Open Access Article
Open Access ArticleAmericium preferred: lanmodulin, a natural lanthanide-binding protein favors an actinide over lanthanides†
Helena
Singer‡
a,
Björn
Drobot‡
 b,
Cathleen
Zeymer
b,
Cathleen
Zeymer
 c,
Robin
Steudtner
c,
Robin
Steudtner
 *b and
Lena J.
Daumann
*b and
Lena J.
Daumann
 *a
*a
aDepartment of Chemistry, Ludwig-Maximilians-University Munich, Butenandtstraße 5 – 13, 81377 München, Germany. E-mail: lena.daumann@lmu.de
bInstitute of Resource Ecology, Helmholtz-Zentrum Dresden-Rossendorf e.V., Bautzner Landstraße 400, 01328 Dresden, Germany. E-mail: r.steudtner@hzdr.de
cDepartment of Chemistry, Technische Universität München, Lichtenbergstraße 4, 85748 Garching, Germany
First published on 26th October 2021
Abstract
The separation and recycling of lanthanides is an active area of research with a growing demand that calls for more environmentally friendly lanthanide sources. Likewise, the efficient and industrial separation of lanthanides from the minor actinides (Np, Am–Fm) is one of the key questions for closing the nuclear fuel cycle; reducing costs and increasing safety. With the advent of the field of lanthanide-dependent bacterial metabolism, bio-inspired applications are in reach. Here, we utilize the natural lanthanide chelator lanmodulin and the luminescent probes Eu3+ and Cm3+ to investigate the inter-metal competition behavior of all lanthanides (except Pm) and the major actinide plutonium as well as three minor actinides neptunium, americium and curium to lanmodulin. Using time-resolved laser-induced fluorescence spectroscopy we show that lanmodulin has the highest relative binding affinity to Nd3+ and Eu3+ among the lanthanide series. When equimolar mixtures of Cm3+ and Am3+ are added to lanmodulin, lanmodulin preferentially binds to Am3+ over Cm3+ whilst Nd3+ and Cm3+ bind with similar relative affinity. The results presented show that a natural lanthanide-binding protein can bind a major and various minor actinides with high relative affinity, paving the way to bio-inspired separation applications. In addition, an easy and versatile method was developed, using the fluorescence properties of only two elements, Eu and Cm, for inter-metal competition studies regarding lanthanides and selected actinides and their binding to biological molecules.
Introduction
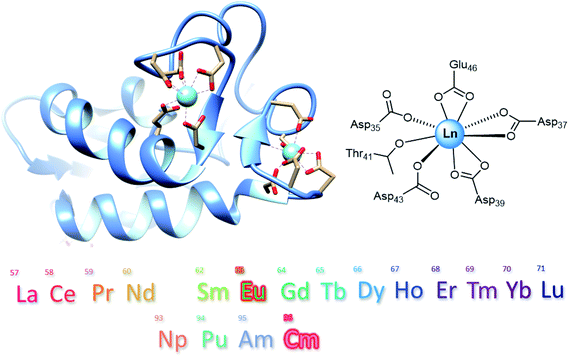
f-Block elements have become indispensable components for modern day life and are found in all high technologies, medicine and agriculture earning them the name Vitamins of Modern Industry.1,2 Primary sources for lanthanides (Ln, or rare earth elements, REE, if Sc and Y are included) are carbonate minerals such as bastnaesite or the phosphate ores monazite or xenotime.3,4 Since only mixtures of REE are available from these natural sources, extensive separation processes with highly engineered ion-exchange and solvent extraction routes are required.1,3 These elaborate procedures are required due to the similar chemical properties (similar ionic radii, oxidation state +III) in the Ln series, which complicates the energy consuming separation and requires a large number of process steps.1 Furthermore, their ores contain the two naturally abundant actinides (An) uranium and thorium, generating a large amount of radioactive waste as by-products during REE mining. Considering the importance of REE for green energy resources and sustainable applications, a more environmentally friendly way would be recovering and recycling REE from end of life items (e.g. e-waste) or coal fly ash.5,6 The separation of REE from An, as they occur together in mixtures in nuclear fuel cycles, is a similarly tricky endeavour and an active area of research.7,8 In addition, efficient chelators for the clean-up of radioactive environmental spills are desired.Nature has designed several ligands and proteins for efficient metal binding, with siderophores at the forefront. Inspired by these natural chelators, such as enterobactin and deferoxamine, researchers have developed highly specific synthetic chelators, for example 3,4,3-LI-1,2-HOPO, a very efficient lanthanide and actinide-binder.9,10 Recent bio-inspired advances to tightly bind certain metal ions also include short peptides sequences known as lanthanide binding tags (LBT),11 and entirely de novo designed metalloproteins with remarkable affinities for Ln.12,13 Ten years ago, it was discovered that some bacteria also utilize Ln. These bacteria have evolved highly specific Ln uptake and Ln transport strategies. We have previously shown that even the isolated natural cofactor pyrroloquinoline quinone (PQQ) of Ln-dependent metalloenzymes (methanol dehydrogenases) is capable to separate Ln by selective precipitation.14 In addition to proposed small molecular weight lanthanophores,15–17 small bacterial proteins, such as lanmodulin (LanM, Fig. 1),17–19 occurring in the Ln-utilizing bacterium Methylorubrum extorquens AM1, have already been shown to be efficient chelators for REE extraction. LanM is an 11.8 kDa protein and has three high affinity sites for REE binding (EF1 to EF3) with picomolar affinity for these metal ions based on EF hand motifs.15,18 Its promise as a selective extractant from electronic waste and lignite leachates has already been demonstrated by Deblonde and Cotruvo.20 During the preparation of this manuscript, a preprint stated that LanM is able to bind actinium with a higher affinity compared to, for example, the REE yttrium.21 Previously, an NMR structure of LanM with Y3+ has been reported, and during the review process of this manuscript a study demonstrating binding of Am and Cm to LanM, was published.19,22 Here, we evaluate the potential of LanM to bind the An neptunium, plutonium, americium and curium and compare the relative binding affinity§ of these An to each other and with the lanthanide series (Fig. 1) at pH 6.7 to mimic physiological conditions. The isolation of americium from lanthanides is of particular interest, as they occur commonly together in nuclear waste and are challenging to separate.23,24 Using time-resolved laser-induced fluorescence spectroscopy (TRLFS) in combination with the excellent luminescent properties of the two f-block elements Cm and Eu, enables inter-metal competition binding studies regarding the whole lanthanide series and selected actinides, revealing a relative affinity trend of LanM for the chosen elements.
Results and discussion
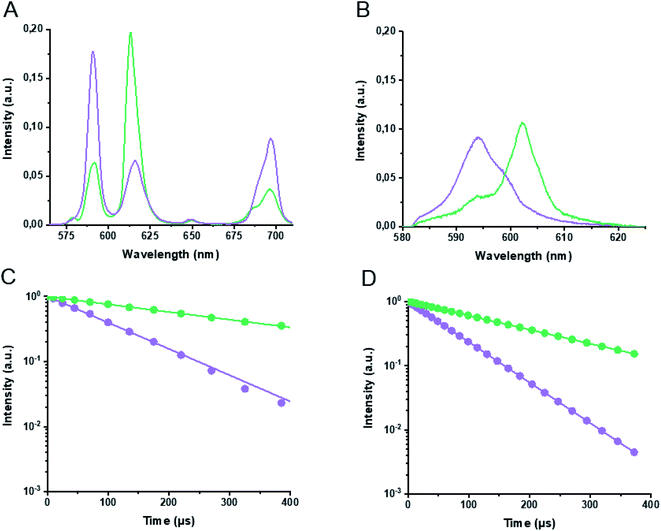
First, Eu3+ and Cm3+ binding to LanM was evaluated with TRLFS and yielded emission spectra (after spectral deconvolution using parallel factor analysis, Fig. S1†). Corresponding luminescence lifetimes of 107 μs and 367 μs after excitation at 394 nm were determined for the Eu3+-aquo ion (purple) and Eu3+-LanM (green), respectively, with the main emission maxima at 591 and 613 nm (Fig. 2A and C). The comparative analysis of the An binding to the LanM system provided the emission spectra of Cm3+-aquo ion (purple) and Cm3+-LanM (green) with distinct emission maxima at 594 and 604 nm and lifetimes of 69 μs and 198 μs after excitation at 396 nm (Fig. 2B and D). The luminescence lifetimes for the Eu3+-and Cm3+-aquo ions are in good accordance to previously reported ones and the spectral characteristics of the formed Eu3+– and Cm3+–LanM species match reported species of the calcium-binding protein calmodulin.25,26 Due to the greater sensitivity and quantum yields of Cm3+ luminescence compared to Eu3+,27 a lower concentration of 18 nM LanM and 100 nM Cm3+ was utilized, while for Eu3+ experiments a final concentration of 1 μM of LanM and 5.5 μM Eu3+ was necessary. For both experiments, 5.5 equivalents of Eu3+ or Cm3+ were required to obtain a 100% occupation of all 4 EF hands of LanM which also corresponded with CD experiments (Fig. S6†).Second, for inter-metal competition studies of LanM with different Ln and An, the lanthanide series with Eu3+ and An series (+Nd3+) with Cm3+ was investigated using TRLFS (Fig. 1). Two different experiments were conducted for each Ln and An series. First – Ln addition – (Fig. 3) where LanM (1 μM) was incubated and saturated with Eu3+ (5.5 μM) for 10 minutes and then subsequently a second lanthanide (5.5 μM) was added. Known from previous studies and experiments (see ESI†), LanM was able to bind Eu3+ immediately after addition without time delay. However, 10 minutes were chosen as equilibrium time (Eu3+ and LanM incubation) before the second Ln3+ was added due to experimental setup and handling of samples as well as to ensure a proper mixing of the sample. Data acquisition was started immediately after adding the competing metal ion and the decrease in Eu3+–LanM signal intensity was observed over 50 min (Fig. 3A, individual spectra can be found in Fig. S2†). In particular, with the earlier Ln (reddish to green), a still persistent decrease of the Eu3+–LanM signal was detected – while with the later Ln (blue to purple) such as Lu3+ a stagnating signal seemed to be already reached within the 50 minutes. After 28 hours (Fig. 3B), the final intensity of the Eu3+–LanM complex was still remaining but had stagnated, indicating that thermodynamic equilibrium between Ln and LanM was reached. In a second experiment – Ln competition – a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of Eu3+ and a second lanthanide (5.5 μM each) were premixed and added simultaneously to 1 μM of LanM. Incubation for 24 hours at room temperature was chosen to ensure that the thermodynamic equilibrium had been reached and the amount of the residual Eu3+–LanM complex was spectroscopically determined. By spectral deconvolution of all Eu3+–LanM emission spectra, the intensity ratio of Eu3+ towards the treated second Ln was calculated (Fig. 4). Based on these two experiments (Ln addition, Ln competition), we obtain two pieces of information overall: the time constants describing how fast the thermodynamic equilibrium is reached (Fig. 3C) and secondly, how strong the capability is of each Ln to replace the Eu from the Eu3+–LanM complex representing the individual relative affinities (Fig. 4). Interestingly, the obtained time constants (Table S1†) and the relative affinities do not follow the same trend and thus the time constants are not solely determined by the relative affinity of the respective metal ions, thus other physical factors are likely at play here. Based on the time constants determination it can be concluded: the smaller the ionic radii of the Ln, the faster the exchange against the Eu3+ in Eu3+–LanM complex. The trend correlates with the typical Ln contraction and is most likely due to a kinetic effect based only on the size and Lewis acidity of the Ln. The trend regarding the relative affinity of the formed Ln3+–LanM complex does not linearly follow the Ln series, but rather peaks at Nd3+, meaning that Nd3+ binds with equal relative affinity to LanM as Eu3+ (which was added for clarity as a theoretical point and set to 50% of Eu3+–LanM complex when 11 equivalents of Eu3+ are added to one equivalent of LanM, resulting in 50% of free Eu3+-aquo species and 50% of Eu3+–LanM complex). It was shown recently that LanM reveals a unique, not yet understood trend regarding the affinities for different Ln (Fig. S5†). Unfortunately, affinities for only selected Ln were reported (Table S4†). While our actual data gives no absolute values of dissociation constants Kd, a trend among the whole series (except Pm) is clearly visible with our developed method.
1 mixture of Eu3+ and a second lanthanide (5.5 μM each) were premixed and added simultaneously to 1 μM of LanM. Incubation for 24 hours at room temperature was chosen to ensure that the thermodynamic equilibrium had been reached and the amount of the residual Eu3+–LanM complex was spectroscopically determined. By spectral deconvolution of all Eu3+–LanM emission spectra, the intensity ratio of Eu3+ towards the treated second Ln was calculated (Fig. 4). Based on these two experiments (Ln addition, Ln competition), we obtain two pieces of information overall: the time constants describing how fast the thermodynamic equilibrium is reached (Fig. 3C) and secondly, how strong the capability is of each Ln to replace the Eu from the Eu3+–LanM complex representing the individual relative affinities (Fig. 4). Interestingly, the obtained time constants (Table S1†) and the relative affinities do not follow the same trend and thus the time constants are not solely determined by the relative affinity of the respective metal ions, thus other physical factors are likely at play here. Based on the time constants determination it can be concluded: the smaller the ionic radii of the Ln, the faster the exchange against the Eu3+ in Eu3+–LanM complex. The trend correlates with the typical Ln contraction and is most likely due to a kinetic effect based only on the size and Lewis acidity of the Ln. The trend regarding the relative affinity of the formed Ln3+–LanM complex does not linearly follow the Ln series, but rather peaks at Nd3+, meaning that Nd3+ binds with equal relative affinity to LanM as Eu3+ (which was added for clarity as a theoretical point and set to 50% of Eu3+–LanM complex when 11 equivalents of Eu3+ are added to one equivalent of LanM, resulting in 50% of free Eu3+-aquo species and 50% of Eu3+–LanM complex). It was shown recently that LanM reveals a unique, not yet understood trend regarding the affinities for different Ln (Fig. S5†). Unfortunately, affinities for only selected Ln were reported (Table S4†). While our actual data gives no absolute values of dissociation constants Kd, a trend among the whole series (except Pm) is clearly visible with our developed method.
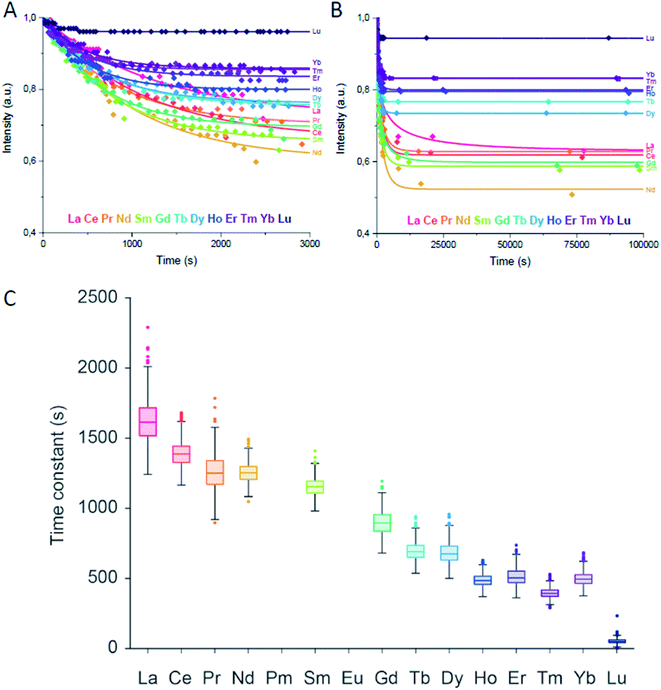 | ||
Fig. 3 Lanthanide addition: (A) intensity of the emission signal of the Eu3+–LanM complex within the first 3000 seconds (50 minutes) after the addition of the second lanthanide based on emission spectra and subsequent PARAFAC analysis. (B) Intensity of the emission signal of the Eu3+–LanM complex up to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 seconds (28 hours) after addition of the second lanthanide based on emission spectra and subsequent PARAFAC analysis. The solid lines are first order decay fits with R2 = La (0.97752), Ce (0.98773), Pr (0.91498), Nd (0.95209), Sm (0.95356), Gd (0.97783), Tb (0.98542), Dy (0.9809), Ho (0.97413), Er (0.96369), Tm (0.97539), Yb (0.97657), Lu (0.84107). Individual traces can be found in Fig. S2.† (C) A Monte Carlo approach was applied to evaluate the robustness of the fit (see ESI 2.3†). The box plot summarizes the fitted time constants of all the 1000 MC runs across the Ln series based on B. For the measurements, an excitation wavelength of 394 nm and concentrations of 1 μM LanM, 5.5 μM Eu3+ with additional 5.5 μM Ln3+ were selected. 000 seconds (28 hours) after addition of the second lanthanide based on emission spectra and subsequent PARAFAC analysis. The solid lines are first order decay fits with R2 = La (0.97752), Ce (0.98773), Pr (0.91498), Nd (0.95209), Sm (0.95356), Gd (0.97783), Tb (0.98542), Dy (0.9809), Ho (0.97413), Er (0.96369), Tm (0.97539), Yb (0.97657), Lu (0.84107). Individual traces can be found in Fig. S2.† (C) A Monte Carlo approach was applied to evaluate the robustness of the fit (see ESI 2.3†). The box plot summarizes the fitted time constants of all the 1000 MC runs across the Ln series based on B. For the measurements, an excitation wavelength of 394 nm and concentrations of 1 μM LanM, 5.5 μM Eu3+ with additional 5.5 μM Ln3+ were selected. | ||
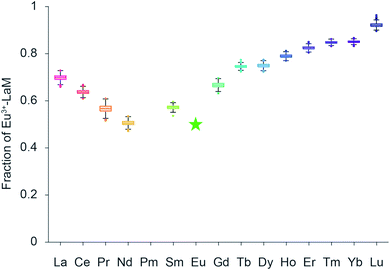 | ||
| Fig. 4 Lanthanide competition: box plots (1000 MC runs, see ESI 2.3†) of the final intensity of the Eu3+–LanM-complex emission 24 hours after simultaneous addition of equimolar amounts of Eu3+ and a second lanthanide ion based on collected emission spectra and subsequent PARAFAC analysis. For the measurements, an excitation wavelength of 394 nm and concentrations of 1 μM LanM, 5.5 μM Eu3+ and 5.5 μM Ln3+ were selected. The green star data point was added for clarity and indicates and intensity of 50% of the Eu3+–LanM complex. | ||
The Ln left of Nd3+ in the series bind with lower relative affinity to LanM, with La3+ occupying approximately 20% of LanM and Eu3+ 80% when added in equimolar amounts. The Ln to the right side of Nd3+ with smaller ionic radii as well bind less efficiently to LanM than Eu3+, with Lu3+ only 10% occupying LanM when Eu3+ is present. These results are both somewhat unexpected, but also in line what has been found for the activity of Ln-dependent methanol dehydrogenase isolated from Methylacidiphilum fumariolicum SolV. Here, adding lanthanides to a partially occupied Eu-MDH yielded the highest activity with Pr3+ and Nd3+, however, this might be just a coincidence.28 Despite various factors such as the gadolinium break (referring to an often observed change in the chemical behavior after this lanthanide in the series29), it has been suggested that the natural abundance of the Ln has an impact and bacterial Ln uptake and metabolism is designed to use the most abundant Ln.
After having experimentally verified that Nd3+ binds equally well as Eu3+ to LanM in our two lanthanide binding studies, we put forward this competitive approach for four trivalent An. The trivalent An – Np3+, Pu3+, Am3+, Cm3+ – were chosen to compete with Nd3+ as the strongest opponent of lanthanides for the LanM relative binding affinity. Cm3+ was used as comparable luminescence probe for the inter-metal competition studies. In the first experiment – An addition – LanM (18 nM) was added to Cm3+ (100 nM) and after 10 min, the equilibrated solution was treated with either Nd3+, Np3+, Pu3+ or Am3+ (100 nM, Fig. 5).
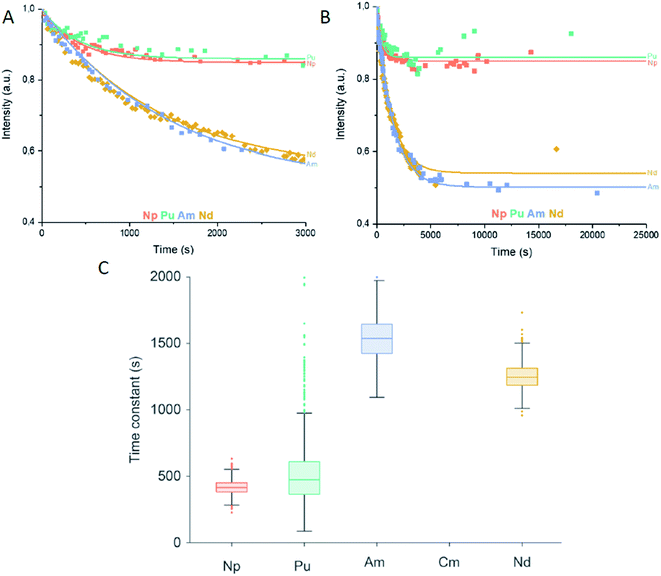 | ||
Fig. 5 Actinide addition: (A) intensity of the emission signal of the Cm3+–LanM complex within the first 3000 seconds (50 minutes) after the addition of the second actinide or Nd3+ based on collected emission spectra and subsequent PARAFAC analysis. (B) Intensity of the emission signal of the Cm3+–LanM complex up to 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 seconds (7 hours) after addition of the second actinide or Nd3+ based on collected emission spectra and subsequent PARAFAC analysis. R2 = Nd (0.97383), Am (0.97056), Pu (0.92988), Np (0.96142). Individual traces can be found in Fig. S3.† (C) Box plots of fitted time constants of the 1000 MC runs based on B (see Fig. S3†). For the measurements, an excitation wavelength of 396 nm and concentrations of 18 nM LanM, 100 nM Cm3+ with additional 100 nM An3+ or Nd3+ were selected. 000 seconds (7 hours) after addition of the second actinide or Nd3+ based on collected emission spectra and subsequent PARAFAC analysis. R2 = Nd (0.97383), Am (0.97056), Pu (0.92988), Np (0.96142). Individual traces can be found in Fig. S3.† (C) Box plots of fitted time constants of the 1000 MC runs based on B (see Fig. S3†). For the measurements, an excitation wavelength of 396 nm and concentrations of 18 nM LanM, 100 nM Cm3+ with additional 100 nM An3+ or Nd3+ were selected. | ||
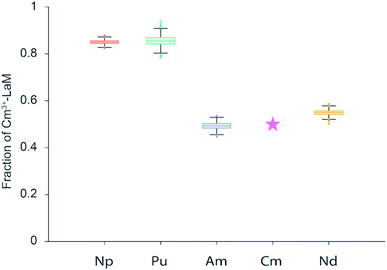
By monitoring the Cm3+ luminescence, the reduction of the Cm3+–LanM intensity over time was clearly visible with a faster leveling for Np3+ and Pu3+ than for Nd3+ and Am3+ (Fig. 5A and B). To obtain a clear picture of the thermodynamic equilibrium (An competition), equimolar amounts (100 nM each) of Cm3+ and respectively Nd3+, Np3+, Pu3+ or Am3+ were added to LanM and the intensity of Cm3+–LanM complex were measured after 24 hours of incubation (Fig. 6).
Again, the theoretical value of 50% for Cm3+ (pink star) was added. In the An experiments we observe clearly two groups based on time constants (Fig. 5C) and on the amount of Cm3+–LanM complex in equilibrium (Fig. 6): Nd–Am and Np–Pu. The results show that the An f-block elements reveal similar relative binding affinities to LanM like that of the Ln f-block elements, which repeatedly underlines their chemical analogy. Secondly, Np and Pu display a smaller relative binding affinity to LanM compared to Cm3+, however, an exchange of Cm3+ with the added An is observable; since the intensity of the Cm3+–LanM complex is distinctly reduced. The initial oxidation state of Np and Pu was +III, but Np and Pu reveal a manifold redox chemistry under certain conditions (Table S3†). In aqueous solution, Np is mainly present in +III to +VII with the most stable pentavalent dioxo form NpO2+, while for Pu, a co-existence in equilibrium of four different oxidation states from +III to +VI has been reported.30 Due to the more versatile redox chemistry of Np and Pu the oxidation to the penta- and hexavalent actinyl (AnO2+ and AnO22+) structures bearing a trans-dioxo motif (Oax![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) An(+V/VI)
An(+V/VI)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) Oax) are easily conceivable. The oxidation state most probably changed by varying the chemical environment of Np and Pu by dilution, as a function of the aqueous speciation or interaction with complexing agent. No spectroscopic method is available to examine the final oxidation state of Np and Pu in the nanomolar concentration range after the incubation with LanM. Reflecting the determined time constants for Np and Pu, LanM binds Np3+ and Pu3+ with lower relative affinities, comparable to the later, or very early Lns. In contrast, the trans-dioxo cations might be not able to effectively bind to LanM. Thirdly and surprisingly, the trivalent f-block element Am3+ exhibits a higher relative affinity to LanM than Cm3+ while Nd3+ and Cm3+ bind very similarly. The high relative binding affinity is most probably due to the same most stable oxidation states and ionic radii of the metal ions (Table S2†). Am3+ and Cm3+ are both stable in the preferred +III oxidation state and have ionic radii that strongly resemble Nd3+.31 A direct comparison between the Eu3+ and Cm3+ systems and experimental setups is ambitious. For the Cm3+ experiments, the concentrations were 50 times lower, and dilution effects, solubility, hydrolysis or carbonate complexation of Ln/An, as well as parameters affecting the kinetics, should only interfere (if at all) with minor contribution. To be able to draw a comparison between both Ln3+–LanM and An3+–LanM studies, Nd3+ was chosen for both setups, enabling an estimation of relative affinity for all selected Lns and Ans. Nd3+ was the best of the Ln series besides Eu3+ to bind to LanM, but Am3+ displayed an even higher relative binding affinity with a faster exchange rate. Finally, with all things considered and under the conditions chosen, LanM, a natural lanthanide-binding protein, favors an actinide over the lanthanides. In addition, we show a unique relative affinity trend of LanM for the Ln series that will also help in the future to develop new innovative bio-inspired strategies for Ln or An-recycling and separation. In addition, an easy and versatile method was developed, using Eu3+ and Cm3+ as comparative luminescence probe, for inter-metal competition studies regarding lanthanides and selected actinides and their binding to biological molecules.
Oax) are easily conceivable. The oxidation state most probably changed by varying the chemical environment of Np and Pu by dilution, as a function of the aqueous speciation or interaction with complexing agent. No spectroscopic method is available to examine the final oxidation state of Np and Pu in the nanomolar concentration range after the incubation with LanM. Reflecting the determined time constants for Np and Pu, LanM binds Np3+ and Pu3+ with lower relative affinities, comparable to the later, or very early Lns. In contrast, the trans-dioxo cations might be not able to effectively bind to LanM. Thirdly and surprisingly, the trivalent f-block element Am3+ exhibits a higher relative affinity to LanM than Cm3+ while Nd3+ and Cm3+ bind very similarly. The high relative binding affinity is most probably due to the same most stable oxidation states and ionic radii of the metal ions (Table S2†). Am3+ and Cm3+ are both stable in the preferred +III oxidation state and have ionic radii that strongly resemble Nd3+.31 A direct comparison between the Eu3+ and Cm3+ systems and experimental setups is ambitious. For the Cm3+ experiments, the concentrations were 50 times lower, and dilution effects, solubility, hydrolysis or carbonate complexation of Ln/An, as well as parameters affecting the kinetics, should only interfere (if at all) with minor contribution. To be able to draw a comparison between both Ln3+–LanM and An3+–LanM studies, Nd3+ was chosen for both setups, enabling an estimation of relative affinity for all selected Lns and Ans. Nd3+ was the best of the Ln series besides Eu3+ to bind to LanM, but Am3+ displayed an even higher relative binding affinity with a faster exchange rate. Finally, with all things considered and under the conditions chosen, LanM, a natural lanthanide-binding protein, favors an actinide over the lanthanides. In addition, we show a unique relative affinity trend of LanM for the Ln series that will also help in the future to develop new innovative bio-inspired strategies for Ln or An-recycling and separation. In addition, an easy and versatile method was developed, using Eu3+ and Cm3+ as comparative luminescence probe, for inter-metal competition studies regarding lanthanides and selected actinides and their binding to biological molecules.
Data availability
TRLFS data and MATLAB code available upon request (from E-mail: b.drobot@hzdr.de)Author contributions
LJD and HS conceptualized the idea and wrote the initial draft of this manuscript, LJD acquired funding for this project, all authors developed the methodology and were involved in review and editing of this manuscript. CZ and HS purified LanM. HS, BD and RS acquired the TRLF data and conducted lanthanide and actinide titrations. BD wrote the code and fitted the TRLF data. LJD, CZ and RS provided the necessary resources and infrastructure for this work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
LJD acknowledges a grant from the Klaus-Tschira Boost Fund and support from the ERC Starting grant Lanthanophore.References
- T. Cheisson and E. J. Schelter, Science, 2019, 363, 489–493 CrossRef CAS PubMed.
- V. Balaram, Geosci. Front., 2019, 10, 1285–1303 CrossRef CAS.
- F. Xie, T. A. Zhang, D. Dreisinger and F. Doyle, Miner. Eng., 2014, 56, 10–28 CrossRef CAS.
- A. Jordens, Y. P. Cheng and K. E. Waters, Miner. Eng., 2013, 41, 97–114 CrossRef CAS.
- T. G. Ambaye, M. Vaccari, F. D. Castro, S. Prasad and S. Rtimi, Environ. Sci. Pollut. Res., 2020, 27, 36052–36074 CrossRef CAS PubMed.
- R. C. Smith, R. K. Taggart, J. C. Hower, M. R. Wiesner and H. Hsu-Kim, Environ. Sci. Technol., 2019, 53, 4490–4499 CrossRef CAS PubMed.
- A. V. Gelis, P. Kozak, A. T. Breshears, M. A. Brown, C. Launiere, E. L. Campbell, G. B. Hall, T. G. Levitskaia, V. E. Holfeltz and G. J. Lumetta, Sci. Rep., 2019, 9, 12842 CrossRef PubMed.
- N. P. Bessen, J. A. Jackson, M. P. Jensen and J. C. Shafer, Coord. Chem. Rev., 2020, 421, 213446 CrossRef CAS.
- A. S. Ivanov, B. F. Parker, Z. Zhang, B. Aguila, Q. Sun, S. Ma, S. Jansone-Popova, J. Arnold, R. T. Mayes, S. Dai, V. S. Bryantsev, L. Rao and I. Popovs, Nat. Commun., 2019, 10, 819 CrossRef CAS PubMed.
- R. J. Abergel, P. W. Durbin, B. Kullgren, S. N. Ebbe, J. Xu, P. Y. Chang, D. I. Bunin, E. A. Blakely, K. A. Bjornstad, C. J. Rosen, D. K. Shuh and K. N. Raymond, Health Phys., 2010, 99, 401–407 CrossRef CAS PubMed.
- T. Hatanaka, N. Kikkawa, A. Matsugami, Y. Hosokawa, F. Hayashi and N. Ishida, Sci. Rep., 2020, 10, 19468 CrossRef CAS PubMed.
- S. J. Caldwell, I. C. Haydon, N. Piperidou, P.-S. Huang, M. J. Bick, H. S. Sjöström, D. Hilvert, D. Baker and C. Zeymer, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 30362 CrossRef CAS PubMed.
- J. A. Mattocks, J. L. Tirsch and J. A. Cotruvo, in Methods Enzymol., ed. J. A. Cotruvo, Academic Press, 2021, vol. 651, pp. 23–61 Search PubMed.
- H. Lumpe, A. Menke, C. Haisch, P. Mayer, A. Kabelitz, K. V. Yusenko, A. Guilherme Buzanich, T. Block, R. Pöttgen, F. Emmerling and L. J. Daumann, Chem. - Eur. J., 2020, 26, 10133–10139 CrossRef CAS PubMed.
- J. A. Cotruvo Jr, ACS Cent. Sci., 2019, 5, 1496–1506 CrossRef PubMed.
- L. J. Daumann, Angew. Chem., Int. Ed., 2019, 58, 12795–12802 CrossRef CAS PubMed.
- A. M. Ochsner, L. Hemmerle, T. Vonderach, R. Nüssli, M. Bortfeld-Miller, B. Hattendorf and J. A. Vorholt, Mol. Microbiol., 2019, 111, 1152–1166 CrossRef CAS PubMed.
- J. A. Cotruvo, E. R. Featherston, J. A. Mattocks, J. V. Ho and T. N. Laremore, J. Am. Chem. Soc., 2018, 140, 15056–15061 CrossRef PubMed.
- E. C. Cook, E. R. Featherston, S. A. Showalter and J. A. Cotruvo Jr, Biochemistry, 2019, 58, 120–125 CrossRef CAS PubMed.
- G. J. P. Deblonde, J. A. Mattocks, D. M. Park, D. W. Reed, J. A. Cotruvo and Y. Jiao, Inorg. Chem., 2020, 59, 11855–11867 CrossRef CAS PubMed.
- G. J. P. Deblonde, J. A. Mattocks, Z. Dong, P. T. Wooddy, J. A. J. Cotruvo Jr and M. Zavarin, Sci. Adv., 2021, 7(43), eabk0273 CrossRef CAS PubMed.
- G. J. P. Deblonde, J. A. Mattocks, H. Wang, E. M. Gale, A. B. Kersting, M. Zavarin and J. A. Cotruvo, J. Am. Chem. Soc., 2021, 143(38), 15769–15783 CrossRef CAS PubMed.
- J. M. Richards and R. Sudowe, Anal. Chem., 2016, 88, 4605–4608 CrossRef CAS PubMed.
- C. Marie, P. Kaufholz, V. Vanel, M.-T. Duchesne, E. Russello, F. Faroldi, L. Baldini, A. Casnati, A. Wilden, G. Modolo and M. Miguirditchian, Solvent Extr. Ion Exch., 2019, 37, 313–327 CrossRef CAS.
- B. Drobot, M. Schmidt, Y. Mochizuki, T. Abe, K. Okuwaki, F. Brulfert, S. Falke, S. A. Samsonov, Y. Komeiji, C. Betzel, T. Stumpf, J. Raff and S. Tsushima, Phys. Chem. Chem. Phys., 2019, 21, 21213–21222 RSC.
- C. Wilke, A. Barkleit, T. Stumpf and A. Ikeda-Ohno, J. Inorg. Biochem., 2017, 175, 248–258 CrossRef CAS PubMed.
- C. Moulin, P. Decambox and P. Mauchien, J. Radioanal. Nucl. Chem., 1997, 226, 135–138 CrossRef CAS.
- H. Lumpe, A. Pol, H. J. M. Op den Camp and L. J. Daumann, Dalton Trans., 2018, 47, 10463–10472 RSC.
- G. Schwarzenbach and R. Gut, Helv. Chim. Acta, 1956, 39, 1589–1599 CrossRef CAS.
- L. R. Morss, N. M. Edelstein, J. Fuger, J. J. Katz and L. Morss, The chemistry of the actinide and transactinide elements, Springer, 2006 Search PubMed.
- G. T. Seaborg and D. E. Hobart, Summary of the properties of the lanthanide and actinide elements, Indian Association of Nuclear Chemists and Allied Scientists, India, 1996 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc04827a |
| ‡ Shared first author. |
| § Here we use the term relative binding affinity, because absolute values cannot be obtained from the competition experiment performed. This relative binding affinity is based on comparing the average affinities of all four LanM EF-hands for different Ln and An under given conditions. |
| This journal is © The Royal Society of Chemistry 2021 |