Open Access Article
Open Access ArticleCleavable and tunable cysteine-specific arylation modification with aryl thioethers†
Jian
Li
,
Jun-Jie
Deng
,
Zhibin
Yin
,
Qi-Long
Hu
,
Yang
Ge
,
Zhendong
Song
,
Ying
Zhang
,
Albert S. C.
Chan
,
Huilin
Li
and
Xiao-Feng
Xiong
 *
*
Guangdong Key Laboratory of Chiral Molecule and Drug Discovery, School of Pharmaceutical Sciences, Sun Yat-sen University, Guangzhou 510006, P. R. China. E-mail: xiongxf7@mail.sysu.edu.cn
First published on 18th February 2021
Abstract
Cysteine represents an attractive target for peptide/protein modification due to the intrinsic high nucleophilicity of the thiol group and low natural abundance. Herein, a cleavable and tunable covalent modification approach for cysteine containing peptides/proteins with our newly designed aryl thioethers via a SNAr approach was developed. Highly efficient and selective bioconjugation reactions can be carried out under mild and biocompatible conditions. A series of aryl groups bearing different bioconjugation handles, affinity or fluorescent tags are well tolerated. By adjusting the skeleton and steric hindrance of aryl thioethers slightly, the modified products showed a tunable profile for the regeneration of the native peptides.
Introduction
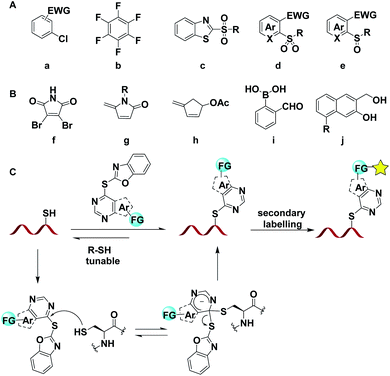
Peptide/protein modification is an important approach for profiling their structures and functions, studying protein–protein interactions (PPIs), monitoring cellular biological processes, and developing therapeutic agents.1 In comparison to de novo peptide synthesis, site-selective modification of existing peptides provides a more straightforward and effective option to diversify peptides for functional studies.2 Despite various methodologies developed to modify native amino acid residues, a tunable, biocompatible, highly selective and efficient protein modification approach is still highly in demand.2c,3 Among the 20 proteogenic amino acids, cysteine represents an attractive target for post-translational modification due to the intrinsic high nucleophilicity of the thiol group and low natural abundance (1.9%), and could be easily incorporated into specific sites by site-directed mutagenesis.4To achieve cysteine-specific modification, numerous efforts have been made towards the development of modification reagents that enable cysteine labelling with good efficiency and selectivity, including halogenoalkanes,5 maleimide,6 alkenes,7 alkynes,4b allenamides,4d and hypervalent iodine compounds.8 Among the established methods, the arylation strategy was underdeveloped to label peptides which have multiple functional groups and a large molecular weight.9 The nucleophilic aromatic substitution (SNAr) strategy provides the possibility for peptide arylation with easy manipulation and no need for extra additives, in comparison with metal- or photo-catalyzed arylation chemistry.10 In recent years, several arylation reagents based on SNAr were developed and showed good efficiency in labelling peptides, including aryl halides,11 perfluoroaromatic molecules,12 heteroaryl methylsulfone and methylsulfoxide reagents (Fig. 1A).13 However, the methods mentioned above were mainly focused on improving the modification efficiency or deliberately optimizing the stability of the Cys-bioconjugated linkage. Cleavable and tunable Cys-selective modification, which could enable the regeneration of the native peptides/proteins, was desired in areas such as antibody–drug conjugates (ADCs) and epigenetic modifications, but less studied.14 Maleimide derivatives f–h, formyl benzeneboronic acid i, and naphthol derivatives j could be applied to regenerate the native peptides or proteins; however the acidic or basic regeneration conditions and the necessity of well-defined triggers/stimuli prevented their further applications (Fig. 1B).6a,15 The pursuit of developing an efficient, biocompatible and regeneration tunable cysteine-specific modification strategy is still in high demand, especially because the modification could facilitate further decoration of peptides or proteins. Herein, we reported a cysteine selective, transition-metal free, regeneration tunable peptide/protein modification approach with aryl thioethers with good chemo-selectivity and reactivity, and the arylation could be applied to secondary decoration of peptides (Fig. 1C).
Results and discussion
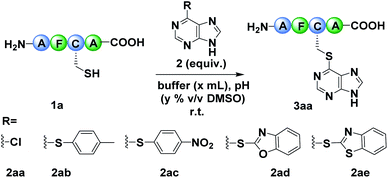
Our research was initiated by testing the reactivity of model peptide 1a with an array of arylation reagents. Commercially available 6-chloro purine 2aa, which could react with mercaptan and presents good skeleton functionalization feasibility, was employed initially. However, no reaction occurred, which might be attributed to the inapposite leaving ability of the chlorine atom within 2aa (Table 1, entry 1).16 Inspired by the work of Swager, which disclosed the dynamic and self-correcting nature of SNAr, we replaced the chloro- with a series of aromatic sulphides and investigated their reactivity with 1a (Table 1, entries 2–5).17 To our delight, 2ad and 2ae could react with 1a in 100 mM PBS buffer (pH = 8.0), affording the desired modified product 3aa with the yields of 43% and 17% after 1 h (Table 1, entries 4–5).18 It worth noting that the free C-terminus and N-terminus did not react with the thioethers 2ad and 2ae, and the chemo-selectivity was determined by 1D 1H NMR spectroscopy (Fig. S1†). Subsequently, different buffers and pH were screened using 2ad as the arylation agent. 100 mM Tris buffer (pH = 8.0) turned out to be the best solvent for the modification, as it gave the product in 47% yield after 1 h (Table 1, entries 6–9 and Table S1†). To further improve the modification efficiency, we adjusted the equivalent of 2ad, the reaction concentration and the ratio of DMSO for the reactions (Table 1, entries 10–13 and Table S1†). The results showed that when reducing the ratio of DMSO, the byproduct generated from the dimerization of 1a was increased accompanied by the decrease of modification yields (Fig. S6†). Interestingly, by fine-tuning the ratio of DMSO and reaction concentration, the dimerized byproduct could be efficiently suppressed and the desired product 3aa was obtained in 90% yield, despite the slightly increased reaction time (Table S1†). Finally, the optimal reaction conditions were established as performing the reaction at room temperature, with a concentration of peptide of 1 mM and 3 equivalents of arylation reagent in 100 mM Tris buffer (pH 8.0, 1% v/v DMSO) (Table 1, entry 13). Based on previous work and time-course analysis of the arylation process (Fig. S215†), we hypothesized that 2ad reacted with 1avia a two-step SNAr mechanism and the first step is rate-limiting (Fig. 1C).13c| Entry | 2 (eq.) | Buffer (pH) | x | y | Yieldb (%) |
|---|---|---|---|---|---|
| a Reaction conditions: 1.0 μmol 1a, 2 (eq.) in 100 mM non-degassed buffer at room temperature for 1 h. b Reported yields are LC-MS yields after 1 h (the yields in parentheses correspond to the yields after a 5 h reaction). c Isolated yield. | |||||
| 1 | 2aa (1.5) | PBS (8.0) | 0.5 | 10 | n.d |
| 2 | 2ab (1.5) | PBS (8.0) | 0.5 | 10 | n.d |
| 3 | 2ac (1.5) | PBS (8.0) | 0.5 | 10 | n.d |
| 4 | 2ad (1.5) | PBS (8.0) | 0.5 | 10 | 43 |
| 5 | 2ae (1.5) | PBS (8.0) | 0.5 | 10 | 17 |
| 6 | 2ad (1.5) | HEPES (8.0) | 0.5 | 10 | 41 |
| 7 | 2ad (1.5) | Tris (8.0) | 0.5 | 10 | 47 |
| 8 | 2ad (1.5) | Tris (7.6) | 0.5 | 10 | 39 |
| 9 | 2ad (1.5) | Tris (7.8) | 0.5 | 10 | 43 |
| 10 | 2ad (3.0) | Tris (8.0) | 0.5 | 10 | 64 (85) |
| 11 | 2ad (3.0) | Tris (8.0) | 0.5 | 5 | 62 (87) |
| 12 | 2ad (3.0) | Tris (8.0) | 0.5 | 1 | 45 (80) |
| 13 | 2ad (3.0) | Tris (8.0) | 1.0 | 1 | 59 (90, 83c) |
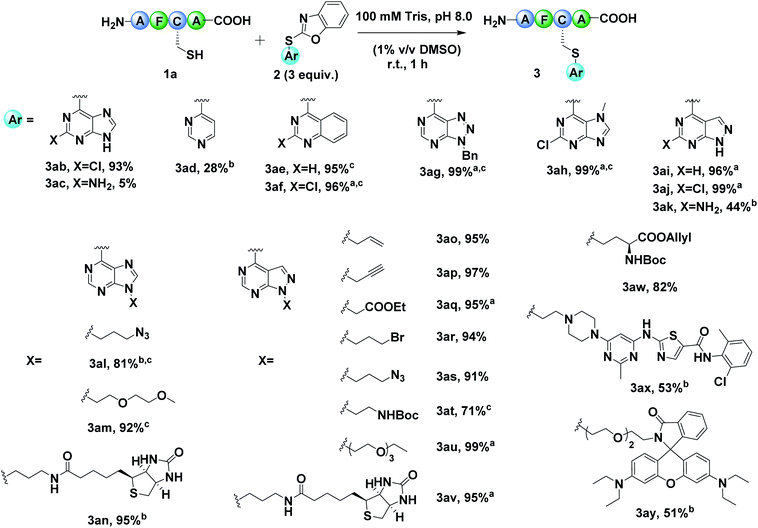
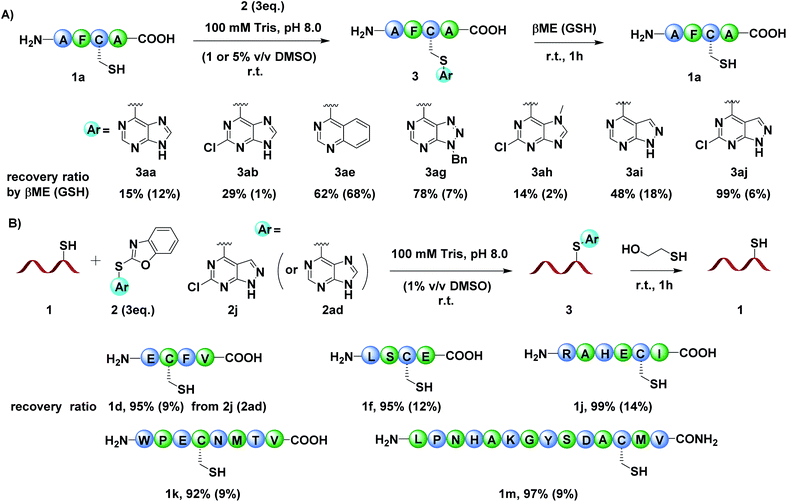
After establishing the optimal reaction conditions, we set out to exam the diversity of the arylation reagents 2 (Fig. S2†). Benzoxazole sulfides 2 with different heterocycle skeletons, including purine (2b–2c, 2h), pyrimidine (2d), quinazoline (2e, 2f), triazole pyrimidine (2g) and pyrazolopyrimidine (2i–2k), all ran smoothly to afford the desired modified products (Fig. 2, 3ab–3ak). Generally, arylation reagents 2 bearing electron-withdrawing groups on the purine (Fig. S2,†2b–2c), quinazoline (2e–2f) or pyrazolopyrimidine (2i–2k) could improve the reaction efficiency while electron-donating groups had reverse effects (Fig. 2, 3ab–3ac, 3ae–3af, and 3ai–3ak). Taking reaction efficiency, synthetic feasibility and functional group compatibility into consideration, we chose purine and pyrazolopyrimidine based benzoxazole sulfides as templates to further diversify the modification reagents. Purine based benzoxazole sulfides with different functional groups could efficiently modify peptide 1a in good yields (Fig. 2 and 3al–3an). The arylation process proceeded more efficiently when replacing the purine skeleton with pyrazolopyrimidine. A variety of widely applied biorelevant groups, including alkyne, azide, poly ethylene glycol (PEG) polymer and affinity label, could be introduced into arylation reagents and gave the corresponding products in almost quantitative yields (Fig. 2 and 3ao–3av). Notably, arylation reagents bearing amino acid fragments, drug molecules or fluorescent tags were also tolerated under the optimized conditions (Fig. 2, 3aw, 3ax and 3ay), demonstrating the potential applications of this method in branched peptide synthesis and cyclization, antibody–drug conjugates (ADCs) and physiological studies. It is worth mentioning that the unreacted modification reagents 2 could be easily removed after the reaction completion by simple filtration due to their poor solubility in water. The reaction kinetics was evaluated by time-course analysis and competition experiments, which showed that 2j reacted with peptide 1a faster than iodoacetamide and fluorinated arenes but slower than maleimide (Fig. S215 and Table S2†).
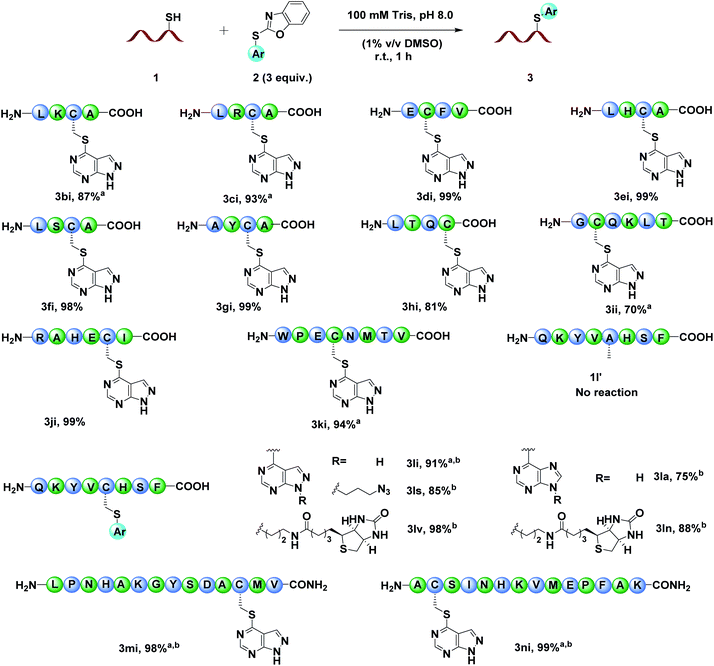
As benzoxazole sulfides 2 bearing pyrazolopyrimidine moieties presented satisfactory reaction efficiency and functionalization feasibility, we chose 2i to further investigate the scope of cysteine containing peptides (Fig. 3). Tetrapeptide substrates with different functional groups were surveyed firstly. The results showed that the peptides with nucleophilic Lys (3bi), Arg (3ci) or Glu (3di) were well tolerated. In the presence of peptides with other nucleophilic amino acids, e.g., His (3ei), Ser (3fi), Tyr (3gi) and Thr (3hi), no side reactions were detected and the desired products were obtained in almost quantitative yields, demonstrating the unique chemo-selectivity of the benzoxazole sulfides to Cys residues. The modification reactivity of benzoxazole sulfide 2i on longer peptides was also examined. Hexapeptides, octapeptides and 14-mer peptide 2m or 2n with various amino acid residues all ran smoothly to afford the desired cysteine modified products in excellent yields. To further explore the potential utility of this method, different functional groups were incorporated into benzoxazole sulfides 2. To our delight, pyrazolopyrimidine and purine based benzoxazole sulphides all reacted with octapeptide 1l and afforded the desired cysteine modified products (Fig. 3, 3li, 3ls, 3lv, 3la and 3ll). The control peptide 1l′, which derived from 1l by replacing Cys with Ala, didn't react with 2i and no arylation product was detected, further demonstrating the excellent chemo-selectivity to Cys over other amino acids. The stability of the modified peptides 3li, 3ls, 3lv, 3la and 3ll to acids, bases, oxidants and external thiols was tested (Fig. S150†). The results showed that all tested peptides were stable in AcOH buffer (pH 4.0), 5 mM K2CO3 solution and Tris buffer (pH 8.0) after 24 h. In 5 mM H5IO6 solution, approximately 50% of 3li, 3ls and 3la remained, while 3lv and 3ln were completely oxidized after 24 h, demonstrating that the modified aryl sulfides were relatively stable compared to alkyl sulfides under oxidizing conditions.10a Most interestingly, modified peptides bearing pyrazolopyrimidine skeletons (3li, 3ls, and 3lv) could be attacked by external thiols and recovered to their native unmodified state, while purine skeleton based modified peptides (3la, 3ln) could not, indicating the possibility of regenerating peptides/proteins after modification by simply adjusting the skeleton structure of aryl thioethers (Fig. S150†).
To verify our hypothesis that the regenerating ability of the modified peptides could be adjusted by changing the skeleton of aryl thioethers, we performed the Cys arylation reaction using model peptide 1a with different arylation reagents under the optimized conditions, followed by adding external thiols mercaptoethanol (βME) or glutathione (GSH) to the reaction mixture, and monitored the process by LC-MS. The results showed that modified peptides bearing different aryl skeletons exhibited distinct regenerating activity (Fig. 4A and Fig. S151†). The purine based products were relatively stable in βME or GSH (3aa, 3ab and 3ah), while quinazoline (3ae), triazole pyrimidine (3ag) and pyrazolopyrimidine (3ai and 3aj) based modified peptides could be easily attacked by βME and recovered to the unmodified state. The introduction of an electron-withdrawing group into the arylation reagents led to an improved native peptide regeneration ability with βME but reduced ability with GSH, which might be attributed to the synchronously improved reactivity to thiols and steric hindrance of the modified peptides 3, thereby accelerating the regeneration process with βME and preventing the native peptide regeneration with GSH (Fig. 4A and S151,†3aa, 3ab, 3ah–3aj). These results clearly demonstrated that a cleavable, tunable and Cys-specific peptide modification could be achieved by simply adjusting the skeletons and steric hindrance of our newly developed aryl thioethers. To demonstrate the generality of the cleavable and tunable Cys modification approach, we chose 2ad and 2i that showed different regeneration abilities to modify peptides, followed by adding βME to the reaction mixture and analysed by LC-MS. The results showed that the 2i modified peptides 3 with a peptide length ranging from 4-mer to 14-mer all could be effective attacked by βME and recovered to the native state 1 with a recovery ratio over 90% after 1 h, while 2ad modified peptides showed a weak regeneration ability with a recovery ratio of less than 14% (Fig. 4B).
Protein modification is vital to explore their structures and functions. To investigate the possibility of our method for protein modification, we tested the reactivity of the developed benzoxazole sulfide on bovine serum albumin (BSA). Benzoxazole sulfide 2v equipped with an affinity label was incubated with BSA to afford the modified product with good efficiency under slightly modified reaction conditions (Fig. 5A, B). Besides, secondary labelling of the modified peptide was also investigated. The modified product 3ls bearing an azide handle could react with fluorescent tag 4 to afford the desired fluorescent product 5via the azide–alkyne click reaction, suggesting the potential application of this method in biochemistry (Fig. 5C).
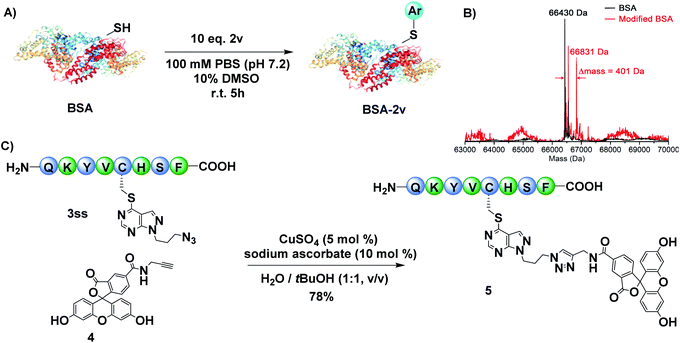 | ||
| Fig. 5 (A) BSA modification with 2v. (B) ESI-MS spectra of native BSA and modified BSA-2v proteins. (C) Secondary labeling of modified peptide 3ss. | ||
Conclusions
In conclusion, we developed a class of aryl thioethers for cysteine-containing peptide/protein modification with excellent chemoselectivity and efficiency via a SNAr approach. This modification strategy has broad substrate scope and functional group compatibility under optimized reaction conditions. A variety of biorelevant groups could be well introduced into modification agents. By simply adjusting the skeleton of aryl thioethers, a cleavable and regeneration tunable modification for peptides/proteins could be achieved. Moreover, secondary labeling of modified peptides was also realized, indicating the broad application of our method in the peptide area.Author contributions
Xiao-Feng Xiong conceived the study and was responsible for the funding acquisition, project administration and writing of the manuscript. Jian Li designed and performed the study, collected and analysed the data, and wrote the original draft. The other authors also assisted in analysing and validating the data and were involved in preparing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge financial support by the National Natural Science Foundation of China (No. 22077144, 81602972, and 81872836), Guangdong Natural Science Funds for Distinguished Young Scholar (No. 2018B030306017), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2018), Fundamental Research Funds for the Central Universities (20ykzd15), and Key Research and Development Program of Guangdong Province (2020B1111110003).Notes and references
- (a) C. D. Spicer and B. G. Davis, Nat. Commun., 2014, 5, 4740 CrossRef CAS; (b) O. Boutureira and G. J. Bernardes, Chem. Rev., 2015, 115, 2174–2195 CrossRef CAS; (c) J. N. deGruyter, L. R. Malins and P. S. Baran, Biochemistry, 2017, 56, 3863–3873 CrossRef CAS PubMed; (d) N. Krall, F. P. da Cruz, O. Boutureira and G. J. Bernardes, Nat. Chem., 2016, 8, 103–113 CrossRef CAS; (e) L.-G. Milroy, T. N. Grossmann, S. Hennig, L. Brunsveld and C. Ottmann, Chem. Rev., 2014, 114, 4695–4748 CrossRef CAS; (f) E. A. Hoyt, P. M. S. D. Cal, B. L. Oliveira and G. J. L. Bernardes, Nat. Rev. Chem., 2019, 3, 147–171 CrossRef CAS; (g) X. Li, S. Chen, W.-D. Zhang and H.-G. Hu, Chem. Rev., 2020, 120, 10079–10144 CrossRef CAS PubMed.
- (a) C. P. Hackenberger and D. Schwarzer, Angew. Chem., Int. Ed., 2008, 47, 10030–10074 CrossRef CAS PubMed; (b) W. Wang, M. M. Lorion, J. Shah, A. R. Kapdi and L. Ackermann, Angew. Chem., Int. Ed., 2018, 57, 14700–14717 CrossRef CAS PubMed; (c) Q.-L. Hu, K.-Q. Hou, J. Li, Y. Ge, Z.-D. Song, A. S. C. Chan and X.-F. Xiong, Chem. Sci., 2020, 11, 6070–6074 RSC; (d) J. C. Vantourout, S. R. Adusumalli, K. W. Knouse, D. T. Flood, A. Ramirez, N. M. Padial, A. Istrate, K. Maziarz, J. N. deGruyter, R. R. Merchant, J. X. Qiao, M. A. Schmidt, M. J. Deery, M. D. Eastgate, P. E. Dawson, G. J. L. Bernardes and P. S. Baran, J. Am. Chem. Soc., 2020, 142, 17236–17242 CrossRef CAS.
- (a) S. R. Adusumalli, D. G. Rawale, U. Singh, P. Tripathi, R. Paul, N. Kalra, R. K. Mishra, S. Shukla and V. Rai, J. Am. Chem. Soc., 2018, 140, 15114–15123 CrossRef CAS; (b) M. J. Matos, B. L. Oliveira, N. Martínez-Sáez, A. Guerreiro, P. M. S. D. Cal, J. Bertoldo, M. Maneiro, E. Perkins, J. Howard, M. J. Deery, J. M. Chalker, F. Corzana, G. Jiménez-Osés and G. J. L. Bernardes, J. Am. Chem. Soc., 2018, 140, 4004–4017 CrossRef CAS; (c) A. F. M. Noisier, M. J. Johansson, L. Knerr, M. A. Hayes, W. J. Drury III, E. Valeur, L. R. Malins and R. Gopalakrishnan, Angew. Chem., Int. Ed., 2019, 58, 19096–19102 CrossRef CAS PubMed; (d) Q. Luo, Y. Tao, W. Sheng, J. Lu and H. Wang, Nat. Commun., 2019, 10, 142 CrossRef PubMed; (e) N. C. Reddy, M. Kumar, R. Molla and V. Rai, Org. Biomol. Chem., 2020, 18, 4669–4691 RSC.
- (a) S. M. Marino and V. N. Gladyshev, J. Mol. Biol., 2010, 404, 902–916 CrossRef CAS; (b) M. Lo Conte, S. Staderini, A. Marra, M. Sanchez-Navarro, B. G. Davis and A. Dondoni, Chem. Commun., 2011, 47, 11086–11088 RSC; (c) D. Abegg, R. Frei, L. Cerato, D. Prasad Hari, C. Wang, J. Waser and A. Adibekian, Angew. Chem., Int. Ed., 2015, 54, 10852–10857 CrossRef CAS PubMed; (d) A. Abbas, B. Xing and T.-P. Loh, Angew. Chem., Int. Ed., 2014, 53, 7491–7494 CrossRef CAS PubMed.
- H. P. Hemantha, S. N. Bavikar, Y. Herman-Bachinsky, N. Haj-Yahya, S. Bondalapati, A. Ciechanover and A. Brik, J. Am. Chem. Soc., 2014, 136, 2665–2673 CrossRef CAS PubMed.
- (a) J. Yu, X. Yang, Y. Sun and Z. Yin, Angew. Chem., Int. Ed., 2018, 57, 11598–11602 CrossRef CAS; (b) N. Lundell and T. Schreitmuller, Anal. Biochem., 1999, 266, 31–47 CrossRef CAS PubMed; (c) M. E. B. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed; (d) Y. Zhang, C. Zang, G. An, M. Shang, Z. Cui, G. Chen, Z. Xi and C. Zhou, Nat. Commun., 2020, 11, 1015 CrossRef CAS PubMed.
- (a) Y. Wang and D. H.-C. Chou, Angew. Chem., Int. Ed., 2015, 54, 10931–10934 CrossRef CAS; (b) R. Huang, Z. Li, Y. Sheng, J. Yu, Y. Wu, Y. Zhan, H. Chen and B. Jiang, Org. Lett., 2018, 20, 6526–6529 CrossRef CAS PubMed.
- (a) R. Tessier, J. Ceballos, N. Guidotti, R. Simonet-Davin, B. Fierz and J. Waser, Chem, 2019, 5, 2243–2263 CrossRef CAS; (b) R. Tessier, R. K. Nandi, B. G. Dwyer, D. Abegg, C. Sornay, J. Ceballos, S. Erb, S. Cianférani, A. Wagner, G. Chaubet, A. Adibekian and J. Waser, Angew. Chem., Int. Ed., 2020, 59, 10961–10970 CrossRef CAS.
- C. Zhang, E. V. Vinogradova, A. M. Spokoyny, S. L. Buchwald and B. L. Pentelute, Angew. Chem., Int. Ed., 2019, 58, 4810–4839 CrossRef CAS PubMed.
- (a) E. V. Vinogradova, C. Zhang, A. M. Spokoyny, B. L. Pentelute and S. L. Buchwald, Nature, 2015, 526, 687–691 CrossRef CAS PubMed; (b) M. S. Messina, J. M. Stauber, M. A. Waddington, A. L. Rheingold, H. D. Maynard and A. M. Spokoyny, J. Am. Chem. Soc., 2018, 140, 7065–7069 CrossRef CAS PubMed; (c) A. J. Rojas, B. L. Pentelute and S. L. Buchwald, Org. Lett., 2017, 19, 4263–4266 CrossRef CAS PubMed; (d) C. Bottecchia, M. Rubens, S. B. Gunnoo, V. Hessel, A. Madder and T. Noel, Angew. Chem., Int. Ed., 2017, 56, 12702–12707 CrossRef CAS PubMed.
- (a) C. M. Johnson, T. W. Linsky, D. W. Yoon, M. D. Person and W. Fast, J. Am. Chem. Soc., 2011, 133, 1553–1562 CrossRef CAS PubMed; (b) N. C. Price, M. Cohn and R. H. Schirmer, J. Biol. Chem., 1975, 250, 644–652 CrossRef CAS PubMed.
- (a) A. M. Embaby, S. Schoffelen, C. Kofoed, M. Meldal and F. Diness, Angew. Chem., Int. Ed., 2018, 57, 8022–8026 CrossRef CAS; (b) A. M. Spokoyny, Y. Zou, J. J. Ling, H. Yu, Y.-S. Lin and B. L. Pentelute, J. Am. Chem. Soc., 2013, 135, 5946–5949 CrossRef CAS PubMed.
- (a) D. Zhang, N. O. Devarie-Baez, Q. Li, J. R. Lancaster Jr and M. Xian, Org. Lett., 2012, 14, 3396–3399 CrossRef CAS PubMed; (b) N. Toda, S. Asano and C. F. Barbas III, Angew. Chem., Int. Ed., 2013, 52, 12592–12596 CrossRef CAS PubMed; (c) C. Zambaldo, E. V. Vinogradova, X. Qi, J. Iaconelli, R. M. Suciu, M. Koh, K. Senkane, S. R. Chadwick, B. B. Sanchez, J. S. Chen, A. K. Chatterjee, P. Liu, P. G. Schultz, B. F. Cravatt and M. J. Bollong, J. Am. Chem. Soc., 2020, 142, 8972–8979 CrossRef CAS PubMed.
- (a) E. Weerapana, G. M. Simon and B. F. Cravatt, Nat. Chem. Biol., 2008, 4, 405–407 CrossRef CAS PubMed; (b) G. C. Adam, B. F. Cravatt and E. J. Sorensen, Chem. Bio., 2001, 8, 81–95 CrossRef CAS; (c) G. J. L. Bernardes, M. Steiner, I. Hartmann, D. Neri and G. Casi, Nat. Protoc., 2013, 8, 2079–2089 CrossRef CAS PubMed; (d) C. Chatterjee and T. W. Muir, J. Biol. Chem., 2010, 285, 11045–11050 CrossRef CAS PubMed.
- (a) H. Faustino, M. Silva, L. F. Veiros, G. J. L. Bernardes and P. M. P. Gois, Chem. Sci., 2016, 7, 5052–5058 RSC; (b) A. Bandyopadhyay, S. Cambray and J. Gao, Chem. Sci., 2016, 7, 4589–4593 RSC; (c) S. Arumugam, J. Guo, N. E. Mbua, F. Friscourt, N. Lin, E. Nekongo, G.-J. Boons and V. V. Popik, Chem. Sci., 2014, 5, 1591–1598 RSC; (d) M. E. Smith, F. F. Schumacher, C. P. Ryan, L. M. Tedaldi, D. Papaioannou, G. Waksman, S. Caddick and J. R. Baker, J. Am. Chem. Soc., 2010, 132, 1960–1965 CrossRef CAS PubMed; (e) D. P. Nguyen, M. Mahesh, S. J. Elsässer, S. M. Hancock, C. Uttamapinant and J. W. Chin, J. Am. Chem. Soc., 2014, 136, 2240–2243 CrossRef CAS PubMed; (f) N. Wu, A. Deiters, T. A. Cropp, D. King and P. G. Schultz, J. Am. Chem. Soc., 2004, 126, 14306–14307 CrossRef CAS PubMed; (g) Y. Zhang, X. Zhou, Y. Xie, M. M. Greenberg, Z. Xi and C. Zhou, J. Am. Chem. Soc., 2017, 139, 6146–6151 CrossRef CAS PubMed.
- (a) A. K. Pathak, V. Pathak, L. E. Seitz, W. J. Suling and R. C. Reynolds, J. Med. Chem., 2004, 47, 273–276 CrossRef CAS PubMed; (b) Y. B. Bhujabal, K. S. Vadagaonkar, A. Gholap, Y. S. Sanghvi, R. Dandela and A. R. Kapdi, J. Org. Chem., 2019, 84, 15343–15354 CrossRef CAS PubMed.
- W. J. Ong and T. M. Swager, Nat. Chem., 2018, 10, 1023–1030 CrossRef CAS PubMed.
- The LC-MS yields were determined as follows: %yield = Sproduct/Stotal, where Sproduct is the peak area of the product and Stotal is the peak area of combined peptide-containing species (product, starting material and by-product).
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc06576e |
| This journal is © The Royal Society of Chemistry 2021 |