Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Homologues of epigenetic pyrimidines: 5-alkyl-, 5-hydroxyalkyl and 5-acyluracil and -cytosine nucleotides: synthesis, enzymatic incorporation into DNA and effect on transcription with bacterial RNA polymerase†
Filip
Gracias
a,
Olatz
Ruiz-Larrabeiti
b,
Viola
Vaňková Hausnerová
b,
Radek
Pohl
a,
Blanka
Klepetářová
a,
Veronika
Sýkorová
a,
Libor
Krásný
*b and
Michal
Hocek
 *ac
*ac
aInstitute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, Flemingovo nam. 2, CZ-16000, Prague 6, Czech Republic. E-mail: hocek@uochb.cas.cz
bLab. of Microbial Genetics and Gene Expression, Institute of Microbiology, Czech Academy of Sciences, Vídeňská 1083, CZ-14220, Prague 4, Czech Republic. E-mail: krasny@biomed.cas.cz
cDepartment of Organic Chemistry, Faculty of Science, Charles University, Hlavova 8, CZ-12843, Prague 2, Czech Republic
First published on 30th June 2022
Abstract
Homologues of natural epigenetic pyrimidine nucleosides and nucleotides were designed and synthesized. They included 5-ethyl-, 5-propyl-, 5-(1-hydroxyethyl)-, 5-(1-hydroxypropyl)- and 5-acetyl- and 5-propionylcytosine and -uracil 2′-deoxyribonucleosides and their corresponding 5′-O-triphosphates (dNXTPs). The epimers of 5-(1-hydroxyethyl)- and 5-(1-hydroxypropyl)pyrimidine nucleosides were separated and their absolute configuration was determined by a combination of X-ray and NMR analysis. The modified dNXTPs were used as substrates for PCR synthesis of modified DNA templates used for the study of transcription with bacterial RNA polymerase. Fundamental differences in transcription efficiency were observed, depending on the various modifications. The most notable effects included pronounced stimulation of transcription from 5-ethyluracil-bearing templates (200% transcription yield compared to natural thymine) and an enhancing effect of 5-acetylcytosine versus inhibiting effect of 5-acetyluracil. In summary, these results reveal that RNA polymerase copes with dramatically altered DNA structure and suggest that these nucleobases could potentially play roles as artificial epigenetic DNA nucleobases.
Introduction
Epigenetic modifications of histones and DNA are important regulators of gene expression.1–3 In eukaryotic genomic DNA, the major epigenetic modification is 5-methylcytosine (5mC),4 which is formed through cytosine methylation by DNA methyltransferases5 and downregulates transcription when present at high levels. The oxidized derivatives,6 5-hydroxymethylcytosine (5hmC),7,8 5-formylcytosine (5fC)9 and 5-carboxycytosine (5caC),10 are rarer modifications formed through oxidation of 5mC by ten-eleven translocation (TET) enzymes.11–13 They are intermediates in an active demethylation process14,15 but also stable epigenetic marks16,17 with their own role in regulation of gene expression.18–20 5-Hydroxymethyluracil (5hmU) is another rare natural DNA modification present in human stem cells,21 cancer cells,22 protozoan parasites23 and the genomes of certain bacteriophages, where 5hmU almost completely replaces thymine;24,25 yet its biological role is not fully understood.26 Another modification, 5-formyluracil (5fU), can be formed in DNA as a product of oxidative damage of thymine and is known to cause mutations due to base-pairing with both A and G.27,28 In our previous systematic study of the influence of non-natural and natural modifications in DNA on transcription with Escherichia coli RNA polymerase (RNAP), we found that some non-natural nucleobase modifications29 can be tolerated by RNAP and, surprisingly, the presence of 5hmU in the Pveg promoter significantly increased the transcription efficiency.30 Later on, we developed transcription switches based on photocaging and the release of 5hmU or 5hmC in DNA.31,32Furthermore, there are even some examples of very rare natural pyrimidine DNA nucleobases bearing even more bulky modifications, e.g. glycine, 2-aminoethyl33 and several types of conjugates of 5hmU or 5hmC with glucose,34–36 amino acids, amines etc.33 The role of these modifications is either unknown or elusive. Recently, the K. Islam group has published37 an intriguing work showing that the TET2 enzyme can even oxidize non-natural 5-ethylC to 5-(1-hydroxyethyl)cytosine, which can be further chemically oxidized to 5-acetylcytosine or enzymatically glucosylated.37 These recent pioneering works prompted us to design and synthesize several more complex homologues of epigenetic pyrimidine nucleotides and study their enzymatic incorporation into DNA and their influence on transcription.
Results and discussion
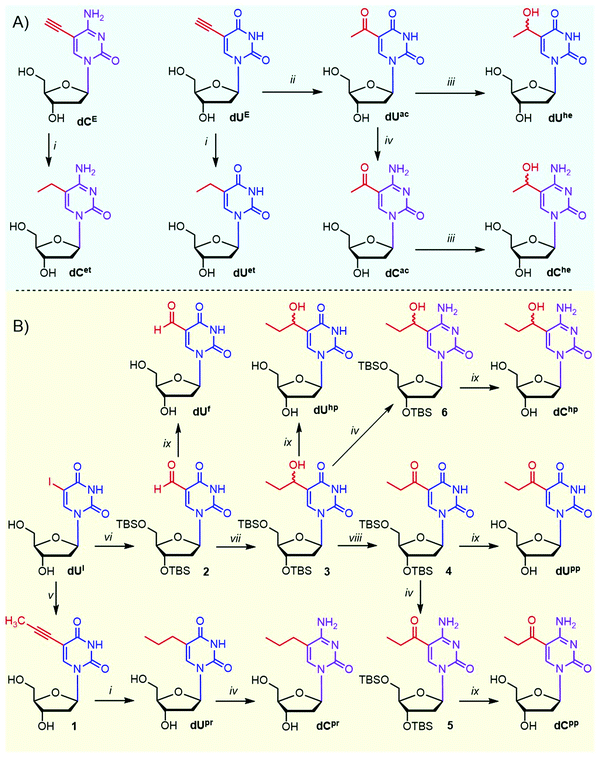
The target homologues of epigenetic pyrimidine nucleotides were derived from 5-ethyl- and 5-propyluracil and -cytosine and included their oxidized congeners, i.e. 1-hydroxyalkyl and 1-oxoalkyl derivatives. For comparison, we also included natural 5-formylpyrimidines38 as well as previously studied 5-vinyl- and 5-ethynylpyrimidines.29,39,40 These modifications were attached to the 5-position of 2′-deoxyuridine (dU) and 2′-deoxycytidine (dC).Although the synthesis of several ethyl-based pyrimidine nucleosides is known,41–43 we prepared some of them in a different and more efficient way (Scheme 1A). Catalytic hydrogenation of 5-ethynylpyrimidine nucleosides dUE (ref. 44) and dCE gave ethyl derivatives dUet and dCet in 84 and 60% yields, respectively. Acid-catalyzed hydration of a terminal triple bond of 5-ethynyl-2′-deoxyuridine dUE with dilute sulfuric acid in methanol gave acetyl derivative dUac (69%)45 that was the key intermediate for the synthesis of other required derivatives. The Luche reduction46 of dUac with NaBH4 and CeCl3 afforded 5-(1-hydroxyethyl)uracil nucleoside dUhe in a 49% yield (34% overall from dUE). Amination47 of dUac at position 4 with (7-azabenzotriazol-1-yloxy)trispyrrolidinophosphonium hexafluorophosphate (PyAOP) and NH4OH in the presence of DBU gave acetylcytosine nucleotide dCac (67% yield) that was reduced to dChe (51% yield) using the Luche reduction (overall yield 24% from dUE).
The propyl-based nucleoside series was synthesized from 5-iodo-2′-deoxyuridine (dUI) in two different pathways (Scheme 1B). The first pathway consisted of the Sonogashira reaction of dUI with generated propyne gas to produce 5-(prop-1-ynyl)-dU (1),48 which was hydrogenated to propyl-dU (dUpr),49 which was further transformed to cytidine derivative dCpr through the above-mentioned amination. The main intermediate of the second pathway was TBS-protected 5-(1-hydroxypropyl)-dU (3) synthesized from dUI by TBS protection, Pd-catalyzed carbonylation yielding the protected formyl-dU (2),50 which was further reacted with EtMgBr. Hydroxypropylpyrimidine intermediate 3 was obtained in an overall 51% yield and was further oxidized by Dess–Martin periodate and either deprotected or aminated and deprotected to give 5-(propionyl)-dU and -dC nucleosides (dUpp and dCpp). The same intermediate 3 was also directly deprotected or aminated and deprotected to give 5-(1-hydroxypropyl)-dU and -dC (dUhp and dChp). Known dUf (ref. 14) was prepared by deprotection of 2.
In the case of hydroxy derivatives dNhe and dNhp, a mixture of two diastereoisomers (epimers) was obtained in each case. To study the influence of each epimer on transcription separately, we separated both epimers by HPLC, using either non-chiral or chiral columns (Fig. 1A and Fig. S13–S16 in ESI†). As the new chiral center is distant from the deoxyribose, the determination of the relative and absolute configuration was non-trivial. Fortunately, we succeeded in crystallization of two cytosine derivatives dCShe and dCShp and determined their configuration by X-ray diffraction (Fig. 1B, Fig. S39 and S40 in ESI†), which also indirectly revealed the configuration of the complementary epimers dCRhe and dCRhp. Uridine derivatives were assigned by an amination reaction (Scheme 1, step iv) on a single epimer of uridine derivative to acquire the cytidine analog (Fig. 1C). The obtained cytidine derivative was then mixed in a single NMR tube with one of the cytidine derivatives of known configuration and the measured 1H NMR showed whether the configuration matched or not (Fig. 1C and D). This procedure was performed for dUShe and dURhp epimers to assign the absolute configuration to all four hydroxyalkyluridine derivatives.
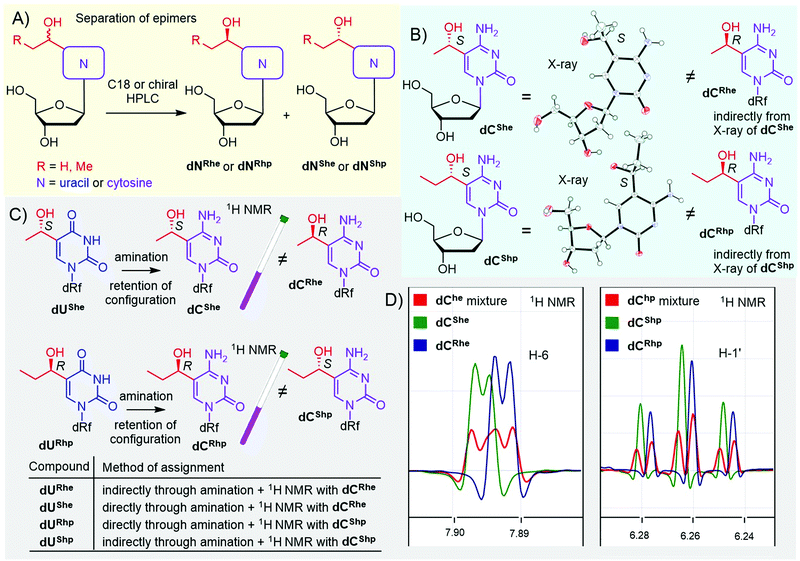 | ||
| Fig. 1 Separation and assignment of epimers of dNhe and dNhp nucleosides. (A) separation of epimers; (B) X-ray assignment of dChe and dChp: CCDC 2166141 – dCShe, CCDC 2166142 – dCShp; (C and D) NMR assignment of dUhe and dUhp nucleosides. | ||
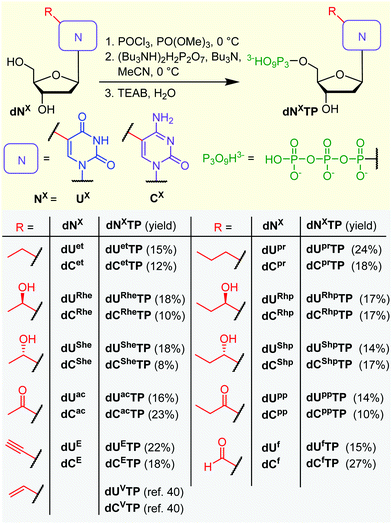
All the prepared modified nucleosides were triphosphorylated to nucleoside triphosphates (dNXTPs) in one pot synthesis using slightly modified standard conditions,51 with yields ranging from 8 to 27% (Scheme 2). In the case of some uridine derivatives, a non-nucleophilic base proton sponge was used. This was particularly important for the prevention of unwanted acid-catalyzed epimerization of the benzylic chiral center during the initial phosphorylation step of dURhp and dUShp (partial epimerization was observed in the absence of the proton sponge, data not shown). Protection of the hydroxyalkyl group at position 5 was not necessary.
With the full series of modified dNXTPs in hand, we tested their substrate activity in enzymatic incorporation into DNA by primer extension reaction (PEX). All the prepared nucleoside triphosphates were good substrates for KOD XL DNA polymerase, as confirmed by both denaturing polyacrylamide gel electrophoresis (PAGE, see Fig. S6 and S7 in ESI†) and mass spectroscopy analysis (see Table S5 and Fig. S17–S38 in ESI†). Subsequently, we used each of them for PCR synthesis of DNA templates for transcription studies. We used a 235-bp template containing the Pveg promoter region as previously described36,52 (Fig. 2A). Almost all the modified nucleotides were successfully incorporated by KOD XL DNA polymerase in PCR (Fig. 2B, Fig. S9 in ESI†), although in most cases some optimization together with higher amounts of dNXTP and DNA polymerase was needed to prepare full length products in sufficient yields. Only in the case of dCETP we used Vent (exo−) polymerase (KOD XL failed to give sufficient amount of PCR amplicon). With three nucleotides: dURheTP, dURhpTP and dUfTP, the PCR did not give the desired amplicons with either KOD XL or other DNA polymerases (Vent (exo−), Pwo or Taq DNA polymerases, data not shown). In the case of dUfTP, this was caused probably by its ability to mispair with both A or G27,28 or by formation of Schiff-base cross-links with the polymerase.53 Therefore, these modifications were not studied further. Interestingly, S-epimers of dU (dUSheTP, dUShpTP) were good substrates for KOD XL, even under PCR conditions, whereas R-epimers (dURheTP, dURhpTP) were very poor substrates and the PCR products were not obtained even after optimization. This stereoselective discrimination in substrate activity was not observed for hydroxyalkylcytidine derivatives, where both epimeric series of nucleotides were successfully incorporated in PCR. At this moment, we do not have any structural explanation for the dichotomy. The PCR products were quantified in order to use them as modified DNA templates for transcription. The quantification was performed with 6-fluorescein-labelled primers or GelRed staining as these methods provide a good balance between accuracy and ease of preparation, avoiding potentially dangerous manipulation with radiolabeled DNA.31,32,36
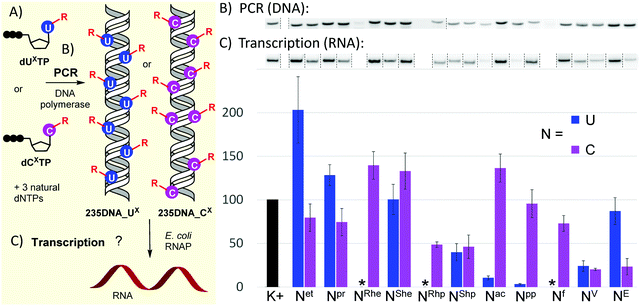 | ||
| Fig. 2 (A) Scheme of PCR synthesis of modified DNA templates and transcription; (B) agarose gel electrophoresis of PCR synthesis of modified 235DNA_NX DNA templates; (C) relative transcription from the modified DNA templates (K+ = non-modified DNA, * = DNA template was not obtained by PCR). For original uncut gels, see the ESI.† | ||
The prepared modified DNA templates containing modified nucleobase UX or CX fully replacing the natural T or C in the whole sequence except for the primers were confirmed by sequencing (see ESI†) and were then tested in multiple round transcription assays, using E. coli RNAP in a reaction supplemented with α-32P-UTP to label the transcript (Fig. 2B and C; for original uncut gels, see Fig. S11 and S12 in ESI†). The transcription products were quantified and the yields normalized for the relative amount of the DNA template and compared to those obtained with a non-modified natural DNA template (K+). Ethyl modification at U (Uet) significantly increased transcription efficiency (ca. 200%), while 5-ethynyl-dC had a slightly suppressing effect (79%). The more bulky propyl modification on U also had a modest stimulatory effect (128%), while on C it moderately decreased the transcription efficiency (74%). The 1-hydroxyethyl modification on U had no apparent effect (UShe, ca. 100%), whereas it slightly enhanced the transcription when attached to C (CRhe at 139%, CShe at 133%). No significant difference was observed between the R- or S-epimer in the deoxycytidine series. A similar lack of stereodiscrimination was observed in 1-hydroxypropyl modification, albeit with a suppressing effect on both U (UShp, 40%) and C (CRhp at 49%, CShp at 46%). The most pronounced effect on transcription was observed with acylpyrimidine derivatives. The presence of an acetyl or propionyl group at U almost completely inhibited transcription (Uac 11%, Upp 3%), while the acetyl group attached to C had a slightly enhancing effect (Cac at 136%) and the propionyl-modified C allowed similar transcription as natural DNA (Cpp at 96%). The formyl group at C exerted only a minor suppression effect (Cf, 73%). Both vinyl- and ethynyl-modified templates were previously studied with a longer 339-bp template containing the same Pveg promoter,29,30 so they were also included in this study for comparison. Vinyl modification of both U and C had a strong suppressing effect on transcription (UV, 24%; CV, 20%), whereas the ethynyl group at C or U showed differential effects: strong suppression when present at C (CE, 23%) and only weak suppression when present at U (UE, 87%).
In conclusion, we have designed and prepared 5-ethyl-, 5-propyl-, epimeric 5-(1-hydroxyethyl)- and 5-(1-hydroxypropyl)-, as well as 5-acetyl and 5-propionyl-uracil and -cytosine 2′-deoxyribonucleosides and their corresponding dNXTPs as homologues of natural epigenetic pyrimidines derived from 5mC and its oxidized congeners. We have also successfully separated and identified individual epimers of 5-(1-hydroxyethyl)- and 5-(1-hydroxypropyl)pyrimidine nucleosides based on X-ray and NMR analysis. Most of the dNXTPs were good substrates for DNA polymerases and were used in PCR synthesis of modified 235-bp DNA templates. Finally, we systematically studied their effect on transcription with bacterial RNAP.
Although the studied pyrimidine modifications have not been detected among the natural epigenetic modifications in genomic DNA (at least not yet), this work brings several important and biologically relevant insights and suggests some prospective applications. We revealed how amazingly robust RNAP is in its ability to interact with DNA decorated with complex modifications and identified both stimulatory and inhibitory effects of some modifications. The surprisingly strong enhancing effect of 5-ethyuracil (200% transcription compared to T) could be then used in biotechnology to increase the transcription efficiency from modified plasmids, possibly even in production of certain therapeutic RNAs. Moreover, the 5-ethyl-2′-deoxyuridine nucleoside could be a potential epigenetic regulator with the opposite effect to 5-aza-dC. Interestingly, some of these modifications are similar in size and functionality to some recently discovered rare DNA nucleobases, so the effects described here may correspond with their roles in Nature. The dichotomy in the effect of acetylpyrimidine derivatives opens the possibility of turning OFF transcription through deamination of CAc to UAc by activation-induced cytidine deaminase previously shown to deaminate Che.37 Conversely, the ten-eleven translocation 2 (TET2) enzyme that was recently shown37 to oxidize Cet to Che could be used to slightly increase the transcription. These potential applications, studies of mechanistic aspects of the effects of the modified nucleobases on transcription, and studies of these modifications in eukaryotic transcription systems will be further pursued in our labs.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Czech Science Foundation (20-00885X to F. G. and M. H. and 22-12023S to L. K.) and by the European Regional Development Fund; OP RDE (No. CZ.02.1.01/0.0/0.0/16_019/0000729 to V. S.). O. R.-L. was supported by the postdoctoral grant from Basque Government (No. POS_2019_1_0033). The PhD scholarship for F. G. from the Department of Organic Chemistry of the University of Chemistry and Technology, Prague is also acknowledged.References
- K. Chen, B. S. Zhao and C. He, Cell Chem. Biol., 2016, 23, 74–85 CrossRef CAS PubMed.
- M. K. Bilyard, S. Becker and S. Balasubramanian, Curr. Opin. Chem. Biol., 2020, 57, 1–7 CrossRef CAS PubMed.
- E.-A. Raiber, R. Hardisty, P. van Delft and S. Balasubramanian, Nat. Rev. Chem., 2017, 1, 0069 CrossRef CAS.
- C. Luo, P. Hajkova and J. R. Ecker, Science, 2018, 361, 1336–1340 CrossRef CAS PubMed.
- J. A. Law and S. E. Jacobsen, Nat. Rev. Genet., 2010, 11, 204–220 CrossRef CAS PubMed.
- T. Carell, M. Q. Kurz, M. Müller, M. Rossa and F. Spada, Angew. Chem., Int. Ed., 2018, 57, 4296–4312 CrossRef CAS PubMed.
- M. Münzel, D. Globisch and T. Carell, Angew. Chem., Int. Ed., 2011, 50, 6460–6468 CrossRef PubMed.
- D. Globisch, M. Münzel, M. Müller, S. Michalakis, M. Wagner, S. Koch, T. Brückl, M. Biel and T. Carell, PLoS One, 2010, 5, e15367 CrossRef CAS PubMed.
- E.-A. Raiber, P. Murat, D. Y. Chirgadze, D. Beraldi, B. F. Luisi and S. Balasubramanian, Nat. Struct. Mol. Biol., 2015, 22, 44–49 CrossRef CAS PubMed.
- L. Wang, Y. Zhou, L. Xu, R. Xiao, X. Lu, L. Chen, J. Chong, H. Li, C. He, X.-D. Fu and D. Wang, Nature, 2015, 523, 621–625 CrossRef CAS PubMed.
- X. Lu, B. S. Zhao and C. He, Chem. Rev., 2015, 115, 2225–2239 CrossRef CAS PubMed.
- Y.-F. He, B.-Z. Li, Z. Li, P. Liu, Y. Wang, Q. Tang, J. Ding, Y. Jia, Z. Chen, L. Li, Y. Sun, X. Li, Q. Dai, C.-X. Song, K. Zhang, C. He and G.-L. Xu, Science, 2011, 333, 1303–1307 CrossRef CAS PubMed.
- L. Hu, J. Lu, J. Cheng, Q. Rao, Z. Li, H. Hou, Z. Lou, L. Zhang, W. Li, W. Gong, M. Liu, C. Sun, X. Yin, J. Li, X. Tan, P. Wang, Y. Wang, D. Fang, Q. Cui, P. Yang, C. He, H. Jiang, C. Luo and Y. Xu, Nature, 2015, 527, 118–122 CrossRef CAS PubMed.
- S. Schiesser, T. Pfaffeneder, K. Sadeghian, B. Hackner, B. Steigenberger, A. S. Schröder, J. Steinbacher, G. Kashiwazaki, G. Höfner, K. T. Wanner, C. Ochsenfeld and T. Carell, J. Am. Chem. Soc., 2013, 135, 14593–14599 CrossRef CAS PubMed.
- A. Schön, E. Kaminska, F. Schelter, E. Ponkkonen, E. Korytiaková, S. Schiffers and T. Carell, Angew. Chem., Int. Ed., 2020, 59, 5591–5594 CrossRef PubMed.
- M. Bachman, S. Uribe-Lewis, X. Yang, H. E. Burgess, M. Iurlaro, W. Reik, A. Murrell and S. Balasubramanian, Nat. Chem. Biol., 2015, 11, 555–557 CrossRef CAS PubMed.
- M. Su, A. Kirchner, S. Stazzoni, M. Müller, M. Wagner, A. Schröder and T. Carell, Angew. Chem., Int. Ed., 2016, 55, 11797–11800 CrossRef CAS PubMed.
- L. Lercher, M. A. McDonough, A. H. El-Sagheer, A. Thalhammer, S. Kriaucionis, T. Brown and C. J. Schofield, Chem. Commun., 2014, 50, 1794–1796 RSC.
- A. Perera, D. Eisen, M. Wagner, S. K. Laube, A. F. Künzel, S. Koch, J. Steinbacher, E. Schulze, V. Splith, N. Mittermeier, M. Müller, M. Biel, T. Carell and S. Michalakis, Cell Rep., 2015, 11, 283–294 CrossRef CAS PubMed.
- N. Kitsera, J. Allgayer, E. Parsa, N. Geier, M. Rossa, T. Carell and A. Khobta, Nucleic Acids Res., 2017, 45, 11033–11042 CrossRef CAS PubMed.
- T. Pfaffeneder, F. Spada, M. Wagner, C. Brandmayr, S. K. Laube, D. Eisen, M. Truss, J. Steinbacher, B. Hackner, O. Kotljarova, D. Schuermann, S. Michalakis, O. Kosmatchev, S. Schiesser, B. Steigenberger, N. Raddaoui, G. Kashiwazaki, U. Müller, C. G. Spruijt, M. Vermeulen, H. Leonhardt, P. Schär, M. Müller and T. Carell, Nat. Chem. Biol., 2014, 10, 574–581 CrossRef CAS PubMed.
- Z. Djuric, L. K. Heilbrun, M. S. Simon, D. Smith, D. A. Luongo, P. M. LoRusso and S. Martino, Cancer, 1996, 77, 691–696 CrossRef CAS PubMed.
- F. Kawasaki, D. Beraldi, R. E. Hardisty, G. R. McInroy, P. van Delft and S. Balasubramanian, Genome Biol., 2017, 18, 23 CrossRef PubMed.
- P. Weigele and E. A. Raleigh, Chem. Rev., 2016, 116, 12655–12687 CrossRef CAS PubMed.
- G. Hutinet, Y.-J. Lee, V. de Crécy-Lagard and P. R. Weigele, EcoSal Plus, 2021, 9, eESP00282019 CrossRef PubMed.
- R. Olinski, M. Starczak and D. Gackowski, Mutat. Res., 2016, 767, 59–66 CAS.
- K. Fujikawa, H. Kamiya and H. Kasai, Nucleic Acids Res., 1998, 26, 4582–4587 CrossRef CAS PubMed.
- D. K. Rogstad, J. Heo, N. Vaidehi, W. A. Goddard, A. Burdzy and L. C. Sowers, Biochemistry, 2004, 43, 5688–5697 CrossRef CAS PubMed.
- V. Raindlová, M. Janoušková, M. Slavíčková, P. Perlíková, S. Boháčová, N. Milisavljevič, H. Šanderová, M. Benda, I. Barvík, L. Krásný and M. Hocek, Nucleic Acids Res., 2016, 44, 3000–3012 CrossRef PubMed.
- M. Janoušková, Z. Vaníková, F. Nici, S. Boháčová, D. Vítovská, H. Šanderová, M. Hocek and L. Krásný, Chem. Commun., 2017, 53, 13253–13255 RSC.
- Z. Vaníková, M. Janoušková, M. Kambová, L. Krásný and M. Hocek, Chem. Sci., 2019, 10, 3937–3942 RSC.
- A. Chakrapani, V. Vaňková Hausnerová, O. Ruiz-Larrabeiti, R. Pohl, L. Krásný and M. Hocek, Org. Lett., 2020, 22, 9081–9085 CrossRef CAS PubMed.
- Y.-J. Lee, N. Dai, S. I. Müller, C. Guan, M. J. Parker, M. E. Fraser, S. E. Walsh, J. Sridar, A. Mulholland, K. Nayak, Z. Sun, Y.-C. Lin, D. G. Comb, K. Marks, R. Gonzalez, D. P. Dowling, V. Bandarian, L. Saleh, I. R. Corrêa and P. R. Weigele, Nucleic Acids Res., 2022, 50, 3001–3017 CrossRef CAS PubMed.
- S. R. Kornberg, S. B. Zimmerman and A. Kornberg, J. Biol. Chem., 1961, 236, 1487–1493 CrossRef CAS PubMed.
- J. Tomaschewski, H. Gram, J. W. Crabb and W. Rüger, Nucleic Acids Res., 1985, 13, 7551–7568 CrossRef CAS PubMed.
- A. Chakrapani, O. Ruiz-Larrabeiti, R. Pohl, M. Svoboda, L. Krásný and M. Hocek, Chem. – Eur. J., 2022, 28, e202200911 CrossRef CAS PubMed.
- S. Kavoosi, B. Sudhamalla, D. Dey, K. Shriver, S. Arora, S. Sappa and K. Islam, Chem. Sci., 2019, 10, 10550–10555 RSC.
- B. Steigenberger, S. Schiesser, B. Hackner, C. Brandmayr, S. K. Laube, J. Steinbacher, T. Pfaffeneder and T. Carell, Org. Lett., 2013, 15, 366–369 CrossRef CAS PubMed.
- H. Macíčková-Cahová, R. Pohl and M. Hocek, ChemBioChem, 2011, 12, 431–438 CrossRef PubMed.
- M. Mačková, R. Pohl and M. Hocek, ChemBioChem, 2014, 15, 2306–2312 CrossRef PubMed.
- T. Kulikowski and D. Shugar, J. Med. Chem., 1974, 17, 269–273 CrossRef CAS PubMed.
- A. S. Jones, M. J. Slater and R. T. Walker, J. Chem. Soc., Perkin Trans. 1, 1987, 1325–1329 RSC.
- A. Chentsova, E. Kapourani and A. Giannis, Beilstein J. Org. Chem., 2014, 10, 7–11 CrossRef PubMed.
- M. J. Robins and P. J. Barr, J. Org. Chem., 1983, 48, 1854–1862 CrossRef CAS.
- S. A. Ingale, H. Mei, P. Leonard and F. Seela, J. Org. Chem., 2013, 78, 11271–11282 CrossRef CAS PubMed.
- J.-L. Luche, L. Rodriguez-Hahn and P. Crabbé, J. Chem. Soc., Chem. Commun., 1978, 601–602 RSC.
- X.-A. Zheng, H.-S. Huang, R. Kong, W.-J. Chen, S.-S. Gong and Q. Sun, Tetrahedron, 2018, 74, 7095–7101 CrossRef CAS.
- B. C. Froehler, S. Wadwani, T. J. Terhorst and S. R. Gerrard, Tetrahedron Lett., 1992, 33, 5307–5310 CrossRef CAS.
- J. L. Ruth and D. E. Bergstrom, J. Org. Chem., 1978, 43, 2870–2876 CrossRef CAS.
- G. J. Crouch and B. E. Eaton, Nucleosides Nucleotides, 1994, 13, 939–944 CrossRef CAS.
- J. Ludwig, Acta Biochim. Biophys. Hung., 1981, 16, 131–133 CAS.
- L. Sojka, T. Kouba, I. Barvík, H. Šanderová, Z. Maderová, J. Jonák and L. Krásný, Nucleic Acids Res., 2011, 39, 4598–4611 CrossRef CAS PubMed.
- E.-A. Raiber, G. Portella, S. Martínez Cuesta, R. Hardisty, P. Murat, Z. Li, M. Iurlaro, W. Dean, J. Spindel, D. Beraldi, Z. Liu, M. A. Dawson, W. Reik and S. Balasubramanian, Nat. Chem., 2018, 10, 1258–1266 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2166141 and 2166142. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2cb00133k |
| This journal is © The Royal Society of Chemistry 2022 |