Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of 4-thiouridines with prodrug functionalization for RNA metabolic labeling†
Sarah
Moreno
a,
Melanie
Brunner
b,
Isabel
Delazer
 b,
Dietmar
Rieder
b,
Dietmar
Rieder
 c,
Alexandra
Lusser
c,
Alexandra
Lusser
 *b and
Ronald
Micura
*b and
Ronald
Micura
 *a
*a
aInstitute of Organic Chemistry, Center for Molecular Biosciences Innsbruck, University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria. E-mail: ronald.micura@uibk.ac.at
bInstitute of Molecular Biology, Biocenter, Medical University of Innsbruck, Innrain 80-82, 6020 Innsbruck, Austria. E-mail: alexandra.lusser@i-med.ac.at
cInstitute of Bioinformatics, Biocenter, Medical University of Innsbruck, Innrain 82, 6020 Innsbruck, Austria
First published on 25th February 2022
Abstract
Metabolic labeling has emerged as a powerful tool to endow RNA with reactive handles allowing for subsequent chemical derivatization and processing. Recently, thiolated nucleosides, such as 4-thiouridine (4sU), have attracted great interest in metabolic labeling-based RNA sequencing approaches (TUC-seq, SLAM-seq, TimeLapse-seq) to study cellular RNA expression and decay dynamics. For these and other applications (e.g. PAR-CLIP), thus far only the naked nucleoside 4sU has been applied. Here we examined the concept of derivatizing 4sU into a 5′-monophosphate prodrug that would allow for cell permeation and potentially improve labeling efficiency by bypassing the rate-limiting first step of 5′ phosphorylation of the nucleoside into the ultimately bioactive 4sU triphosphate (4sUTP). To this end, we developed robust synthetic routes towards diverse 4sU monophosphate prodrugs. Using metabolic labeling assays, we found that most of the newly introduced 4sU prodrugs were well tolerated by the cells. One derivative, the bis(4-acetyloxybenzyl) 5′-monophosphate of 4sU, was also efficiently incorporated into nascent RNA.
Introduction
Metabolic labeling has emerged as a powerful tool to endow RNA with reactive handles in vivo allowing for subsequent chemical derivatization for a broad range of applications.1–6 Thereby, alkyne-, vinyl-, or azide-modified nucleosides are taken up as cell-permeable metabolic precursors that can directly enter the nucleotide salvage pathway for conversion to nucleotide triphosphates (NTPs).7–9 After incorporation into different types of RNA during transcription or during polyadenylation, click chemistry is used as the labeling of choice.10–14 Moreover, RNA labeled with 4sU has been widely used to capture RNA-binding proteins, due to the intrinsic propensity of 4sU to form covalent links with interacting RNA binding proteins upon UV 365 nm irradiation.15–17 Recently, thiolated nucleosides, in particular 4-thiouridine (4sU), have attracted much interest in metabolic labeling-based RNA sequencing approaches (TUC-seq, SLAM-seq, TimeLapse-seq) to study cellular RNA dynamics, such as synthesis and degradation rates.18–23 4sU uptake into cells is mediated by two distinct families of nucleoside transporters.24 While cell lines, such as HEK293 or HeLa cells, show robust expression of the major uridine transporters SLC29A1 and SLC29A2, and therefore readily take up 4sU, many other cell types and cell lines do not (e.g. ref. 25; see also The Human Protein Atlas26 for transporter expression). To broaden the applicability of 4sU for RNA expression dynamics or PAR-CLIP analyses in such cell types, the use of 4sU monophosphate prodrugs may be a promising approach,27–31 superior to uptake of the naked nucleoside.32–41 Although the exact mechanisms by which nucleoside monophosphate prodrugs enter the cell are not well understood,42 it is often implied that passive absorption is involved,43 or that they interact with other cellular drug transporters.44,45In addition, delivery of 4sU 5′-monophosphate derivatives into the cell allows to circumvent the rate-limiting step of nucleoside monophosphorylation, thus enabling late-stage entry into the nucleotide salvage pathway. Therefore, processing time towards the bioactive di- and triphosphates (4sUDP and 4sUTP) can be shortened which may increase time resolution of RNA labeling experiments.2,10 Interestingly, despite the importance and wide-spread use of 4sU metabolic labeling (Fig. 1), 4sU prodrugs or prodrug-like derivatives have not yet been tested, and to the best of our knowledge, the chemical synthesis of 4sU prodrugs has not yet been reported either. Therefore, we set out to create the synthetic foundation for 4sU monophosphate prodrugs and to test their performance in metabolic labeling experiments.
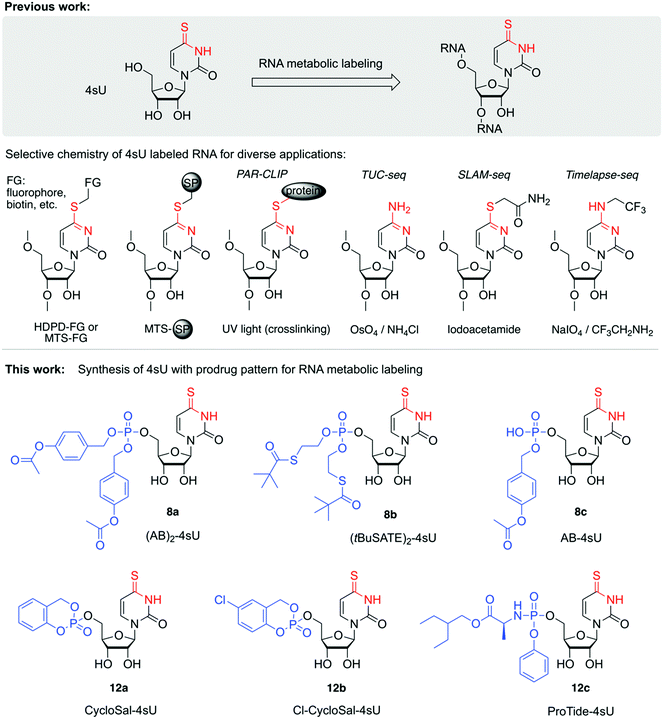 | ||
| Fig. 1 Background and overview of the work described. Upper panel: 4sU is applied for metabolic labeling of RNA to enable thiol-selective chemistry for RNA derivatization. Applications range from covalent labeling with prosthetic groups, fluorophores, biotin for RNA pulldown and enrichment to PAR-CLIP (PhotoActivatable Ribonucleoside-enhanced CrossLinking and ImmunoPrecipitation) to novel RNA sequencing approaches that utilize 4sU derivatization or conversion for defined mutational signatures that shed light on cellular RNA dynamics. Typical reagents are: N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio) propionamide (biotin-HPDP) for streptavidin pulldown or thiosulfonate (MTS) modifed magnetic beads (SP) for one-step enrichment procedure captures 4sU-RNA (see e.g. ref. 69). Additionally, this reagent type is widely used for fluorescent labeling of 4sU containing tRNAs (see e.g. ref. 70). The RNA sequencing approaches TUC-seq, SLAM-seq and TimeLapse-seq rely on chemical 4sU transformations using distinct reagents, thereby converting 4sU into cytosine (TUC-seq; osmium tetroxide (OsO4) and ammonia), alkylated uridine (SLAM-seq; iodoacetamide) or trifluoroethylated cytidine (TimeLapse; 2,2,2-trifluorethylamine (TFEA) in combination with meta-chloroperoxybenzoic acid (mCPBA) or sodium periodate (NaIO4)). Lower panel: Structures of novel 4sU derivatives with monophosphate prodrug pattern that have been synthesized in this study and tested exemplarily in metabolic labeling experiments on HEK293T cells with readout by TUC-seq. | ||
Results and discussion
Design of 4sU prodrugs
Most prodrug strategies for nucleoside analogs rely on 5′-O-phosphate derivatives. The rationale is to shortcut the stepwise conversion path into the biologically active nucleoside triphosphate because the kinases involved are rather ineffective for the modified nucleoside due to high specificity for their native substrates.27 In particular the first phosphorylation is often rate limiting.2,10 However, due to the increase in polarity inhibiting penetration of cellular membranes, the use of free nucleoside monophosphates for metabolic labeling is not possible. To circumvent this problem the negatively charged oxygen atoms of the 5′-O-monophosphate group are masked with 'biolabile' protecting groups which results in membrane-permeable compounds.With respect to 4sU, we focused on established prodrug patterns, including 4-acetyloxybenzyl (AB),29,46S-pivaloyl-2-thioethyl (tBuSATE),47S-acetylthioethyl (SATE),48 and cycloSaligenyl (CycloSal) phosphates,31 as well as phosphoramidates consisting of an amino acid ester promoiety linked via P–N bond to the nucleoside aryl phosphate (ProTide).28,49 Depending on the prodrug pattern, different cellular pathways to release the free 5′-monophosphate nucleotide have been described.50 Additionally, we considered 4sU 5′-O-monophosphates that carry a biolabile moiety at the S4 atom as potential prodrug candidates and set out for their chemical synthesis.
Synthesis of 4sU prodrugs
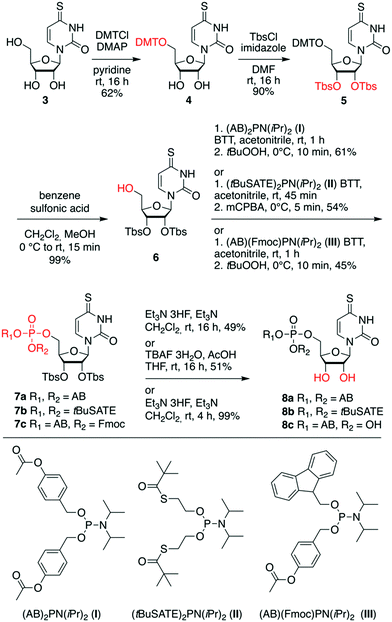
Starting from uridine, we synthesized 4-thiouridine (4sU) in three steps,51,52via acetylation of the hydroxyl groups (compound 1), transformation of the 4-oxo into the 4-thio group using the Lawesson reagent (compound 2), followed by deacetylation with ammonia in methanol to obtain the free 4sU nucleoside 3 in high yields (Scheme S1, ESI†).Targeting the first set of 4sU prodrugs, namely the 4-acetyloxybenzyl (AB)- and S-pivaloyl-2-thioethyl (tBuSATE) 5′-O-monophosphates 8a–c (Scheme 1), we protected the 5′-OH group of 4sU as 4,4′-dimethoxytrityl (DMT) ether using 4,4′-dimethoxytrityl chloride and pyridine to give compound 4. Then, the protection of 2′ and 3′-OH groups was accomplished by reaction with tert-butyldimethylsilyl chloride in the presence of imidazole to yield nucleoside 5. Removal of the 5′-O-DMT moiety under acidic conditions afforded the crucial precursor 6 for subsequent introduction of the phosphotriester moiety. This step was performed using mild phosphor(III)amidite chemistry catalyzed by 5-(benzylthio)-1H-tetrazole. The required reagents bis-(4-acetyloxybenzyl) N,N-diisopropylaminophosphoramidite I,29,53 bis-(S-pivaloyl-2-thioethyl) N,N-diisopropylaminophosphoramidite II,53,54 and 4-[({[bis(propan-2-yl)amino](9H-fluoren-9-ylmethoxy)phosphanyl}oxy)methyl]phenyl acetate III,55,56 were synthesized as described in the ESI,† largely following known procedures that we optimized further. Importantly, selective oxidation of the phosphite P(III) triester intermediates to the required phosphate triester P(V) species was accomplished using equimolar amounts of tert-butyl hydroperoxide (for 7a and 7c) or 3-chlorperbenzoic acid (for 7b) at low temperature and short reaction times. Potential oxidation of the sulfur moiety was not observed under these conditions. Finally, the 2′-O and 3′-O silyl ethers were cleaved by treatment with triethylamine trihydrofluoride to obtain prodrugs 8a and 8c, or by tetrabutylammonium fluoride trihydrate in tetrahydrofuran for prodrug 8b.
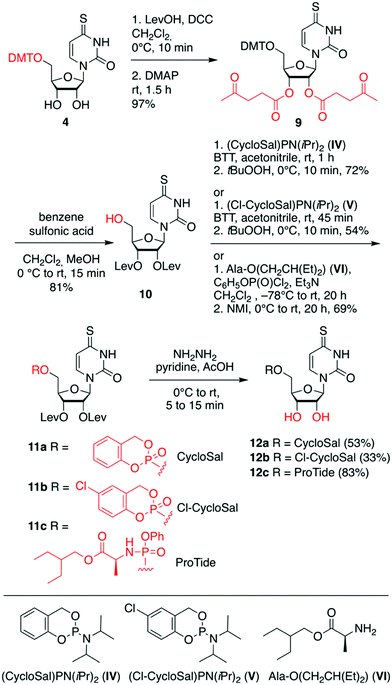
For the second set of 4sU prodrugs (Scheme 2), namely the saligenyl (CycloSal, Cl-CycloSal) and phosphoramidate (ProTide) compounds 12a–c, we started with the 5′-O-tritylated nucleoside 4 and protected the 2′ and 3′-OH groups as levulinyl esters to yield compound 9. Detritylation gave nucleoside 10 which was reacted with either saligenyl-N,N-diisopropylaminophosphoramidite IV57–60 or 5-chlorosaligenyl-N,N-diisopropylaminophosphoramidite V;58,59,61,62 subsequently, oxidation using one equivalent of tert-butyl hydroperoxide gave nucleoside 11a and 11b, respectively. The desired prodrugs 12a and 12b were finally obtained using fast treatment with aqueous hydrazine in pyridine and acetic acid and immediate workup. The levulinyl esters were readily cleaved under these conditions leaving the sensitive saligenyl- and chlorosaligenyl moieties intact. To obtain the 4sU ProTide compound 12c (Scheme 2), the required phosphorylation reagent was prepared in situ by mixing phenyl dichlorophosphate and 2-ethylbutyl (S)-2-aminopropanoate hydrochloride VI,63 activated with triethylamine. The resulting phosphoryl chloride was reacted with precursor 10 to furnish nucleotide 11c, which was further transformed into the desired prodrug 12c; both steps were high yielding.
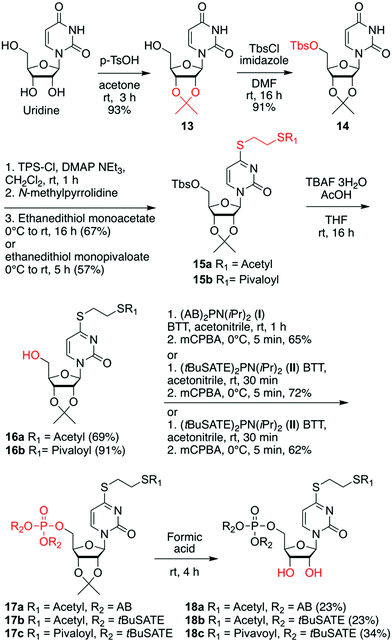
For the third subset of 4sU prodrugs (Scheme 3), namely S4-(S-pivaloyl-2-thioethyl)- or S4-(S-acetylthioethyl) functionalized AB and tBuSATE prodrugs 18a–c, the synthesis started with simultaneous protection of the 2′ and 3′-OH groups as isopropylidene acetal to yield nucleoside 13, followed by 5′-OH protection as Tbs ether to give derivative 14. Subsequently, the O4 functionality was derivatized as sulfonium ester which was then substituted by either ethanedithiol monoacetate or its pivaloyl analog. The resulting compounds 15a and 15b were then unmasked to provide precursors 16a and 16b. Functionalization of the free 5′-OH group was performed using bis-(4-acetyloxybenzyl) N,N-diisopropylaminophosphoramidite I29,53 to obtain intermediate 17a or bis-(S-pivaloyl-2-thioethyl) N,N-diisopropylaminophosphoramidite II53,54 to obtain intermediates 17b and 17c after selective oxidation. Finally, cleavage of the 2′-O, 3′-O cyclic acetal with formic acid provided the desired prodrugs 18a–c.
In vivo evaluation of 4sU monophosphate prodrugs
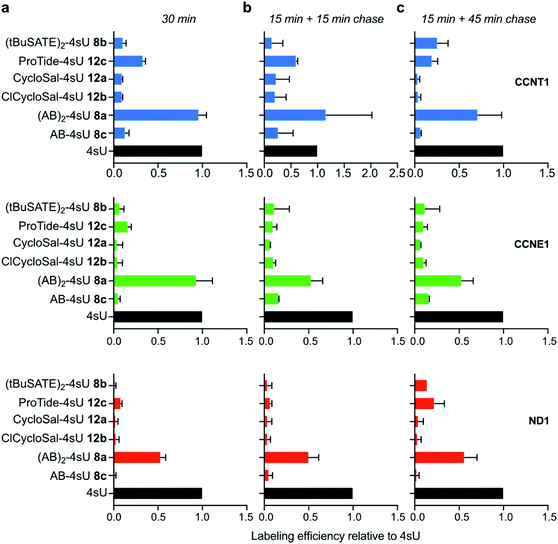
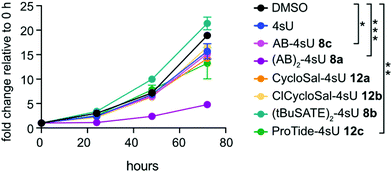
To examine the in vivo applicability of the synthesized prodrugs, we performed metabolic labeling experiments with HEK293T cells. In a first step, we tested the incorporation efficiency into nascent RNA. To this end, we employed our recently developed TUC-seq method that allows for detection of labeled RNA by chemical conversion of 4sU to cytosine using OsO4-mediated chemistry.20 Specific target mRNAs were amplified by PCR and subjected to amplicon sequencing to identify labeled transcripts.23 We first used a 30 min labeling regime with each prodrug at a concentration of 50 μM and measured incorporation into the two nuclear encoded mRNAs cyclin T1 (CCNT1) and cyclin E1 (CCNE1) as well as the mitochondrially transcribed NADH:ubiquinone oxidoreductase core subunit 1 (ND1) mRNA. For comparison, cells were labeled with 4sU. Of all tested prodrugs, (AB)2-4sU 8a performed best with an incorporation efficiency comparable to 4sU (Fig. 2a and Fig. S1, ESI†), while the other reagents (8b,c and 12a,b,c) exhibited clearly lower labeling efficiency. For the high turn-over mRNA CCNT1 (ref. 24 and Fig. S1, ESI†), 4sU incorporation was also detected with the ProTide-4sU 12c, albeit at only ∼30% of 4sU. We then shortened the labeling time to 15 min and extracted RNA either 15 or 45 min after replacing the labeling medium with normal medium. The outcome was comparable to the 30 min labeling approach, except that the longer chase period led to increased incorporation of ProTide-4sU 12c as well as (tBuSATE)2-4sU 8b in all three mRNA targets (Fig. 2b, c and Fig. S1, ESI†). These results suggest that the bioavailability of 4sU released from those prodrugs may be delayed compared to (AB)2-4sU 8a. Thus, (AB)2-4sU 8a appears to be the most efficient among all tested prodrugs, reaching almost the same levels of incorporation as 4sU.To determine cytotoxicity of the diverse 4sU prodrugs, we incubated HEK293T cells at a concentration of 50 μM for 30 min and monitored cell viability over a period of 72 h using an ATP-based assay. Interestingly, the two drugs that exhibited the best 4sU incorporation efficiency ((AB)2-4sU 8a, ProTide-4sU 12c; Fig. 2) had the most adverse impact on cell viability, while the prodrugs with low labeling capacity (8b,c and 12a,b) were tolerated well (Fig. 3). We also examined the effects of the S4-protected 4sU prodrugs (Scheme S2, ESI†) on cell viability. Both AB- as well as tBuSATE-derivatized prodrugs (18a–c) showed strong cytotoxicity after 30 min incubation (Fig. S2, ESI†) and therefore underwent no further labeling tests.
Generally, we suspect that certain metabolites released from the prodrugs – e.g. p-quinone methide in case of (AB)2-4sU 8a – cause the observed adverse effects.64,65 AB-4sU 8c which, in contrast to (AB)2-4sU 8a, retains negative charge on the phosphate moiety, has a moderate effect on cell viability but at the same time does not lead to 4sU incorporation suggesting that it most likely does not permeate the cell membrane efficiently. For CycloSal (12a) ClCycloSal (12b), and (tBuSATE)2 (8b) derivatives, the labeling and viability results indicate that 4sU is either very slowly released in the cell or that the compounds do not enter the cells. Studies with CycloSal derivatives of d4T monophosphate rather point towards the former explanation since cycloSal-d4T compounds proved to be stable in RPMI cell culture medium containing 10% fetal calf serum with halflives of t1/2 = 1.4 to 10.8 hours.66,67 Likewise, tBuSATE-derivatives of 3′-azido-2′,3′-dideoxythymidine (AZT) had high stability in buffer,58 suggesting that the uptake of intact CycloSal (12a,b) and tBuSATE 4sU (8b) prodrugs should be possible. By contrast, it was shown that the last step of hydrolysis of tBuSATE AZT by cellular enzymes proved to be rate limiting and was in the order of t1/2 = 3.9 to 25 h.68
Conclusions
In summary, we developed robust synthetic routes towards 5′-O-phosphate 4-thiouridines with diverse prodrug masking groups by mastering protection concepts and reaction conditions that allowed for P(III)-to-P(V) conversions under preservation of the oxidation-sensitive 4-thio moiety. In our set of metabolic labeling experiments limited to HEK293T cells, only the bis(4-acetyloxybenzyl) 5′-monophosphate derivative of 4sU (8a) showed incorporation efficiencies comparable to 4sU, albeit at cost of increased cytotoxicity. Hence, while the monophosphate prodrug derivatives studied here contribute no immediate advantage to metabolic labeling of cells that are amenable to labeling with the 4sU nucleoside, some of the novel prodrugs might be beneficial for the labeling of cells with low expression of the major nucleoside transporters. In those cases, labeling times will likely have to be extended to allow for efficient cell entry and subsequent release of the active compound.Experimental
Synthesis of 4sU prodrugs
Synthetic procedures and analysis data for the synthesis of compounds 1 to 18 and reagents I to VI are described in the ESI.† All compounds synthesized are >95% pure by NMR analysis, and all NMR spectra are depicted in the ESI.†Metabolic labeling
1.5 × 106 HEK293T cells were seeded into 3 cm round cell culture dishes and grown for six hours at 37 °C and 5% CO2 in culture medium (DMEM/Ham's F-12 + 10% fetal calf serum and GlutaMAX; PAN Bioteck, Gibco). Medium was replaced with culture medium supplemented with 50 μM 4-thiouridine (4sU; Jena Bioscience), 50 μM of the different synthesized prodrugs or vehicle (DMSO). Cells were incubated for 30 min and subsequently harvested. For pulse-chase experiments, labeling medium was replaced with culture medium after 15 min incubation, and cells were further incubated for 15 or 45 min before collection.TUC treatment
Total RNA was isolated from labeled and DMSO-treated cells using the innuPREP RNA Mini Kit 2.0 (Analytik Jena) according to the manufacturer's instructions. RNA was incubated with 0.45 mM OsO4 and 180 mM NH4Cl for 1 h at 40 °C. The reaction was stopped by precipitation with 6 volumes of ice cold ethanol and 2 volumes precipitation buffer (185 mM NaOAc pH 5.2 and 0.25 mg ml−1 glycogen).20Amplicon sequencing and data analysis
TUC-treated RNA was reverse transcribed with GoScript Reverse Transcriptase (Promega) with random hexamer primers according to the manufacturer's instructions. Amplicons of CCNT1 (296 bp), CCNE1 (417 bp) and ND1 (353 bp) were generated by PCR with barcoded primers, purified from 1% agarose gels and pooled at equimolar ratio (primers are listed in Table S1, ESI†). Library preparation from the amplicon pool and sequencing using the Illumina HighSeq platform was performed by Eurofins (Ebersberg, Germany). Processing of raw reads, determination of U-to-C conversion frequency and calculation of labeling efficiency was performed exactly as described,20,24 with the addition that we filtered all reads that had >1 unspecific T conversion (T → A|G).Viability assay
HEK293T cells were labeled for 30 min as described above followed by replacing the labeling medium with culture medium. Cells were further cultivated for 72 h. Viability was measured at 0, 24, 48 and 72 h using the CellTiter-Glo 2.0 Assay (Promega) according to the manufacturer's instructions. Measurements were performed with technical triplicates and three independent experiments were carried out. All values were expressed as fold change over the 0 h timepoint for each labeling reagent. Statistical analysis was performed by one-way ANOVA using Graphpad Prism 9.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Stefan Ameres (University of Vienna) for discussions, Marco Oberlechner (University of Innsbruck) for synthetic contributions, and Daniel Fellner (University of Innsbruck) and Ulrike Schober (University of Innsbruck) for technical support. This work was supported by the Austrian Science Fund FWF (P31691, F8011-B) to R. M.; P33936-B, F8009-B to A. L.), the Austrian Research Promotion Agency FFG [West Austrian BioNMR 858017], and the Wiener Wissenschafts-, Forschungs- und Technologiefonds (WWTF LS17-003).Notes and references
- L. Dölken, Z. Ruzsics, B. Rädle, C. C. Friedel, R. Zimmer, J. Mages, R. Hoffmann, P. Dickinson, T. Forster, P. Ghazal and U. H. Koszinowski, High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay, RNA, 2008, 14, 1959–1972 CrossRef PubMed.
- N. Muthmann, K. Hartstock and A. Rentmeister, Chemo-enzymatic treatment of RNA to facilitate analyses, Wiley Interdiscip. Rev.: RNA, 2019, 11, e1561 Search PubMed.
- A. Messikommer, K. Seipel, S. Byrne, P. J. M. Valk, T. Pabst and N. W. Luedtke, RNA Targeting in Acute Myeloid Leukemia, ACS Pharmacol. Transl. Sci., 2020, 3, 1225–1232 CrossRef CAS PubMed.
- J. Russo, A. M. Heck, J. Wilusz and C. J. Wilusz, Metabolic labeling and recovery of nascent RNA to accurately quantify mRNA stability, Methods, 2017, 120, 39–48 CrossRef CAS PubMed.
- K. Kawata, H. Wakida, T. Yamada, K. Taniue, H. Han, M. Seki, Y. Suzuki and N. Akimitsu, Metabolic labeling of RNA using multiple ribonucleoside analogs enables the simultaneous evaluation of RNA synthesis and degradation rates, Genome Res., 2020, 30, 1481–1491 CrossRef CAS PubMed.
- E. X. Szabo, P. Reichert, M. K. Lehniger, M. Ohmer, M. de Francisco Amorim, U. Gowik, C. Schmitz-Linneweber and S. Laubinger, Metabolic labeling of RNAs uncovers hidden features and dynamics of the arabidopsis transcriptome, Plant Cell, 2020, 32, 871–887 CrossRef CAS PubMed.
- S. Nainar, B. J. Cuthbert, N. M. Lim, W. E. England, K. Ke, K. Sophal, R. Quechol, D. L. Mobley, C. W. Goulding and R. C. Spitale, An optimized chemical-genetic method for cell-specific metabolic labeling of RNA, Nat. Methods, 2020, 17, 311–318 CrossRef CAS PubMed.
- M. Kubota, S. Nainar, S. M. Parker, W. England, F. Furche and R. C. Spitale, Expanding the scope of RNA metabolic labeling with vinyl nucleosides and inverse electron-demand Diels–Alder chemistry, ACS Chem. Biol., 2019, 14, 1698–1707 CrossRef CAS PubMed.
- D. Wang, Y. Zhang and R. E. Kleiner, Cell- and polymerase-selective metabolic labeling of cellular RNA with 2′-azidocytidine, J. Am. Chem. Soc., 2020, 142, 14417–14421 CrossRef CAS PubMed.
- N. Klöcker, F. P. Weissenboeck and A. Rentmeister, Covalent labeling of nucleic acids, Chem. Soc. Rev., 2020, 49, 8749–8773 RSC.
- M. S. Siegrist, B. M. Swarts, D. M. Fox, S. A. Lim and C. R. Bertozzi, Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface, FEMS Microbiol. Rev., 2015, 39, 184–202 CrossRef PubMed.
- Z. He, S. Peng, Q. Wei, S. Jia, S. Guo, K. Chen and X. Zhou, Metabolic labeling and imaging of cellular RNA via bioorthogonal cyclopropene–tetrazine ligation, CCS Chem., 2020, 2, 89–97 CrossRef CAS.
- L. Meng, Y. Guo, Q. Tang, R. Huang, Y. Xie and X. Chen, Metabolic RNA labeling for probing RNA dynamics in bacteria, Nucleic Acids Res., 2020, 48, 12566–12576 CrossRef CAS PubMed.
- S. Kath-Schorr, Cycloadditions for studying nucleic acids, Top. Curr. Chem., 2016, 374, 4 CrossRef PubMed.
- M. Hafner, M. Landthaler, L. Burger, M. Khorshid, J. Hausser, P. Berninger, A. Rothballer, M. Ascano Jr., A. C. Jungkamp, M. Munschauer, A. Ulrich, G. S. Wardle, S. Dewell, M. Zavolan and T. Tuschl, Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP, Cell, 2010, 141, 129–141 CrossRef CAS PubMed.
- J. Spitzer, M. Hafner, M. Landthaler, M. Ascano, T. Farazi, G. Wardle, J. Nusbaum, M. Khorshid, L. Burger, M. Zavolan and T. Tuschl, PAR-CLIP (photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation): A step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins, Methods Enzymol., 2014, 539, 113–361 CAS.
- M. Hafner, M. Katsantoni, T. Köster, J. Marks, J. Mukherjee, D. Staiger, J. Ule and M. Zavolan, CLIP and complementary methods, Nat. Rev. Methods Primers, 2021, 1, 20 CrossRef CAS.
- M. A. P. Baptista and L. Dölken, RNA dynamics revealed by metabolic RNA labeling and biochemical nucleoside conversions, Nat. Methods, 2018, 15, 171–172 CrossRef CAS PubMed.
- M. Singha, L. Spitalny, K. Nguyen, A. Vandewalle and R. C. Spitale, Chemical methods for measuring RNA expression with metabolic labeling, Wiley Interdiscip. Rev.: RNA, 2021, 12, e1650 CAS.
- C. Riml, T. Amort, D. Rieder, C. Gasser, A. Lusser and R. Micura, Osmium-mediated transformation of 4-thiouridine to cytidine as key to study RNA dynamics by sequencing, Angew. Chem., Int. Ed., 2017, 56, 13479–13483 CrossRef CAS PubMed.
- V. A. Herzog, B. Reichholf, T. Neumann, P. Rescheneder, P. Bhat, T. R. Burkard, W. Wlotzka, A. von Haeseler, J. Zuber and S. L. Ameres, Thiol-linked alkylation of RNA to assess expression dynamics, Nat. Methods, 2017, 14, 1198–1204 CrossRef CAS PubMed.
- J. A. Schofield, E. E. Duffy, L. Kiefer, M. C. Sullivan and M. D. Simon, TimeLapse-seq: Adding a temporal dimension to rnA sequencing through nucleoside recoding, Nat. Methods, 2018, 15, 221–225 CrossRef CAS PubMed.
- A. Lusser, C. Gasser, L. Trixl, P. Piatti, I. Delazer, D. Rieder, J. Bashin, C. Riml, T. Amort and R. Micura, Thiouridine-to-cytidine conversion sequencing (TUC-Seq) to measure mRNA transcription and degradation rates, Methods Mol. Biol., 2020, 2062, 191–211 CrossRef CAS PubMed.
- J. D. Young, S. Y. Yao, J. M. Baldwin, C. E. Cass and S. A. Baldwin, The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29, Mol. Aspects Med., 2013, 34, 529–547 CrossRef CAS PubMed.
- F. Hinze, P. Drewe-Boss, M. Milek, U. Ohler, M. Landthaler and M. Gotthardt, M: Expanding the map of protein-RNA interaction sites via cell fusion followed by PAR-CLIP, RNA Biol., 2018, 15, 359368 Search PubMed.
- The Human Protein Atlas, https://www.proteinatlas.org/ (accessed January 2022).
- U. Pradère, E. C. Garnier-Amblard, S. J. Coats, F. Amblard and R. F. Schinazi, Synthesis of nucleoside phosphate and phosphonate prodrugs, Chem. Rev., 2014, 114, 9154–9218 CrossRef PubMed.
- M. Maiti, L. Persoons, G. Andrei, R. Snoeck, J. Balzarini and P. Herdewijn, Synthesis and anti-herpetic activity of phosphoramidate ProTides, ChemMedChem, 2013, 8, 985–993 CrossRef CAS PubMed.
- H. J. Jessen, T. Schulz, J. Balzarini and C. Meier, Bioreversible protection of nucleoside diphosphates, Angew. Chem., Int. Ed., 2008, 47, 8719–8722 CrossRef CAS PubMed.
- C. Zhao, S. Weber, D. Schols, J. Balzarini and C. Meier, Prodrugs of γ-alkyl-modified nucleoside triphosphates: Improved inhibition of HIV reverse transcriptase, Angew. Chem., Int. Ed., 2020, 59, 22063–22071 CrossRef CAS PubMed.
- C. Meier, U. Görbig, C. Müller and J. Balzarini, CycloSal-PMEA and cycloAmb-PMEA: Potentially new phosphonate prodrugs based on the cycloSal-pronucleotide approach, J. Med. Chem., 2005, 48, 8079–8086 CrossRef CAS PubMed.
- C. Gasser, I. Delazer, E. Neuner, K. Pascher, K. Brillet, S. Klotz, L. Trixl, M. Himmelstoß, E. Ennifar, D. Rieder, A. Lusser and R. Micura, Thioguanosine conversion enables mRNA-lifetime evaluation by RNA sequencing using double metabolic labeling (TUC-seq DUAL), Angew. Chem., Int. Ed., 2020, 59, 6881–6886 CrossRef CAS PubMed.
- M. Rabani, J. Z. Levin, L. Fan, X. Adiconis, R. Raychowdhury, M. Garber, A. Gnirke, C. Nusbaum, N. Hacohen, N. Friedman, I. Amit and A. Regev, Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells, Nat. Biotechnol., 2011, 29, 436–442 CrossRef CAS PubMed.
- L. S. Borowski and R. J. Szczesny, Measurement of mitochondrial RNA stability by metabolic labeling of transcripts with 4-thiouridine, Methods Mol. Biol., 2014, 1125, 277–286 CrossRef CAS PubMed.
- A. J. Rutkowski and L. Dölken, High-resolution gene expression profiling of RNA synthesis, processing, and decay by metabolic labeling of newly transcribed RNA using 4-thiouridine, Methods Mol. Biol., 2017, 1507, 129–140 CrossRef CAS PubMed.
- W. Sun and W. Chen, Metabolic labeling of newly synthesized RNA with 4sU to in parallel assess RNA transcription and decay, Methods Mol. Biol., 2018, 1720, 25–34 CrossRef CAS PubMed.
- M. J. Marzi and F. Nicassio, Uncovering the stability of mature miRNAs by 4-thio-uridine metabolic labeling, Methods Mol. Biol., 2018, 1823, 141–152 CrossRef CAS PubMed.
- U. Sharma, F. Sun, C. C. Conine, B. Reichholf, S. Kukreja, V. A. Herzog, S. L. Ameres and O. J. Rando, Small RNAs are trafficked from the epididymis to developing mammalian sperm, Dev. Cell, 2018, 46, 481–494 CrossRef CAS PubMed.
- G.-J. Hendriks, L. A. Jung, A. J. M. Larsson, M. Lidschreiber, O. Andersson Forsman, K. Lidschreiber, P. Cramer and R. Sandberg, NASC-seq monitors RNA synthesis in single cells, Nat. Commun., 2019, 10, 3138 CrossRef PubMed.
- B. Schwalb, M. Michel, B. Zacher, K. Frühauf, C. Demel, A. Tresch, J. Gagneur and P. Cramer, TT-seq maps the human transient transcriptome, Science, 2016, 352, 1225–1228 CrossRef CAS PubMed.
- L. H. Gregersen, R. Mitter and J. Q. Svejstrup, Using TTchem-seq for profiling nascent transcription and measuring transcript elongation, Nat. Protoc., 2020, 15, 604–627 CrossRef CAS PubMed.
- L. W. Peterson and C. E. McKenna, Prodrug approaches to improving the oral absorption of antiviral nucleotide analogues, Expert Opin. Drug. Delivery, 2009, 6, 405–420 CrossRef CAS PubMed.
- M. Slusarczyk, M. Serpi and F. Pertusati, Phosphoramidates and phosphonamidates (ProTides) with antiviral activity, Antivir. Chem. Chemother., 2018, 26, 1–31 Search PubMed.
- A. T. Nies, J. König, U. Hofmann, C. Kölz, M. F. Fromm and M. Schwab, Interaction of remdesivir with clinically relevant hepatic drug uptake transporters, Pharmaceutics, 2021, 13, 369 CrossRef CAS PubMed.
- A. Telbisz, C. Ambrus, O. Mozner, E. Szab, G. Varady, E. Bakos, B. Sarkadi and C. Özvegy-Laczka, Interactions of potential anti-COVID-19 compounds with multispecific ABC and OATP drug transporters, Pharmaceutics, 2021, 13, 81 CrossRef CAS PubMed.
- M. Tera, S. M. K. Glasauer and N. W. Luedtke, In vivo incorporation of azide groups into DNA by using membrane-permeable nucleotide triesters, ChemBioChem, 2018, 19, 1939–1943 CrossRef CAS PubMed.
- S. Peyrottes, C. Périgau, A. M. Aubertin, G. Gosselin and J. L. Imbach, Influence of a Cα-substitution on the S-pivaloyl-2-thioethyl chain on the anti-HIV activity and stability of the resulting zidovudine mononucleoside phosphotriester, Antivir. Chem. Chemother., 2001, 12, 223–232 CrossRef CAS PubMed.
- N. Schlienger, S. Peyrottes, T. Kassem, J. L. Imbach, G. Gosselin, A. M. Aubertin and C. Périgaud, S-Acyl-2-thioethyl aryl phosphotriester derivatives as mononucleotide prodrugs, J. Med. Chem., 2000, 43, 4570–4574 CrossRef CAS PubMed.
- Y. Mehellou, H. S. Rattan and J. Balzarini, The ProTide prodrug technology: From the concept to the clinic, J. Med. Chem., 2018, 61, 2211–2226 CrossRef CAS PubMed.
- C. R. Wagner, V. V. Iyer and E. J. McIntee, Pronucleotides: Toward the in vivo delivery of antiviral and anticancer nucleotides, Med. Res. Rev., 2000, 20, 417–451 CrossRef CAS PubMed.
- J. J. Fox, D. V. Praag, I. Wempen, I. L. Doerr, L. Cheong, J. E. Knoll, M. L. Eidinoff, A. Bendich and G. B. Brown, J. Am. Chem. Soc., 1959, 81, 178–187 CrossRef CAS.
- A. El-Tayeb, A. Qi, R. A. Nicholas and C. E. Mueller, Structural modifications of UMP, UDP, and UTP leading to subtype-selective agonists for P2Y2, P2Y4, and P2Y6 receptors, J. Med. Chem., 2011, 54, 2878–2890 CrossRef CAS PubMed.
- E. J. Nurminen, J. K. Mattinen and H. Lönnberg, Kinetics and mechanism of tetrazole-catalyzed phosphoramidite alcoholysis, J. Chem. Soc., Perkin Trans. 2, 1998, 1621–1628 RSC.
- I. Lefebvre, C. Perigaud, A. Pompon, A.-M. Aubertin, J.-L. Girardet, A. Kirn, G. Gosselin and J.-L. Imbach, Mononucleoside phosphotriester derivatives with S-acyl-2-thioethyl bioreversible phosphate-protecting groups: Intracellular delivery of 3′-azido-2′,3′-dideoxythymidine 5′-monophosphate, J. Med. Chem., 1995, 38, 3941–3950 CrossRef CAS PubMed.
- A. Routledge, I. Walker, S. Freeman, A. Hay and N. Mahmood, Synthesis, bioactivation and anti-HIV activity of 4-acyloxybenzyl bis (nucleosid-5′-yl)phosphates, Nucl. Nucl. Nucl. Acids, 1995, 14, 1545–1558 CAS.
- I. Pavlovic, D. T. Thakor, L. Bigler, M. S. C. Wilson, D. Laha, G. Schaaf, A. Saiardi and H. J. Jessen, Prometabolites of 5-diphospho-myo-inositol pentakisphosphate, Angew. Chem., Int. Ed., 2015, 54, 9622–9626 CrossRef CAS PubMed.
- C. Ducho, U. Görbig, S. Jessel, N. Gisch, J. Balzarini and C. Meier, Bis-cyclosal-d4T-monophosphates: Drugs that deliver two molecules of bioactive nucleotides, J. Med. Chem., 2007, 50, 1335–1346 CrossRef CAS PubMed.
- A. Wissner, M. L. Carroll, K. E. Green, S. S. Kerwar, W. C. Pickett, R. E. Schaub, L. W. Torley, S. Wrenn and C. A. Kohler, Analogs of platelet activating factor. 6. Mono-and bis-aryl phosphate antagonists of platelet activating factor, J. Med. Chem., 1992, 35, 1650–1662 CrossRef CAS PubMed.
- C. Meier, E. De Clercq and J. Balzarini, Nucleotide delivery from cycloSaligenyl-3′-azido-3′-deoxythymidine monophosphates (cycloSal-AZTMP), Eur. J. Org. Chem., 1998, 837–846 CrossRef CAS.
- M. Bergner, D. C. Duquette, L. Chio and B. M. Stoltz, Exceedingly efficient synthesis of (±)-grandifloracin and acylated analogues, Org. Lett., 2015, 17, 3008–3010 CrossRef CAS PubMed.
- I. Sarac and C. Meier, Efficient automated solid-phase synthesis of DNA and RNA 5′-triphosphates, Chem. – Eur. J., 2015, 21, 16421–16426 CrossRef CAS PubMed.
- D. S. Kemp, R. I. Carey, J. C. Dewan, N. G. Galakatos, D. Kerkman and S. L. Leung, Rational design of templates for intramolecular O,N-acyl transfer via medium-sized cyclic intermediates derived from L-cysteine. Definition of an experimental maximum in effective molarity through the study of tunable templates, J. Org. Chem., 1989, 54, 1589–1603 CrossRef CAS.
- M. Wang, L. Zhang, X. Huo, Z. Zhang, Q. Yuan, P. Li, J. Chen, Y. Zou, Z. Wu and W. Zhang, Catalytic asymmetric synthesis of the anti-COVID-19 drug Remdesivir, Angew. Chem., Int. Ed., 2020, 59, 20814–20819 CrossRef CAS PubMed.
- N. Hulsman, J. P. Medema, C. Bos, A. Jongejan, R. Leurs, M. J. Smit, I. J. de Esch, D. Richel and M. Wijtmans, Chemical insights in the concept of hybrid drugs: The antitumor effect of nitric oxide-donating aspirin involves a quinone methide but not nitric oxide nor aspirin, J. Med. Chem., 2007, 50, 2424–2431 CrossRef CAS PubMed.
- C. Courtens, M. Risseeuw, G. Caljon, P. Cos and S. Van Calenbergh, Acyloxybenzyl and alkoxyalkyl prodrugs of a fosmidomycin surrogate as antimalarial and antitubercular agents, ACS Med. Chem. Lett., 2018, 9, 986–989 CrossRef CAS PubMed.
- C. Meier, M. Lorey, E. De Clercq and J. Balzarini, Cyclo-saligenyl-2′,3′-dideoxy-2′,3′-didehydroythymidinemonophosphate (CycloSal-d4TMP) — A new pro-nucleotide approach, Nucleosides Nucleotides, 1997, 16, 1303–1306 CrossRef CAS.
- C. Meier and J. Balzarini, Application of the cycloSal-prodrug approach for improving the biological potential of phosphorylated biomolecules, Antiviral Res., 2006, 71, 282–292 CrossRef CAS PubMed.
- S. Peyrottes, N. Schlienger, T. Beltran, I. Lefebvre, A. Pompon, G. Gosselin, A.-M. Aubertin, J.-L. Imbach and C. Périgaud, Design of new mononucleotide prodrugs: Aryl (SATE) phosphotriester derivatives, Nucl. Nucl. Nucl. Acids, 2001, 20, 315–321 CrossRef CAS PubMed.
- E. E. Duffy, D. Canzio, T. Maniatis and M. D. Simon, Solid phase chemistry to covalently and reversibly capture thiolated RNA, Nucleic Acids Res., 2018, 46, 6996–7005 CrossRef CAS PubMed.
- S. C. Blanchard, R. L. Gonzalez, H. D. Kim, S. Chu and J. D. Puglisi, tRNA selection and kinetic proofreading in translation, Nat. Struct. Mol. Biol., 2004, 11, 1008–1014 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2cb00001f |
| This journal is © The Royal Society of Chemistry 2022 |