (Fig. 6 and Fig. S5, ESI†). Given the sequence similarity between the Kac![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) Kac motif of PDIA1 and K12ac
Kac motif of PDIA1 and K12ac![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) K16ac of H4, we surmised that BRD1 may bind PDIA1 peptide 6 with similar affinity as histone peptide 2. In an ITC-based binding assay, BRD1-BD indeed recognized peptide 6 with a Kd value of 24.5 ± 11.7 μM, close to that of canonical H4 peptide 2 with Kd of 17.3 ± 3.6 μM (Fig. 6 and Table S2, ESI†). Remarkably, a non-acetylated PDIA1 peptide AKAAGKLKA 7 failed to bind BRD1-BD, confirming that the acetyllysine residues are critical for interactome recognition by the bromodomain (Fig. 6 and Fig. S5, ESI†). As measured previously, BRD4-BD1 binds peptide 6 with a Kd of 353 μM, which is about 15-fold higher compared to that of BRD1-BD.36 Although further studies are required to gain molecular insights into the mode of recognition, our result suggests that the BRD1-PDIA1 interaction is likely more relevant in regulating biological processes. This proof-of-concept demonstration of acetylated PDIA1 as a novel partner of BRD1 suggests that systematic binding experiments will validate several other proteins identified in the current proteomic study as authentic interacting partners of BRD1. Future biochemical and structural studies are expected to uncover new binding modes between the BRD1 bromodomain and acetyllysine motifs present in these non-histone partners of BRD1 and their roles in dictating the biological functions of BRD1. Furthermore, systematic analysis of the proteomic results will help identify the crosslinked species and uncover non-canonical sequence motifs as a novel BRD1 interactome.
K16ac of H4, we surmised that BRD1 may bind PDIA1 peptide 6 with similar affinity as histone peptide 2. In an ITC-based binding assay, BRD1-BD indeed recognized peptide 6 with a Kd value of 24.5 ± 11.7 μM, close to that of canonical H4 peptide 2 with Kd of 17.3 ± 3.6 μM (Fig. 6 and Table S2, ESI†). Remarkably, a non-acetylated PDIA1 peptide AKAAGKLKA 7 failed to bind BRD1-BD, confirming that the acetyllysine residues are critical for interactome recognition by the bromodomain (Fig. 6 and Fig. S5, ESI†). As measured previously, BRD4-BD1 binds peptide 6 with a Kd of 353 μM, which is about 15-fold higher compared to that of BRD1-BD.36 Although further studies are required to gain molecular insights into the mode of recognition, our result suggests that the BRD1-PDIA1 interaction is likely more relevant in regulating biological processes. This proof-of-concept demonstration of acetylated PDIA1 as a novel partner of BRD1 suggests that systematic binding experiments will validate several other proteins identified in the current proteomic study as authentic interacting partners of BRD1. Future biochemical and structural studies are expected to uncover new binding modes between the BRD1 bromodomain and acetyllysine motifs present in these non-histone partners of BRD1 and their roles in dictating the biological functions of BRD1. Furthermore, systematic analysis of the proteomic results will help identify the crosslinked species and uncover non-canonical sequence motifs as a novel BRD1 interactome.
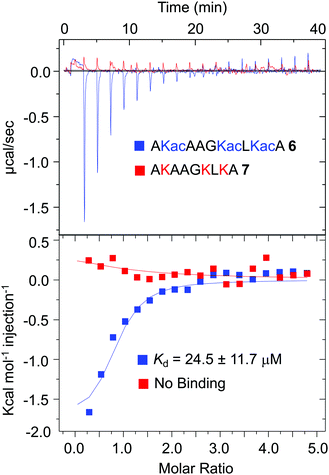 | ||
| Fig. 6 Isothermal titration calorimetry-based measurements of dissociation constants of PDIA1 peptides (acetylated 6 and non-acetylated 7) from wild type BRD1 (NB = no binding). | ||
Conclusions
Bromodomains bind acetyllysine residues in histone and non-histone proteins in sequence-specific manners to regulate a myriad of cellular processes. Bromodomains play a central role in chromatin-dependent events such as transcription and DNA repair and are validated drug targets for a range of human diseases. Given that bromodomains often coexist with other protein interaction modules, it has been a challenge to decipher the bromodomain-specific interactome. Furthermore, non-histone lysine acetylation is emerging as a major regulatory mechanism for metabolic and transcriptional processes in mammalian cells. However, the link between bromodomains and non-histone acetylation in the context of these processes is largely unexplored.BRD1, a multi-domain protein with a bromodomain, is known to form transient complexes with several transcriptional regulators in an isoform-specific manner. It has been implicated in human brain development and neurodegenerative conditions such as schizophrenia and bipolar disorder. To better understand the function of BRD1-BD, we intended to identify its acetylated non-histone interacting partners in an unbiased manner by employing a chemoproteomic approach called IBPP. Towards this end, we developed a BRD1 mutant by replacing R585 with a photo-crosslinkable amino acid AzF in the hydrophobic cavity of the bromodomain. The R585AzF mutant recapitulated the activity of wild type BRD1, such as binding with acetylated histone peptide and full-length protein, in biochemical assays. More importantly, the mutant underwent efficient crosslinking with the cognate interacting partner histone H4 upon photo-irradiation. Subsequently, R585AzF was employed to capture novel binding partners from HEK293T cells. Tandem mass spectrometric characterization of the enriched proteins revealed a series of novel BRD1 binding partners, extending the canonical functions of BRD1 beyond chromatin remodeling.
A survey of the primary literature and proteomic databases indicated that several of the newly identified proteins are acetylated at lysine residues. The functional relevance of many of these acetylation events in mammalian cells is currently unknown. By identifying BRD1 as a potential reader of these proteins, our work provides insights into how the non-histone acetylation-BRD1 axis regulates diverse signaling pathways. As a representative example, we identified several 14-3-3 proteins such as YWHAE, YWHAQ and YWHAZ as the BRD1 interactome. 14-3-3 proteins facilitate localization of transcriptional regulators such as the ATP-dependent PBAF complex to a specific genomic locus via interaction with phosphorylated histone H3S10 and H3S28, providing mechanistic insights into how BRD1 and 14-3-3 interaction may regulate chromatin remodeling and transcription.41,42 Genetic studies with ywhae+/− and ywhaz−/− mice have implicated 14-3-3 genes, similar to BRD1, in neurodevelopmental and schizophrenia-associated phenotypes.43–47 By identifying YWHAE and YWHAZ as interacting partners of BRD1, our study provides a potential mechanistic link between BRD1-BD-mediated recognition of YWHAE and YWHAZ and these disease conditions. We anticipate that future biochemical and cell-based validation studies with the interacting partners identified by the IBPP platform will lead to detailed understanding of BRD1 in the context of transcription, erythropoiesis, brain development and deregulated processes, such as cancer and schizophrenia. The newly revealed interacting motifs will be crucial to design and develop potent and specific small-molecule inhibitors of BRD1, a therapeutic target for several human conditions.48,49
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the University of Pittsburgh and the National Institutes of Health (R01GM123234) for financial support, the UPCI Cancer Biomarkers Facility (supported in part by award P30CA047904) for LC-MS/MS analysis, K. Hinkelman and N. Hartos for reagents, and Dr D. Chakraborty of the manuscript.Notes and references
- T. Kouzarides, Cell, 2007, 128, 693–705 CrossRef CAS PubMed.
- C. D. Allis and T. Jenuwein, Nat. Rev. Genet., 2016, 17, 487–500 CrossRef CAS PubMed.
- A. Dhall and C. Chatterjee, ACS Chem. Biol., 2011, 6, 987–999 CrossRef CAS PubMed.
- S. D. Taverna, H. Li, A. J. Ruthenburg, C. D. Allis and D. J. Patel, Nat. Struct. Mol. Biol., 2007, 14, 1025–1040 CrossRef CAS PubMed.
- C. A. Musselman, M. E. Lalonde, J. Cote and T. G. Kutateladze, Nat. Struct. Mol. Biol., 2012, 19, 1218–1227 CrossRef CAS PubMed.
- B. M. Dancy and P. A. Cole, Chem. Rev., 2015, 115, 2419–2452 CrossRef CAS PubMed.
- P. Filippakopoulos, S. Picaud, M. Mangos, T. Keates, J. P. Lambert, D. Barsyte-Lovejoy, I. Felletar, R. Volkmer, S. Muller, T. Pawson, A. C. Gingras, C. H. Arrowsmith and S. Knapp, Cell, 2012, 149, 214–231 CrossRef CAS PubMed.
- S. G. Smith and M. M. Zhou, ACS Chem. Biol., 2016, 11, 598–608 CrossRef CAS PubMed.
- P. Filippakopoulos and S. Knapp, FEBS Lett., 2012, 586, 2692–2704 CrossRef CAS PubMed.
- K. T. Smith and J. L. Workman, Nat. Biotechnol., 2009, 27, 917–919 CrossRef CAS PubMed.
- J. P. Lambert, S. Picaud, T. Fujisawa, H. Hou, P. Savitsky, L. Uuskula-Reimand, G. D. Gupta, H. Abdouni, Z. Y. Lin, M. Tucholska, J. D. R. Knight, B. Gonzalez-Badillo, N. St-Denis, J. A. Newman, M. Stucki, L. Pelletier, N. Bandeira, M. D. Wilson, P. Filippakopoulos and A. C. Gingras, Mol. Cell, 2019, 73(621-638), e617 Search PubMed.
- Y. Mishima, S. Miyagi, A. Saraya, M. Negishi, M. Endoh, T. A. Endo, T. Toyoda, J. Shinga, T. Katsumoto, T. Chiba, N. Yamaguchi, I. Kitabayashi, H. Koseki and A. Iwama, Blood, 2011, 118, 2443–2453 CrossRef CAS PubMed.
- P. McCullagh, T. Chaplin, J. Meerabux, D. Grenzelias, D. Lillington, R. Poulsom, A. Gregorini, V. Saha and B. D. Young, Oncogene, 1999, 18, 7442–7452 CrossRef CAS PubMed.
- M. Nyegaard, J. E. Severinsen, T. D. Als, A. Hedemand, S. Straarup, M. Nordentoft, A. McQuillin, N. Bass, J. Lawrence, S. Thirumalai, A. C. P. Pereira, R. Kandaswamy, G. J. Lydall, P. Sklar, E. Scolnick, S. Purcell, D. Curtis, H. M. D. Gurling, P. B. Mortensen, O. Mors and A. D. Børglum, Am. J. Med. Genet., Part B, 2010, 153b, 582–591 CrossRef CAS PubMed.
- J. E. Severinsen, C. R. Bjarkam, S. Kiaer-Larsen, I. M. Olsen, M. M. Nielsen, J. Blechingberg, A. L. Nielsen, I. E. Holm, L. Foldager, B. D. Young, W. J. Muir, D. H. Blackwood, T. J. Corydon, O. Mors and A. D. Børglum, Mol. Psychiatry, 2006, 11, 1126–1138 CrossRef CAS PubMed.
- A. P. Rajkumar, P. Qvist, J. G. Donskov, R. Lazarus, J. Pallesen, N. Nava, G. Winther, N. Liebenberg, S. H. Cour, V. Paternoster, T. Fryland, J. Palmfeldt, K. Fejgin, A. Mørk, M. Nyegaard, B. Pakkenberg, M. Didriksen, J. R. Nyengaard, G. Wegener, O. Mors, J. H. Christensen and A. D. Børglum, Transl. Psychiatry, 2020, 10, 239 CrossRef CAS PubMed.
- P. Qvist, S. F. Eskildsen, B. Hansen, M. Baragji, S. Ringgaard, J. Roovers, V. Paternoster, S. Molgaard, T. J. Corydon, H. Stødkilde-Jørgensen, S. Glerup, O. Mors, G. Wegener, J. R. Nyengaard, A. D. Børglum and J. H. Christensen, Sci. Rep., 2018, 8, 16486 CrossRef PubMed.
- B. J. Klein, M. E. Lalonde, J. Côté, X. J. Yang and T. G. Kutateladze, Epigenetics, 2014, 9, 186–193 CrossRef CAS PubMed.
- T. M. Weaver, E. A. Morrison and C. A. Musselman, Molecules, 2018, 23 Search PubMed.
- M. Zhang, M. Lei, S. Qin, A. Dong, A. Yang, Y. Li, P. Loppnau, T. R. Hughes, J. Min and Y. Liu, Biochim. Biophys. Acta, Gene Regul. Mech., 2021, 1864, 194688 CrossRef CAS PubMed.
- S. Qin, L. Jin, J. Zhang, L. Liu, P. Ji, M. Wu, J. Wu and Y. Shi, J. Biol. Chem., 2011, 286, 36944–36955 CrossRef CAS PubMed.
- A. Vezzoli, N. Bonadies, M. D. Allen, S. M. Freund, C. M. Santiveri, B. T. Kvinlaug, B. J. Huntly, B. Göttgens and M. Bycroft, Nat. Struct. Mol. Biol., 2010, 17, 617–619 CrossRef CAS PubMed.
- L. You, K. Yan, J. Zou, H. Zhao, N. R. Bertos, M. Park, E. Wang and X. J. Yang, J. Biol. Chem., 2015, 290, 11349–11364 CrossRef CAS PubMed.
- K. Yan, L. You, C. Degerny, M. Ghorbani, X. Liu, L. Chen, L. Li, D. Miao and X. J. Yang, J. Biol. Chem., 2016, 291, 2647–2663 CrossRef CAS PubMed.
- H. I. Cho, M. S. Kim and Y. K. Jang, Exp. Cell Res., 2016, 346, 30–39 CrossRef CAS PubMed.
- A. Poplawski, K. Hu, W. Lee, S. Natesan, D. Peng, S. Carlson, X. Shi, S. Balaz, J. L. Markley and K. C. Glass, J. Mol. Biol., 2014, 426, 1661–1676 CrossRef CAS PubMed.
- T. Fryland, J. H. Christensen, J. Pallesen, M. Mattheisen, J. Palmfeldt, M. Bak, J. Grove, D. Demontis, J. Blechingberg, H. S. Ooi, M. Nyegaard, M. E. Hauberg, N. Tommerup, N. Gregersen, O. Mors, T. J. Corydon, A. L. Nielsen and A. D. Børglum, Genome Med., 2016, 8, 53 CrossRef PubMed.
- N. D. Pham, R. B. Parker and J. J. Kohler, Curr. Opin. Chem. Biol., 2013, 17, 90–101 CrossRef CAS PubMed.
- H. Neumann, P. Neumann-Staubitz, A. Witte and D. Summerer, Curr. Opin. Chem. Biol., 2018, 45, 1–9 CrossRef CAS PubMed.
- T. A. Nguyen, M. Cigler and K. Lang, Angew. Chem., Int. Ed., 2018, 57, 14350–14361 CrossRef CAS PubMed.
- Y. Tian, M. P. Jacinto, Y. Zeng, Z. Yu, J. Qu, W. R. Liu and Q. Lin, J. Am. Chem. Soc., 2017, 139, 6078–6081 CrossRef CAS PubMed.
- J. Liu, S. Li, N. A. Aslam, F. Zheng, B. Yang, R. Cheng, N. Wang, S. Rozovsky, P. G. Wang, Q. Wang and L. Wang, J. Am. Chem. Soc., 2019, 141, 9458–9462 CrossRef CAS PubMed.
- C. C. Liu and P. G. Schultz, Annu. Rev. Biochem., 2010, 79, 413–444 CrossRef CAS PubMed.
- H. Rannversson, J. Andersen, L. Sorensen, B. Bang-Andersen, M. Park, T. Huber, T. P. Sakmar and K. Stromgaard, Nat. Commun., 2016, 7, 11261 CrossRef PubMed.
- Y. Zheng, M. J. Gilgenast, S. Hauc and A. Chatterjee, ACS Chem. Biol., 2018, 13, 1137–1141 CrossRef CAS PubMed.
- B. Sudhamalla, D. Dey, M. Breski, T. Nguyen and K. Islam, Chemical Science, Royal Society of Chemistry, 2010, 2017, vol. 8, pp. 4250–4256 Search PubMed.
- S. Wagner, B. Sudhamalla, P. Mannes, S. Sappa, S. Kavoosi, D. Dey, S. Wang and K. Islam, Chem. Commun., 2020, 56, 3641–3644 RSC.
- J. O. Obi, M. Y. Lubula, G. Cornilescu, A. Henrickson, K. McGuire, C. M. Evans, M. Phillips, S. P. Boyson, B. Demeler, J. L. Markley and K. C. Glass, Curr. Res. Struct. Biol., 2020, 2, 104–115 CrossRef PubMed.
- A. Dhall, S. Wei, B. Fierz, C. L. Woodcock, T. H. Lee and C. Chatterjee, J. Biol. Chem., 2014, 289, 33827–33837 CrossRef CAS PubMed.
- C. Choudhary, C. Kumar, F. Gnad, M. L. Nielsen, M. Rehman, T. C. Walther, J. V. Olsen and M. Mann, Science, 2009, 325, 834–840 CrossRef CAS PubMed.
- S. Healy, D. H. Khan and J. R. Davie, Discovery Med., 2011, 11, 349–358 Search PubMed.
- B. Drobic, B. Pérez-Cadahía, J. Yu, S. K. Kung and J. R. Davie, Nucleic Acids Res., 2010, 38, 3196–3208 CrossRef CAS PubMed.
- R. Bell, J. Munro, C. Russ, J. F. Powell, A. Bruinvels, R. W. Kerwin and D. A. Collier, Am. J. Med. Genet., 2000, 96, 736–743 CrossRef CAS PubMed.
- P. S. Cheah, H. S. Ramshaw, P. Q. Thomas, K. Toyo-Oka, X. Xu, S. Martin, P. Coyle, M. A. Guthridge, F. Stomski, M. van den Buuse, A. Wynshaw-Boris, A. F. Lopez and Q. P. Schwarz, Mol. Psychiatry, 2012, 17, 451–466 CrossRef CAS PubMed.
- M. Ikeda, T. Hikita, S. Taya, J. Uraguchi-Asaki, K. Toyo-oka, A. Wynshaw-Boris, H. Ujike, T. Inada, K. Takao, T. Miyakawa, N. Ozaki, K. Kaibuchi and N. Iwata, Hum. Mol. Genet., 2008, 17, 3212–3222 CrossRef CAS PubMed.
- P. Qvist, J. H. Christensen, I. Vardya, A. P. Rajkumar, A. Mørk, V. Paternoster, E. M. Füchtbauer, J. Pallesen, T. Fryland, M. Dyrvig, M. E. Hauberg, B. Lundsberg, K. Fejgin, M. Nyegaard, K. Jensen, J. R. Nyengaard, O. Mors, M. Didriksen and A. D. Børglum, Biol. Psychiatry, 2017, 82, 62–76 CrossRef CAS PubMed.
- P. Qvist, A. P. Rajkumar, J. P. Redrobe, M. Nyegaard, J. H. Christensen, O. Mors, G. Wegener, M. Didriksen and A. D. Børglum, Neurobiol. Learn. Mem., 2017, 141, 44–52 CrossRef CAS PubMed.
- K. Klein, M. Kato, M. Frank-Bertoncelj, C. Kolling, A. Ciurea, S. Gay and C. Ospelt, Sci. Rep., 2018, 8, 11125 CrossRef PubMed.
- L. Bouché, C. D. Christ, S. Siegel, A. E. Fernández-Montalván, S. J. Holton, O. Fedorov, A. Ter Laak, T. Sugawara, D. Stöckigt, C. Tallant, J. Bennett, O. Monteiro, L. Díaz-Sáez, P. Siejka, J. Meier, V. Pütter, J. Weiske, S. Müller, K. V. M. Huber, I. V. Hartung and B. Haendler, J. Med. Chem., 2017, 60, 4002–4022 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2cb00043a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |