Open Access Article
Open Access ArticleConstruction and evaluation of detachable bone-targeting MOF carriers for the delivery of proteasome inhibitors†
Hongbing Yang‡
abc,
Zhenyan Yu‡a,
Shuaishuai Jia,
Jie Yana,
Lei Hanc,
Yang Liu f,
Yanjuan Wangd,
Yimin Niu*bd,
Qiang Huo*a and
Ming Xu*ce
f,
Yanjuan Wangd,
Yimin Niu*bd,
Qiang Huo*a and
Ming Xu*ce
aSchool of Pharmacy, Bengbu Medical College, Bengbu 233030, Anhui, China. E-mail: huoqiang@bbmc.edu.cn
bDepartment of Pharmacy, Zhongda Hospital, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China. E-mail: yiminniu@seu.edu.cn
cDepartment of Occupational Disease Prevention, Jiangsu Provincial Center for Disease Control and Prevention, Nanjing 210009, Jiangsu, China. E-mail: sosolou@jscdc.cn
dDepartment of Neurology, Zhongda Hospital, School of Medicine, Southeast University, Nanjing 210009, Jiangsu, China
eSchool of Public Health, Nanjing Medical University, Nanjing 211166, Jiangsu, China
fSchool of Pharmacy, Nanjing Medical University, Nanjing 211166, Jiangsu, China
First published on 16th May 2022
Abstract
Tumor bone metastasis is an important cause of tumor recurrence and death. Although bone-targeting nanoparticles decorated with targeting ligands have shown good affinity for bone tissues with the properties of adhesion to the bone matrix, it is not easy to detach from the surface of the bone matrix in the tumor-bone microenvironment, attributed to the robust coordination force between the targeting ligands, such as bisphosphates with bone-deposited calcium. This may hinder the transport of nanoparticles from bone tissue to bone metastatic tumors. In this research, we designed a bone-targeting nanocarrier with detachable bone-targeting character for the therapy of bone metastases. The nanoparticles were constructed by using ZIF-8 and bone-targeting and MMP enzyme sensitive polypeptide-modified hyaluronic acid as a carrier and proteasome inhibitor Bortezomib (BTZ) as cargo. The results show that the constructed D8-M3-HA-ZIF8@BTZ nanoparticles possessed several favorable properties such as good colloidal stability, acid-sensitive drug release, D8 peptide mediated bone targeting and MMP enzyme-responsive desorption. Besides, nanoparticle endocytosis and cytotoxicity were enhanced through HA-mediated targeting to CD44 over-expressing tumor cells. Altogether, this study provides a potential cascade targeting strategy for improving the delivery effects of bone targeted nanoparticles for the delivery of proteasome inhibitors.
1. Introduction
Tumor metastasis is one of the main factors in the death of tumor patients.1 The metastasis sites mainly include bone, lung, liver and other tissues.2 Among them, due to the special structure and physiological microenvironment of bone tissue, it is a very suitable microenvironment for the dormancy and resurrection of disseminated tumor cells (DTCs).3,4 Hence, bone metastases have been one of the most common metastatic sites of breast cancer, lung cancer, prostate cancer and other malignant tumors.5 Many advantages have been shown by using bone-targeting carriers to deliver drugs to bone metastases.6 For example, nanocarriers with bone-targeting ligands such as bisphosphates,7 acidic oligopeptides,8 tetracyclines9 and macrocyclic anthraquinones were reported by different groups.Although bone-targeting nanoparticles (NPs) have good affinity for bone tissue,10 unfortunately, it is difficult for NPs to be detached from the surface of bone matrix after accumulated in the malignant niches owing to that the bone targeted ligands such as bisphosphates,11 etc. are closely combined with inorganic calcium salt inside bone matrix, and thus it hinders the transport of NPs from bone to bone-metastatic tumor niches.12 This in turn result in the deficiency of sufficient concentration of anti-tumor drugs to inhibit tumor cell growth. In recent years, although there are some reports about stimulus-responsive drug release carriers to increase local drug concentration for bone metastases,13 there are few studies to address the issue of the dilemma between bone targeting and subsequent bone desorption of NPs.
Bortezomib (BTZ) is the first proteasome inhibitor approved by FDA.14 It can specifically inhibit the activity of proteasome 26S subunit and significantly reduce the degradation of NF-kB inhibitory protein (IKB).15 IKB can inhibit the activity of nuclear factor kB (NF-kB), so as to selectively inhibit the expression of growth-related genes and eventually lead to tumor cell apoptosis. BTZ is generally used in the treatment of multiple myeloma and mantle cell lymphoma clinically.16,17 Although BTZ shows robust inhibitory effects on multiple tumor cells,18 its clinical applications were hindered as some characteristics such as low water-solubility, instability and tumor targeting insufficient.19 Besides, BTZ may cause some side effects such as hypotension, reversible posterior leukoencephalopathy syndrome (RPLS), peripheral neuropathy, etc. with a certain probability.20,21 In brief, these issues have greatly limited the clinical outcomes of BTZ.
Metal organic frameworks (MOFs) materials are novel functional materials formed by the coordination of metal ions with organic ligands.22 Due to their large porosity, specific surface area, they have huge potential in the fields of drug delivery.23,24 Zeolite imidazole skeleton-8 (ZIF-8) is one of its subtypes with displayed good biocompatibility and constructed with transition metal divalent zinc ions (Zn2+) and imidazole as organic ligand.25,26 However, ZIF-8 NPs are incapability of tumor active targeting and easy to aggregate in aqueous solution.27
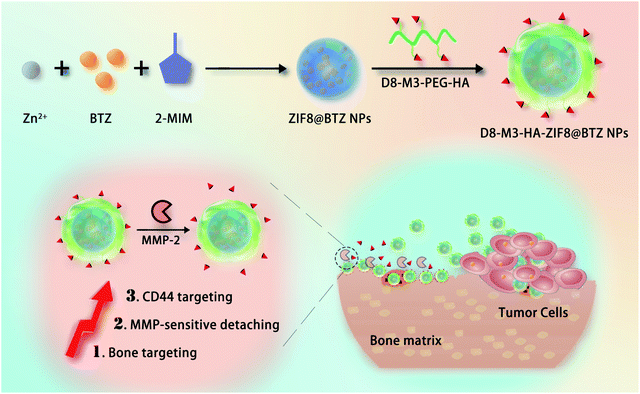
Based on the above analysis, to solve the problem of bone-desorption of bone-targeting NPs and improve the therapeutic effects of BTZ, herein, we designed detachable bone-targeting ZIF-8 carriers to delivery BTZ for the treatment of tumor bone metastases. As depicted in Scheme 1, the vectors were constructed with matrix metalloprotein (MMP) enzyme-sensitive bone-targeting polypeptide ligands, CD44-targeted hyaluronic acid and BTZ-loaded ZIF-8 NPs. The results shown that the constructed D8-M3-HA-ZIF8@BTZ NPs had D8 peptide mediated bone targeting, MMP enzyme-responsive desorption and HA-mediated targeting to CD44 over-expressed tumor cells.28 Our study provides a valuable reference in cascade targeting strategy to delivery of proteasome inhibitors.
2. Materials and methods
2.1 Materials
2-Methylimidazole (2-MIM, M50850), zinc nitrate hexahydrate (J1819123), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). MMP-2 enzyme (MM2-H5225) was purchased from ACRO Biosystems (Beijing, China). Bortezomib was purchased from Macklin (Shanghai, China). Maleimide polyethylene glycol amino (MAL-PEG2000-NH2, Mw = 2 kD, 95%) was obtained from Ponsure Biotechnology (Shanghai, China). Hyaluronic acid (HA, Mw = 90 kD, 95%) and Ham's F-12 medium (with L-glutamine) was purchased from Meilun Biotechnology (Dalian, China). Cell counting kit-8 (CCK-8), Hoechst33342, Lyso-tracker red probe and BCA protein assay kits were obtained from Beyotime (Shanghai, China). Peptides containing MMP enzyme cleavage sites and control peptides were custom synthesized by Bankpeptide biological technology (Hefei, China). FITC was purchased from Fushen Biological Technology (Shanghai, China). Cy5.5 NHS ester was purchased from Aladdin (Shanghai, China). The high crystalline hydroxyapatite tablets were obtained from Huaqiao Technology (Suzhou, China). The synthetic polypeptide sequence is shown in Table 1.| Particles | Sizea (nm) | PDI | Zeta potential (mV) | EEb (%) | DLC (%) |
|---|---|---|---|---|---|
| a The particle size and zeta potential were investigated using a Mastersizer particle size analyzer; PDI: polydispersity index.b Drug loading content was measured by ultraviolet spectrophotometer. EE: entrapment efficiency; DLC: drug loading contents. | |||||
| ZIF8 | 107.9 ± 5.4 | 0.105 ± 0.034 | 13.5 ± 1.1 | — | — |
| ZIF8@BTZ | 124.5 ± 7.2 | 0.282 ± 0.027 | 21.3 ± 0.9 | 61.1 ± 0.97 | 4.66 ± 0.33 |
| HA-ZIF8@BTZ | 157.9 + 7.9 | 0.086 + 0.031 | −22.1 ± 1.1 | — | 4.12 ± 0.42 |
| D8-M3-HA-ZIF8@BTZ | 164.9 ± 9.2 | 0.150 ± 0.025 | −24.3 ± 0.9 | — | 4.06 ± 0.28 |
2.2 Cell lines and animals
The human non-small-cell lung carcinoma (NSCLC) A549 cell line was purchased from Cell Resource Center of Institute of Biological Sciences, Chinese Academy of Sciences (Shanghai, China) and cultured in Ham's F-12 medium (with L-glutamine) supplemented with 10% FBS and 1% double antibiotics. Male SD rats were obtained from Cavens Laboratory Animal (Changzhou, China) and the relative animal experiments were in accordance with the guidelines for care and use of laboratory animals of Bengbu Medical College (Bengbu, China), and approved by the Institutional Animal Care and Use Committee (IACUC) of Bengbu Medical College.2.3 Screening of MMP-2 enzyme sensitive peptides
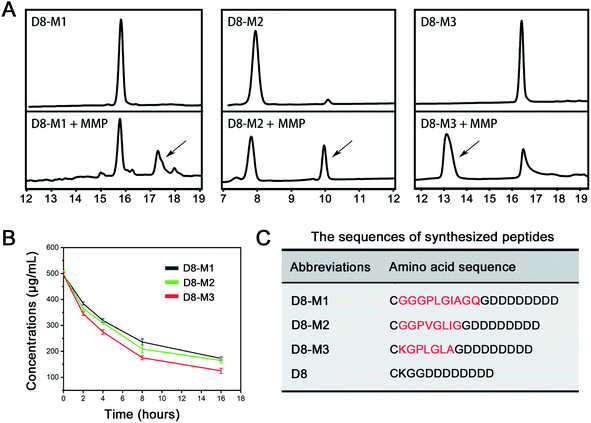
1 mg of D8-M1, D8-M2 and D8-M3 peptides were well dissolved in 1 mL of PBS (pH 7.4, 0.1 M), respectively. Then MMP-2 solution (final concentration was 2.4 μg mL−1) were added into the peptide solutions. The mixtures were put into a constant temperature shake at 37 °C. At a certain point in time (0, 2, 4, 8, 16 hours), the samples were taken out, followed with an equal volume of methanol was added immediately to inactivate the enzyme. The peptides solutions were determined with HPLC using UV detector at 214 nm.2.4 Copolymer synthesis
The synthesis of D8-Pep-PEG-HA was completed in two steps, and the synthesis route is shown in Fig. 1. Firstly, 100 mg of HA and 12.5 mg of HOBt were dissolved in 20 mL of MES buffer (pH 6.5, 0.1 M) and 2 mL of methanol, respectively. Then the HOBt solutions was added into the HA solution under vigorous stirring, followed with be added into 19.25 mg of EDC·HCl and 21.75 mg of sulfo-NHS. After stirring for another 3.5 hours, 40 mg of MAL-PEG2000-NH2 dissolved in 5 mL of MES buffer (pH 6.5, 0.1 M) were slowly added into the mixture and the reaction was continued for 24 hours at room temperature. Next, the products were purified with ultrafiltration tube (Millipore, MWCO: 5 kDa) and washed with MES buffer (pH 6.5, 0.1 M) repeatedly. The solids (MAL-PEG-HA) were obtained with freeze-drying (yield, 88.4%). Subsequently, 102 mg of MAL-PEG-HA was dissolved in 20 mL of MES buffer (pH 6.5, 0.1 M) and stirred for 2 hours. Then, 1 mg of peptides of different formulas were dissolved in 4 mL of MES buffer and slowly added into the above solution. The mixture was allowed to be stirred for 24 hours at room temperature and then were purified with an ultrafiltration tube (MWCO: 5 kDa). The product was obtained by freeze-drying (yield, 90.7–92.3%). The products were characterized by 1H-NMR and FT-IR. As a control, D8-PEG-HA was synthesized with similar approaches. The coupling efficiency (CE) of D8-M3 or D8 peptide was detected by BCA method, and the calculation formula was CE (%) = the amounts of coupled peptide to the polymers/the amounts of added peptides × 100%.2.5 Preparation of D8-M3-HA-ZIF8@BTZ NPs
2 mg of BTZ was dissolved in 1 mL ultrapure water and mixed with 0.972 g of 2-MIM dissolved in 4.5 mL ultrapure water at temperature. The mixture was allowed to be stirred for 2 min and then added rapidly with 0.5 mL of Zn (NO3)2 solution (50 mg mL−1). After be stirred with another 3 min, the products were obtained with centrifugation and washed with ethanol and ultrapure water for 3 times. Subsequently, the solids were dispersed in 5 mL of ultrapure water and added slowly into 5 mL of D8-M3-HA solutions (5 mg mL−1) under vigorous stirring. The mixtures were stirred for 1 hour, followed with the solids were collected with centrifugation and washed with ultrapure water repeatedly. The products (D8-M3-HA-ZIF8@BTZ) were dispersed in 1 mL of ultrapure water for further use.2.6 Characterization of NPs
The hydrodynamic particle size, zeta potential and polydispersity index (PDI) of ZIF8@BTZ, HA-ZIF8@BTZ and D8-M3-HA-ZIF8@BTZ NPs were determined with a laser particle size analyzer (DLS, Zetasizer ZS90, Malvern, UK). The morphology features of ZIF8@BTZ, HA-ZIF8@BTZ and D8-M3-HA-ZIF8@BTZ NPs were observed via a transmission electron microscope (TEM, JEM-1400plus, JEOL, Japan).2.7 Determination of drug loading rate and in vitro release from NPs
5.4 mg of ZIF8@BTZ NPs, HA-ZIF8@BTZ NPs or D8-M3-HA-ZIF8@BTZ NPs were destroyed by 1 mL of HCl (0.1 M) to release all the drugs, then 8.5 mL of methanol was added to dissolve BTZ. The solution was precisely diluted to 100 mL with methanol and then the absorbance was measured at 270 nm via an ultraviolet spectrophotometer. The amount of BTZ was calculate according to the calibration curve (Fig. S4†). After that, the drug loading content (DLC) of BTZ in the NPs was calculated according to a formulate of DLC (%) = the amounts of loaded BTZ/the total amounts of NPs × 100%. Besides, the entrapment efficiency (EE) of BTZ was calculated based on a formulate of DLC (%) = the amounts of loaded BTZ/the total amounts of BTZ × 100%.To evaluate the acidic pH-triggered drug release from BTZ-loaded NPs, 9 mg of HA-ZIF8@BTZ or D8-M3-HA-ZIF8@BTZ NPs were dispersed in 15 mL of PBS solution and putted into a dialysis bag (MWCO: 3500 Da). The dialysis bags were immersed in 300 mL PBS buffer (0.01 M, pH 5.0 or pH 7.4, respectively) under gentle shaking at 37 °C. Then, at 0, 0.5, 1, 1.5, 2, 2.5, 3, 3.5, 4, 4.5, 5, 6, 8, 10, 12, 14, 16, 18 and 24 hours, 4 mL of dialysis solution was collected for analysis, followed with equivalent amounts of fresh PBS were replenished to keep at a constant volume of dialysis solution. The absorption at 270 nm of the collected solution was determined by a UV-Vis spectrometer, and the amounts of cumulative released BTZ were calculated according the calibration curve (Fig. S4†).
2.8 Study on the stability and hemolysis of NPs
To detect the stability of NPs, D8-M3-HA-ZIF8@BTZ NPs were dissipated in PBS (0.01 M, pH 7.4) or PBS containing 10% FBS and incubate at 37 °C. At regular time intervals, a certain amounts of NPs solution were taken out and the DLS sizes were assayed with nano-laser particle detector.To evaluate the hemolysis of MOFs NPs, fresh blood was taken from the orbit of SD rats for hemolysis test. Specifically, the fresh blood collected from healthy SD rats were anticoagulated with heparin sodium and then the purified red blood corpuscles (RBCs) were isolated by centrifugation at 4 °C. After that, the RBCs were dissipated in 2 mL of PBS (pH 7.4, 0.01 M) and blended carefully with different concentrations of D8-M3-HA-ZIF8@BTZ NPs solutions at equal volume. The concentrations of BTZ in the D8-M3-HA-ZIF8@BTZ NPs solutions were 20, 40, 80, 100 and 200 μg mL−1, respectively. The mixtures were incubated at 37 °C for 2 hours, followed with centrifugated at 5000 rpm for 5 min to removing the lysed RBCs. 100 μL of the supernatants from different samples were placed in a 96 well plate and the absorbance were recorded via a microplate reader at 570 nm. Meanwhile, ultrapure water or isosmotic PBS solution (pH 7.4, 0.01 M) treated RBCs were set as positive and negative controls, respectively. The relative hemolytic rate (RHR) was counted based on a formulate of RHR (%) = (Asample − APBS)/(Awater − APBS) × 100%.
2.9 Bone-binding assay in vitro
Bioceramic tablets constructed with high crystallinity hydroxyapatite (diameter 10 mm, height 2.5 mm) were placed in 48 well plates and incubated with D8-HA-ZIF8@FITC or D8-M3-HA-ZIF8@FITC solutions for 24 hours. After that, the tablets were taken out from the plate and rinsed carefully with PBS (pH 7.4, 0.01 M) for 10 min to remove the nonspecifically adsorbed nanoparticles, followed with the fluorescence intensity were observed with fluorescence imager. To detect the detachable bone-targeting performance, the NPs binding tablets were incubated with 2 mL of MMP enzyme solution (20 μg mL−1) and gently shaken at 37 °C for 72 hours. After be rinsed with PBS for 10 min, the fluorescence intensity of tablets was detected with inverted fluorescence microscope (Zeiss, Axio Observer Z1, Germany).2.10 NPs uptake by A549 cells
The Cy5.5-fluorescent labeled NPs (ZIF8@Cy5.5, HA-ZIF8@Cy5.5 and D8-HA-ZIF8@Cy5.5) were used to evaluate their cellular uptake behaviors. A549 cells were seeded in confocal dishes at densities of 5 × 103 cells per well and cultured for 12 hours. Then, the culture mediums were replaced with FBS-free mediums containing Cy5.5-labeled NPs and incubated for 4 hours. After be washed with PBS for three times, the cells were fixed with 4% paraformaldehyde at room temperature for 15 min and stained with hoechst33342 for 5 min. Finally, the cells in confocal dish were observed and imaging with a confocal laser microscope (CLSM, FV-1200MPE, Olympus, Japan). Besides, to free HA inhibition experiments, the A549 cells were incubated with free HA dissolved in culture medium for 2 hours prior the D8-HA-ZIF8@Cy5.5 NPs were added.2.11 Time dependent intracellular transport of NPs
A549 cells were inoculated in confocal dishes at a density of 5 × 103 cells per well and cultured overnight. Then, D8-HA-ZIF8@FITC NPs were added into the dishes and co-incubated for 0.5, 1 and 3 hours, respectively. The cells were stained with lysotracker red for 1 h and fixed with 4% paraformaldehyde at room temperature for 15 min, followed with stained with hoechst33258 for 5 min. Finally, the cells were observed by confocal microscope.2.12 Cytotoxicity
The cytotoxicity of ZIF8@BTZ, HA-ZIF8@BTZ, D8-M3-HA-ZIF8@BTZ and free BTZ on A549 cells were examined using CCK-8 assay. A549 cells were seeded into 96 well plates at densities of 5 × 103 cells per well and cultured for another 24 hours. Subsequently, the medium was replaced with 100 μL of test substances in F12 culture medium, and the cells were incubated for another 48 h in cell incubator. The concentrations of BTZ in different formulas were ranged from 0–100 ng mL−1 (n = 6). Afterwards, 10 μL of CCK-8 solution was added into each well for additional 2 hours incubation in cell incubator. The absorbance values at 450 nm of each well were measured with microplate spectrophotometer (Synergy HT, BioTek, Germany). The cell viability (CV) was calculated using the following formula: CV (%) = [(Asample − Ablank)/(Acontrol − Ablank)] × 100%, where Asample, Acontrol and Ablank represent the OD450 values of sample group, drug-free medium group and blank well, respectively. The experiments were performed in triplicate.3. Results and discussions
3.1 Preparation and characterization of D8-M3-HA-ZIF8@BTZ NPs
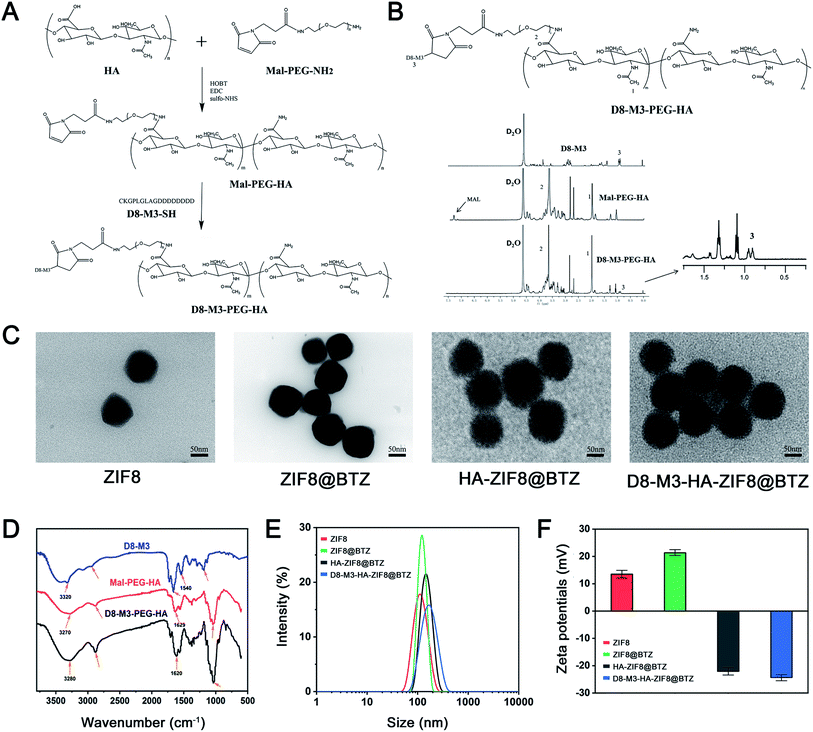
In order to screen vectors with higher MMP enzyme responsive ability, we firstly custom synthesized three different MMP enzyme responsive linker peptides (D8-M1, D8-M2 and D8-M3, Table 1) and investigated their MMP enzyme degradable properties in vitro. As shown in Fig. 1A and B, HPLC assay confirmed that D8-M3 had most conspicuous MMP enzyme sensitivity among three different peptides. Therefore, D8-M3 peptide was selected to synthesis the MMP enzyme responsive polymers.Subsequently, we synthesized D8-M3-PEG-HA polymers by two steps, that is, MAL-PEG-HA was synthesized firstly, and then it was coupled with targeting peptide ligand D8-M3 to get D8-M3-PEG-HA (Fig. 2A). The successful synthesis of the polymers was confirmed by 1H-NMR and FT-IR (Fig. 2B and D). As shown in Fig. 2B, the peak at the chemical shift of 0.95 ppm is the proton characteristic peak of D8-M3, while the peak at 7.02 ppm of MAL groups was disappeared in the spectra of D8-M3-PEG-HA, indicating that the synthesis of D8-M3-PEG-HA was successful. In addition, we synthesized D8-PEG-HA polymers without enzyme digestion site as control (Fig. S1 and S2†). The coupling efficiency of D8-M3 and D8 peptide to MAL-PEG-HA polymers detected by BCA method were 92.1% and 91.8%, respectively. In addition, we furtherly identified the products synthesized by FT-IR. In the FT-IR spectrum of D8-M3-PEG-HA, it can be seen that characteristic peaks appear at 2870 cm−1, 1629 cm−1 and 1030 cm−1, which indicates the existence of D8-M3 groups (Fig. 2D and S3†). In conclusion, these results confirmed the successful synthesis of D8-M3-PEG-HA and D8-PEG-HA polymers.
Next, referring to the literature, we synthesized BTZ-loaded ZIF-8 NPs through one pot method, and then D8-M3-HA molecules was modified on the surface of the ZIF8@BTZ NPs. As shown in TEM images (Fig. 2C), the prepared NPs showed hexagonal like, polygonal like or irregular spherical shapes. Their average particle sizes observed by TEM were approximately 70 nm, 75 nm, 95 nm and 100 nm for ZIF-8, ZIF8@BTZ, HA-ZIF8@BTZ, D8-M3-HA-ZIF8@BTZ, respectively. The particle size of ZIF8@BTZ NPs is larger than that of ZIF8 NPs may be attributed to that the encapsulated BTZ increased the particle size of NPs. Besides, HA were wrapped on the surface of ZIF-8 NPs through electrostatic and coordination forces. From the TEM image, it can be seen that HA wrapping caused slight increase in the particle size. More importantly, the morphology of NPs was changed as film-like substances appeared in the outer layer of HA-ZIF8@BTZ NPs. Furthermore, through DLS analysis, the hydrodynamic particle sizes of ZIF8, ZIF8@BTZ, HA-ZIF8@BTZ, D8-M3-HA-ZIF8@BTZ NPs were 107.9 ± 5.4 nm, 124.5 ± 7.2 nm, 157.9 ± 7.9 nm and 164.9 ± 9.2 nm, respectively (n = 3, Fig. 2E and Table 1). The zeta potentials of ZIF8, ZIF8@BTZ, HA-ZIF8@BTZ, D8-M3-HA-ZIF8@BTZ NPs were 13.5 ± 1.1 mV, 21.3 ± 0.9 mV, −22.1 ± 1.1 mV and −24.3 ± 0.9 mV, respectively (n = 3, Fig. 2F and Table 1). These results exhibited that the particle size of NPs changed from positive to negative along with the encapsulation of HA, which also proved that the surface modification of NPs with HA molecules was successful.
ZIF8 NPs are unstable under acidic conditions as acidic conditions can protonate the 2-MIM organic ligands and result into the breaking of the Zn2+-imidazolium ion coordination bond.29 By decomposing ZIF8 NPs in acid solutions, we detected the drug loading and entrapment efficiency of different BTZ-loaded NPs. As shown in Table 1, the entrapment efficiency of the ZIF8@BTZ nanoparticles was as high as over 60%, and the drug loading capacity of the three nanoparticles was calculated to be over 4%, indicating that the prepared NPs can encapsulate BTZ well.
3.2 Stability and drug release properties of nanoparticles
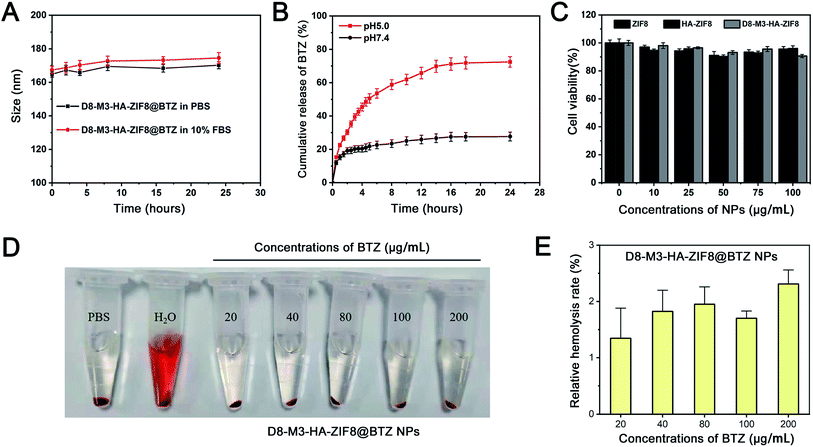
The stability and drug release properties of NPs have an important impact on drug delivery in vivo. As shown in Fig. 3A and B, after be incubated in PBS and PBS containing 10% fetal bovine serum, D8-M3-PEG-HA@BTZ NPs maintained well stability and dispersibility within 24 hours, and the hydrodynamic particle size increased slightly, indicating that the HA-modified NPs have good stability. In addition, as shown in the drug release curves, it can be seen that BTZ was released rapidly from D8-M3-HA-ZIF8@BTZ NPs under acid conditions in the first 12 hours with 72.45 ± 3.15% of the drugs were released from the carrier within 24 hours at pH5.0, while only 27.69 ± 2.65% of the drugs were released from the carrier in 24 hours at pH 7.4 (n = 3), indicating that the drug release of NPs was slow under neutral conditions, while rapidly under acidic conditions. This can be explained that the discomposing of ZIF-8 results in the burst release of cargoes. In short, D8-M3-HA-ZIF8@BTZ NPs have good stability and acid sensitive drug release, which are very conducive for in vivo application and targeted delivery of the loaded drugs.3.3 Uptaking and in vitro antitumor activity of D8-M3-HA-ZIF8@BTZ NPs
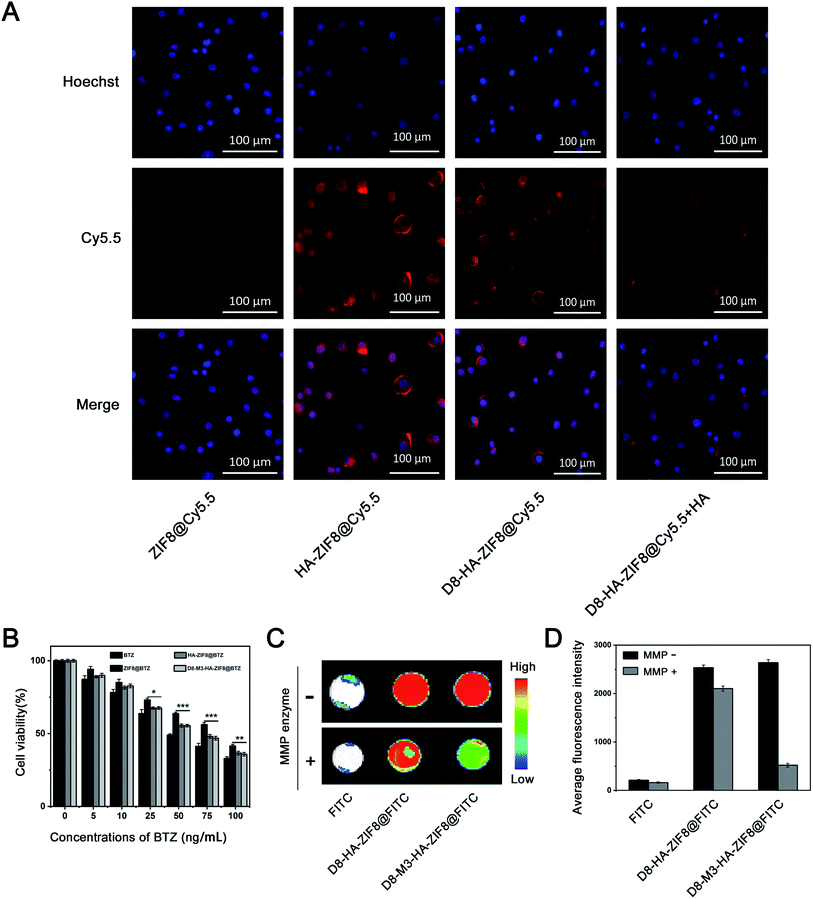
In order to study the biocompatibility of ZIF8 NPs in vitro, drug-free ZIF8, HA-ZIF8, D8-M3-HA-ZIF8 were incubated with A549 cells for 48 hours to evaluate their cytotoxicity in vitro by CCK-8 assay. As shown in the Fig. 3C, A549 cells still maintained a survival rate of more than 90% as the concentration of NPs ranged from 0–100 μg mL−1. Specifically, when the concentrations of ZIF8, HA-ZIF8, D8-M3-HA-ZIF8 were as high as 100 μg mL−1, the survival rates of A549 cells were 95.62 ± 1.79%, 96.01 ± 1.91% and 90.63 ± 1.09% (n = 3), respectively, indicating that ZIF-8 and HA has fine cytocompatibility. In addition, we also investigated the hemolysis of D8-M3-HA-ZIF8@BTZ NPs. The results shown that the hemolysis remained less than 3% although at a higher-concentration of D8-M3-HA-ZIF8@BTZ NPs, indicating fine biosafety at the treatment doses with respect to hemocompatibility.Next, the cellular uptake behavior of NPs was detected by fluorescence labeling method. We used Cy5.5 fluorescein instead of BTZ to prepare various fluorescent labeled NPs. As depicted in Fig. 4A, the uptake of ZIF8@Cy5.5 labeled with red fluorescence by A549 cells was relative weak, while the intercellular HA-ZIF8@Cy5.5 was increased, indicating that HA modification can promote the uptake of NPs. Besides, the uptake of D8-M3-HA-ZIF8@Cy5.5 was slightly affected by polypeptide modification compared with that of HA-ZIF8@Cy5.5, which maybe caused by the steric hindrance of PEG-D8 on the surface of NPs. However, when the CD44 receptor on the surface of A549 cells was saturated by pretreated free HA, the uptake of D8-M3-HA-ZIF8@Cy5.5 NPs was significantly reduced, which further proved the cellular uptake of HA-modified NPs were aided by CD44 protein mediated endocytosis pathway. In addition, we conducted time-dependent uptaking experiment and found that less green fluorescence was distributed in A549 cells at 0.5 hour. With the extension of incubation time, more nanoparticles entered into the cells. More importantly, a lot of green fluorescence was gradually separated from lysotracker red-labeled lysosomes, indicating that green fluorescein molecular was released into the cytoplasm (Fig. S7†).
Furthermore, the toxicity of free BTZ and different types of BTZ-loaded NPs to A549 cells were evaluated by CCK-8 assay. The results shown that the cytotoxic effects of each administration group on A549 cells were obviously in concentration dependent fashion within a certain range (Fig. 4B). For example, when the concentration of BTZ was in 0–10 ng mL−1, there was no significant difference in cell survival among the different groups. However, D8-M3-HA-ZIF8@BTZ and HA-ZIF8@BTZ exhibited stronger cytotoxicity compared with that of ZIF8@BTZ when the concentration of BTZ was in 25–100 ng mL−1. The IC50 value of BTZ, ZIF8@BTZ, HA-ZIF8@BTZ, D8-M3-HA-ZIF8@BTZ were 47.27 ng mL−1, 87.74 ng mL−1, 60.78 ng mL−1 and 59.73 ng mL−1 (n = 6), respectively, indicating that although the inhibitory effect of free BTZ on A549 cells was the strongest, the HA-modification of HA-ZIF8@BTZ and D8-M3-HA-ZIF8@BTZ NPs can enhance the cytotoxicity to A549 cells compared with that of unmodified ZIF-8@BTZ NPs.
3.4 Drug release of D8-M3-HA-ZIF8@BTZ NPs and the detachable bone targeting ability
Finally, we studied the bone-affinity and MMP enzyme-sensitive desorption properties of D8-M3-HA-ZIF8@BTZ NPs. Crystalline hydroxyapatite is one of the main components of bone matrix. Bioceramic tablets made of crystalline hydroxyapatite are widely used orthopaedical biomaterials, which can be used to evaluate the targeted binding ability of NPs to bone matrix. As shown in Fig. 4C and D, fluorescein-labeled D8-HA-ZIF8@FITC and D8-M3-HA-ZIF8@FITC NPs were well bound with bioceramic tablets after incubation for 24 hours, while free FITC exhibited negligible nonspecific binding to bioceramic tablets. Interestingly, after treated with MMP enzyme for 72 hours, the fluorescence intensity of D8-HA-ZIF8@FITC groups did not change significantly, while that of D8-M3-HA-ZIF8@FITC group decreased significantly, indicating that MMP enzyme cut off the enzyme sensitive peptide and resulted in the detachment of abundant D8-M3-HA-ZIF8@FITC NPs. In conclusion, these results confirmed that D8-M3-HA-ZIF8@FITC NPs constructed possessed well MMP enzyme sensitivity and detachable bone-targeting character, thus have great potential to be a bone-targeted vector for bone metastasis therapy.4. Conclusions
Here, we described a D8-M3 peptides conjugated, HA-encapsulated ZIF-8 NPs with protein inhibitors BTZ as cargoes, which has the dual properties of MMP enzyme sensitivity and bone targeting. The constructed D8-M3-HA-ZIF8@BTZ NPs had good colloidal stability and acid-sensitive drug release properties. More importantly, NPs endocytosis and cytotoxicity were enhanced through HA-mediated targeting to CD44 receptor positive tumor cells. Therefore, this cascade targeting strategy, that is the integration of D8 peptide mediated bone targeting, MMP enzyme sensitive desorption and CD44 protein mediated tumor cell targeting, could be a promising route to deliver protein inhibitors, which have the potential in the treatment of bone metastases.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (81901863, 81703201, 81502796, 81801075), the Major Project of Natural Science Research for Colleges and Universities of Anhui Province, China (KJ2020ZD52), the Natural Science Foundation of Jiangsu Province (BK20201485, BK20180379), Jiangsu Provincial 333 Project (BRA2018398), the Lifting Program of Jiangsu Provincial Scientific and Technological Association.References
- N. Maishi and K. Hida, Cancer Sci., 2017, 108, 1921–1926 CrossRef CAS.
- G. M. Sharif and A. Wellstein, Future Oncol., 2015, 11, 3253–3260 CrossRef CAS.
- V. Mittal, T. El Rayes, N. Narula, T. E. McGraw, N. K. Altorki and M. H. Barcellos-Hoff, Adv. Exp. Med. Biol., 2016, 890, 75–110 CrossRef.
- A. S. Gdowski, A. Ranjan and J. K. Vishwanatha, J. Exp. Clin. Cancer Res., 2017, 36, 108 CrossRef PubMed.
- J. Fornetti, A. L. Welm and S. A. Stewart, J. Bone Miner. Res., 2018, 33, 2099–2113 CrossRef CAS.
- R. N. Young and M. D. Grynpas, Curr. Opin. Pharmacol., 2018, 40, 87–94 CrossRef CAS.
- Y. Cong, C. Quan, M. Liu, J. Liu, G. Huang, G. Tong, Y. Yin, C. Zhang and Q. Jiang, J. Biomater. Sci., Polym. Ed., 2015, 26, 629–643 CrossRef CAS PubMed.
- B. Jiang, J. Cao, J. Zhao, D. He, J. Pan, Y. Li and L. Guo, Drug Delivery, 2012, 19, 317–326 CrossRef CAS.
- H. Wang, J. Liu, S. Tao, G. Chai, J. Wang, F. Q. Hu and H. Yuan, Int. J. Nanomed., 2015, 10, 5671–5685 CAS.
- M. Korani, S. Korani, E. Zendehdel, M. R. Jaafari, T. Sathyapalan and A. Sahebkar, Anti-Cancer Agents Med. Chem., 2020, 20, 643–650 CrossRef CAS PubMed.
- L. Xing, F. H. Ebetino, R. K. Boeckman Jr, V. Srinivasan, J. Tao, T. K. Sawyer, J. Li, Z. Yao and B. F. Boyce, Bone, 2020, 138, 115492 CrossRef CAS PubMed.
- N. Ochiai, Y. Nakachi, T. Yokoo, T. Ichihara, T. Eriksson, Y. Yonemoto, T. Kato, H. Ogata, N. Fujimoto, Y. Kobayashi, N. Udagawa, S. Kaku, T. Ueki, Y. Okazaki, N. Takahashi and T. Suda, Commun. Biol., 2019, 2, 86 CrossRef PubMed.
- G. Liu, J. F. Lovell, L. Zhang and Y. Zhang, Int. J. Mol. Sci., 2020, 21(17), 6380 CrossRef CAS.
- Q. P. Dou and J. A. Zonder, Curr. Cancer Drug Targets, 2014, 14, 517–536 CrossRef CAS PubMed.
- R. Oerlemans, C. R. Berkers, Y. G. Assaraf, G. L. Scheffer, G. J. Peters, S. E. Verbrugge, J. Cloos, J. Slootstra, R. H. Meloen, R. H. Shoemaker, B. A. C. Dijkmans, R. J. Scheper, H. Ovaa and G. Jansen, Invest. New Drugs, 2018, 36, 797–809 CrossRef CAS PubMed.
- C. R. C. Tan, S. Abdul-Majeed, B. Cael and S. K. Barta, Clin. Pharmacokinet., 2019, 58, 157–168 CrossRef CAS PubMed.
- S. Zhang, A. A. Kulkarni, B. Xu, H. Chu, T. Kourelis, R. S. Go, M. L. Wang, V. Bachanova and Y. Wang, Blood Cancer J., 2020, 10, 33 CrossRef.
- P. de la Puente, M. J. Luderer, C. Federico, A. Jin, R. C. Gilson, C. Egbulefu, K. Alhallak, S. Shah, B. Muz, J. Sun, J. King, D. Kohnen, N. N. Salama, S. Achilefu, R. Vij and A. K. Azab, J. Controlled Release, 2018, 270, 158–176 CrossRef CAS.
- S. Shen, X. J. Du, J. Liu, R. Sun, Y. H. Zhu and J. Wang, J. Controlled Release, 2015, 208, 14–24 CrossRef CAS PubMed.
- S. K. Kumar, S. J. Jacobus, A. D. Cohen, M. Weiss, N. Callander, A. K. Singh, T. L. Parker, A. Menter, X. Yang, B. Parsons, P. Kumar, P. Kapoor, A. Rosenberg, J. A. Zonder, E. Faber Jr, S. Lonial, K. C. Anderson, P. G. Richardson, R. Z. Orlowski, L. I. Wagner and S. V. Rajkumar, Lancet Oncol., 2020, 21, 1317–1330 CrossRef CAS.
- G. Cengiz Seval and M. Beksac, Expert Opin. Drug Saf., 2018, 17, 953–962 CrossRef CAS.
- X. Fang, B. Zong and S. Mao, Nano-Micro Lett., 2018, 10, 64 CrossRef.
- J. Zhong, R. K. Kankala, S. B. Wang and A. Z. Chen, Polymers, 2019, 11(10), 1627 CrossRef CAS.
- M. Xu, Y. Hu, W. Ding, F. Li, J. Lin, M. Wu, J. Wu, L. P. Wen, B. Qiu, P. F. Wei and P. Li, Biomaterials, 2020, 258, 120308 CrossRef CAS.
- R. Xie, P. Yang, S. Peng, Y. Cao, X. Yao, S. Guo and W. Yang, J. Mater. Chem. B, 2020, 8, 6128–6138 RSC.
- X. Wang, H. Wang, L. Guo, G. Chen, R. Kong, F. Qu and L. Xia, Analyst, 2020, 145, 1362–1367 RSC.
- N. Yuan, X. Gong, W. Sun and C. Yu, Chemosphere, 2021, 267, 128863 CrossRef CAS PubMed.
- Y. Bae, Y. Kim and E. S. Lee, Molecules, 2021, 26(12), 3547 CrossRef CAS PubMed.
- C. Zheng, Y. Wang, S. Z. F. Phua, W. Q. Lim and Y. Zhao, ACS Biomater. Sci. Eng., 2017, 3, 2223–2229 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra00051b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |