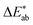Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A data-driven perspective on the colours of metal–organic frameworks†
Kevin Maik
Jablonka
 a,
Seyed Mohamad
Moosavi
a,
Seyed Mohamad
Moosavi
 a,
Mehrdad
Asgari
a,
Mehrdad
Asgari
 bc,
Christopher
Ireland
bc,
Christopher
Ireland
 a,
Luc
Patiny
a,
Luc
Patiny
 d and
Berend
Smit
d and
Berend
Smit
 *a
*a
aLaboratory of Molecular Simulation, Institut des Sciences et Ingénierie Chimiques, École Polytechnique Fédérale de Lausanne (EPFL), Rue de l'Industrie 17, CH-1951 Sion, Switzerland. E-mail: berend.smit@epfl.ch
bInstitute of Mechanical Engineering (IGM), School of Engineering (STI), École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland
cInstitut des Sciences et Ingénierie Chimiques, École Polytechnique Fédérale de Lausanne (EPFL), Rue de l'Industrie 17, CH-1951 Sion, Valais, Switzerland
dInstitute of Chemical Sciences and Engineering, École Polytechnique Fédérale de Lausanne (EPFL), CH-1015 Lausanne, Switzerland
First published on 28th December 2020
Abstract
Colour is at the core of chemistry and has been fascinating humans since ancient times. It is also a key descriptor of optoelectronic properties of materials and is often used to assess the success of a synthesis. However, predicting the colour of a material based on its structure is challenging. In this work, we leverage subjective and categorical human assignments of colours to build a model that can predict the colour of compounds on a continuous scale. In the process of developing the model, we also uncover inadequacies in current reporting mechanisms. For example, we show that the majority of colour assignments are subject to perceptive spread that would not comply with common printing standards. To remedy this, we suggest and implement an alternative way of reporting colour—and chemical data in general. All data is captured in an objective, and standardised, form in an electronic lab notebook and subsequently automatically exported to a repository in open formats, from where it can be interactively explored by other researchers. We envision this to be key for a data-driven approach to chemical research.
Introduction
Colours have been attracting the attention of humans for a long time and are one key aspect that makes chemistry interesting.1 Chemists have some intuition within compound classes how they can tune the colours. For organic compounds, group-contribution methods like the Woodward rules found wide acceptance.2 In other cases, chemists might have an intuition if certain transitions are allowed or forbidden, e.g., based on Laporte's rules in metal complexes.3 However, colours can be influenced by very subtle effects which led some authors to conclude that “the prediction of the colouring properties of yet unsynthesised compounds is a very risky business which still remains in the realm of art rather than of science”.1 Even though we can use quantum chemical calculations to estimate band gaps,4,5 or even the full dielectric function and absorption spectrum,6,7 those calculations are computationally prohibitive for large unit cells and require careful consideration of non-ideal effects such as defects.8In this work, we focus on the colours of metal–organic frameworks (MOFs). MOFs are crystalline materials with a unique chemical tunability.9 By changing the metal node and the organic linker, we can synthesise millions of possible materials. These materials have many interesting applications, ranging from gas storage and separations,10 (photo)catalysis,11 to sensing,12 and luminescence.13 The chemical toolbox, like the substitution of metal and linkers, with which the optical properties of MOFs can be tuned has been exploited in several works in the past.14–17 For applications of MOFs that rely on the optical properties of MOFs (e.g., photocatalysis, sensing, luminescence, optoelectronics) selecting a MOF with the right colour is important. Additionally, the colour is also of importance to assess the success of their synthesis, work-up, and activation—this is one of the reasons why the colour of the products is usually reported in method sections.
In the Cambridge Structure Database (CSD)18 some experimental groups report together with the structure also the colour of the MOF (circa 9000 structures of the more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures19 in the MOF subset20 of the CSD). In this work, we show that this data can be harvested using machine learning to arrive at a tool that can efficiently predict the colour of a MOF. It is important to realise that at present the CSD is the only data source that we have at disposal to develop a model to predict optical properties of MOFs as we are not aware of any large dataset that reports reliable optical gaps or other optical properties of MOFs. In this database, the amount of data is relatively small, and the naming of colours is unfortunately rather subjective. From a machine learning point of view, such little and subjective data pose an interesting scientific question, which we attempt to answer here: how we can use such data to build a surrogate model for the optical properties of MOFs, what we learn by doing so, and more importantly, how could we improve this situation moving forward?
000 structures19 in the MOF subset20 of the CSD). In this work, we show that this data can be harvested using machine learning to arrive at a tool that can efficiently predict the colour of a MOF. It is important to realise that at present the CSD is the only data source that we have at disposal to develop a model to predict optical properties of MOFs as we are not aware of any large dataset that reports reliable optical gaps or other optical properties of MOFs. In this database, the amount of data is relatively small, and the naming of colours is unfortunately rather subjective. From a machine learning point of view, such little and subjective data pose an interesting scientific question, which we attempt to answer here: how we can use such data to build a surrogate model for the optical properties of MOFs, what we learn by doing so, and more importantly, how could we improve this situation moving forward?
The idea of machine learning is that similar MOFs will have similar colours. However, the concept of similarity depends on the property we are interested in.21 It is therefore important to develop a machine learning approach that closely follows the chemical intuition that chemists have developed over many decades in what makes two MOFs give a similar colour. Hence, in our machine learning approach, we ensure that our descriptors can capture all possible ways of electronic transitions that can lead to different colours. This includes information about the metal to be able to describe metal-centred transitions, information about the ligands, and the functional groups to be able to describe ligand-centred transitions, as well as information about the interaction between metal and ligands to be able to describe metal–ligand or ligand–metal transitions. Only if we ensure that our featurisation has the expressivity to describe these phenomena efficiently and effectively, we can combine our chemical insights into what is important in determining a colour with a relatively small database of materials of which we know the colour.
The other interesting point is that a colour can be a well-defined property. One can precisely specify the colour of a material by its tristimulus values in some colour space, similar to how the human eye perceives colour with three types of cones.22 Such high-quality data for MOFs is scarce. In fact, the data in the CSD are the (subjective) names given by the groups that have synthesised the material. If the name is blue or red, it might be clear to which colour (range) the group is referring to, but if the given colour of a MOF reads “straw yellow” or “claybank” it becomes more difficult as we have to take into account that every person has a distinct perception of colours. This is, the colour intended can not easily be inferred from the reported colour name.23 To address this issue we have conducted a survey that allows us to map the colour names to a distribution of coordinates in a colour space (representing the estimates of the likelihood of the colour intended) and also gives us an idea of the perceptive spread for different colour names. This survey also revealed that the current way colour is reported in chemistry is inadequate. It hampers data-driven approaches to chemistry and also limits the reproducibility.
In addition, we also present some very practical tools. One is a web application that allows uploading a structure and the app predicts the colour together with an uncertainty estimate. The other app allows measuring the colour of a MOF based on a picture of the synthesised material, such that chemists can report the tristimulus values (like coordinates in RGB space) together with their favourite name for the colour of the material. We hope that the latter can help to improve the reporting of colours in chemistry. Furthermore, we demonstrate how an electronic lab notebook can be used to capture and share data in standardised and digital from, enabling interactive, and digital, Supporting Information (SI) documents† (see https://go.epfl.ch/colourSI for an example) that are much more accessible to data mining efforts than classical SI documents in portable document format (PDF). Importantly, these interactive electronic SI documents are not limited to our particular application. We envision them to be an important part of chemical publishing in the future.
Colours and their perception
Fig. 1 illustrates some words that are used in the CSD to describe the colour of MOFs. From a machine learning point of view, such discrete and subjective data are of limited use. First, using the names of the colours we cannot easily encode that confusing orange with yellow is not as bad as confusing black and white. Further, if for some colours the spread of the perception is wide, we also would not be surprised if the model is unsure about the colour. | ||
| Fig. 1 Words that are used in the CSD to describe colours. The words are coloured using the median colour (unweighted average in RGB space) from the survey. The size of the words is proportional to their frequency. Figure generated using the WordCloud library.24 | ||
The perception of colour has already been studied in a widely known survey conducted by Randall Munroe (creator of the webcomic xkcd).25 Previous approaches for mapping colours to colour names were mostly focused on human-curated dictionaries. In contrast to that, for Munroe's dataset, nearly half a million participants named colours which they were shown. The large number of participants made this dataset an important reference for data-driven approaches to natural language processing.23,26–30 For our purpose, we are interested in the reverse question, mimicking how chemists would try to assess if they successfully reproduced a colour reported in the literature: given a name what is the colour one would associate with this name and how large is the spread of these colours associated with a colour name? This question, that is also important for natural language understanding, is less widely studied than the reverse one,23,31,32 and information we cannot easily obtain from the xkcd survey. First, because one-third of the colours that are used in the CSD to describe the colour of a MOF are not represented in the xkcd survey and, second, because we are also interested in the spread of responses to get a baseline of how well we can expect our model to perform in different parts of colour space. To obtain some insight into this question we carried out a survey resulting in 4184 assignments of colours to one of the 162 names that occurred in the CSD for colours of MOFs. In the ESI,† the details of the survey are given. Note that in contrast to other works23,30,33,34 based on the xkcd survey we did not attempt to build a general model that maps colour names to the tristimulus coordinates of the intended colour but rather want to infer the likelihood of the intended colour for all colour names that are used for MOFs in the CSD.
Perceptive spread of colours and the current way of reporting colours
The first question to pose is if our survey results can give us meaningful insights, i.e., whether the statistics are good enough, and our data are representative. One way to estimate this is to compare our results with the ones from the xkcd survey for the colours that overlap between both surveys. That is, we ask if the median of the colour distribution obtained from our survey corresponds to the colour that has been given this name in the xkcd survey. From ESI Fig. 8† we see that our findings, in general, agree well with the ones from the xkcd survey. Still, when we analyse the individual submissions, we find that there is a considerable spread in the colours the users selected—even after filtering out outliers (for example, we discarded submissions if the colour was picked in less than 5 s). In Fig. 2 we show the spread in the responses for some colours. It is instructive to quantify the spread in colours. A widely used metric to quantify differences between colours is the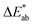 score (using the CIEDE2000 formula),35 which takes into account that the human eye is more sensitive to certain colours. For professional prints one typically expects36
score (using the CIEDE2000 formula),35 which takes into account that the human eye is more sensitive to certain colours. For professional prints one typically expects36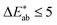 and a
and a 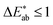 is said to be undetectable for the human eye.37 Notably, we found in our survey only black to have a median
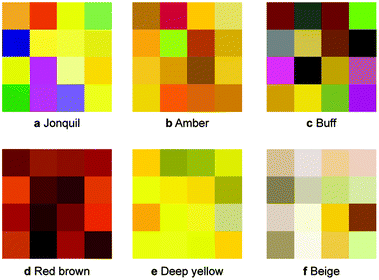
is said to be undetectable for the human eye.37 Notably, we found in our survey only black to have a median 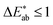 and only five colours in total (black, red, white, whitish colourless, yellow, corresponding to less than four per cent of all colours in our survey) have a median difference between the responses in the survey that would satisfy common printing standards. The overall median of the differences is approximately 10 (mean: 12). This implies that if we filtered out all high variance colours, we would have too little data to train our model (see ESI Fig. 10†). Still, we observe that for some colour names like “jonquil” or “buff” the spread is so large that it is not practical for use in training a model (more discussion in Section 2 of the ESI†).
and only five colours in total (black, red, white, whitish colourless, yellow, corresponding to less than four per cent of all colours in our survey) have a median difference between the responses in the survey that would satisfy common printing standards. The overall median of the differences is approximately 10 (mean: 12). This implies that if we filtered out all high variance colours, we would have too little data to train our model (see ESI Fig. 10†). Still, we observe that for some colour names like “jonquil” or “buff” the spread is so large that it is not practical for use in training a model (more discussion in Section 2 of the ESI†).
For our current study, our simple survey allows us to replace the discrete names, like “cherry red”, with a distribution of colour coordinates in some colour space which we can use as data in our machine learning approach. This approach is sufficient to demonstrate the potential of machine learning in predicting colours. But the fact that we have to use a survey to quantify colour does illustrate that the current way of reporting colours is inadequate—especially in the face of the variance which we observe in the survey results. Clearly, the problems with colour reporting go beyond natural language. For example, the concept of colour constancy (the way in which our brain resolves inconsistent colour signals when the illumination changes) was suggested as an explanation for the different colours humans perceived for “the dress” that went viral in 2015.38,39 Since in science we want to record information in a way that is invariant to subjective perception, we need a new way to record and report colours in chemistry.
Colour reporting and integration with an ELN
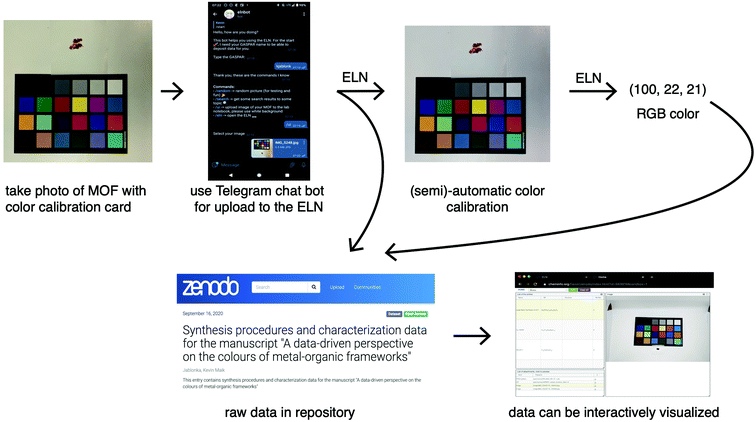
For testing of our machine learning approach with some recently synthesised MOFs, we used a more objective and accurate way of recording colours. The idea is to take a photo of the material together with a colour rendition card. Such an image can then be automatically uploaded to an electronic lab notebook40 (ELN, see Fig. 3). This image can then, with all the characterisation data, be shared in digital, and standardised, form via a repository from where it can be accessed for data mining. A dedicated website can be used to visualise the data deposited in the repository (using the same code that is also used for data visualisation in the ELN). Importantly, our ELN makes it possible to perform this export and publication of findable, accessible, interoperable and reusable (FAIR)41 data for any kind of characterisation method and not only for this specific case. For example, the repository entry for this work also contains the X-ray diffraction patterns, thermogravimetric analysis or UV/Vis spectra for some materials.Since the images we take of the MOFs also contain a colour rendition chart, we can perform colour calibration. By means of the colour calibration, the colour profile can be standardised, which can then be harnessed for more accurate colour measurements. In principle, one could also use a spectrophotometer to quantify the colour of a material. We decided to use images as we found it to be faster (also for small amounts of activated compounds). Moreover, the image records additional important information like the morphology, or reflexive properties, of the sample. Ideally though, one would record as much information as possible.
To facilitate this first step towards a good practice of accurate reporting of the colour of a material we have developed a web application. Our web application (https://go.epfl.ch/colorcalibrator) uses a fully automatic routine that automatically detects the colour rendition chart.42 The user only needs to upload a photo of the MOF with a colour rendition chart and select an area over which the colour averaging should be performed.
The implication of this infrastructure, which directly connects the capture of the data with the publication, is that if it were used by many groups, we would create much more valuable data that would make works more comparable and machine learning methods thrive. Also, we could replace Supporting Information documents in portable document format (PDF) with data that is alive and reusable. As we did for this article, researchers could just report the digital object identifier (DOI) for their repository entry instead of, or in addition to, providing the PDF.
Model development
To build a robust model it is instrumental that two materials that have structures that are close in terms of their colours are also close in terms of the descriptors. The intuition here is to encode the nodes, the linkers, and the functional groups separately by using correlations on a structure graph coloured with some chemically sensible heuristics such as the electronegativity or polarisability. This is, the model will be able to recognise “colouring” functional groups by their characteristic autocorrelation functions. To achieve this, we use the revised autocorrelation (RAC) function43 formalism which was used in the past to predict electronic properties of metal complexes44,45 and recently adapted for MOFs.46 RACs are discrete correlations between heuristics (e.g., Pauling electronegativity) of atoms on the structure graph which are then pooled together for small fragments of different size. For MOFs, we calculate those descriptors separately for linkers, functional groups and the nodes. We augment this set of features with additional descriptors for the linkers, such as the number of aromatic rings, aromatic bond, or double bonds, that we anticipate having a high association with the colour of the compound (see Section 4.1 of the ESI† for more details).For making the predictions based on those descriptors, we use a gradient boosted decision tree (GBDT) model, which is an ensemble of decision trees that are iteratively fitted on the residual of the previous decision tree to predict tristimulus values that are close to the median colour coordinates we extracted from our survey for the colour name of a given MOF. We found this method to perform best across a range of other models we tested (see ESI Section 4†). We built our model based on 6423 structures from the structures in the MOF subset with a colour attribute, from which we dropped duplicates to avoid data leakage (see ESI Section 3†).
Model evaluation
To allow for evaluation of our model, we held out a test set of structures which we did not use for training or hyperparameter tuning. Our model achieves a good predictive performance for those structures, as shown in some examples of randomly picked predictions in ESI Fig. 13† and in numerical metrics [mean absolute error (MAE) = 0.14 (0.13, 0.15), r2 = 0.54 (0.50, 0.57), a mean baseline gives MAE = 0.31 (0.31, 0.32), r2 = 0 (0, 0)] calculated over the full test set. One may wonder how these numbers, i.e. the performance of our model, compares to the perceptive spread we observed in the survey. Above, we calculated the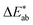 differences for each colour in our survey and did the same for our (baseline) models (Fig. 4). That is, a small
differences for each colour in our survey and did the same for our (baseline) models (Fig. 4). That is, a small 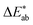 indicates that the colour predicted by our model is close to the median tristimulus values that we extracted for a given MOF colour name using the survey. We observe that the distribution of colour differences for our model has a more pronounced tail of larger differences, and also a larger median of 17 (16, 18), compared to a median of 10 (mean: 12) for the in-survey differences. But the fact that our median is close the median of the in survey errors reflects that our model is mostly limited by the inherent variance of the data (given that the learning curves in ESI Fig. 20† did not saturate). Interestingly, about 28% of our predictions are less than 5
indicates that the colour predicted by our model is close to the median tristimulus values that we extracted for a given MOF colour name using the survey. We observe that the distribution of colour differences for our model has a more pronounced tail of larger differences, and also a larger median of 17 (16, 18), compared to a median of 10 (mean: 12) for the in-survey differences. But the fact that our median is close the median of the in survey errors reflects that our model is mostly limited by the inherent variance of the data (given that the learning curves in ESI Fig. 20† did not saturate). Interestingly, about 28% of our predictions are less than 5 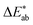 units (the tolerance used for printing) from the median of the survey.
units (the tolerance used for printing) from the median of the survey.
By training models to predict also the quantiles, i.e., the error bars around the median, we observed that the model often is uncertain about the intensity of the colour, e.g., the 90th percentile is frequently close to colourless. This points to another problem with the reporting of colours—the colour string often gives no information about the chromatic intensity. Indeed, if we analyse the colour names in the CSD we observe that only one-third of all colour strings have intensity information such as “light” or “dark” in the name—and even then the exact position on the continuum of intensities is not well-defined.
Test on experimental compounds
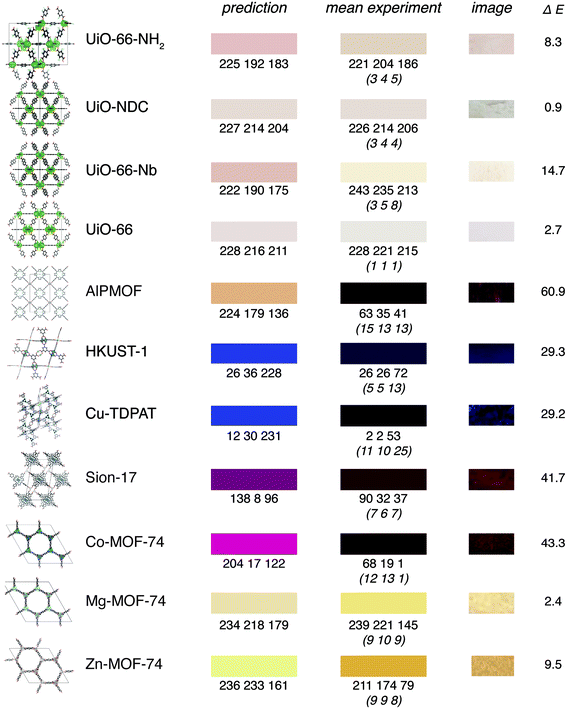
For some compounds that our experimental colleagues had recently prepared for testing of our model, we recorded the colour as outlined in Fig. 3 and used our model to predict the colour. For all compounds, we ensured that we include no other too similar compound within some distance in the feature space in the training set (see ESI Section 6.2†).We can observe that even though the predictions might be quantitatively not perfect, given the uncertainties in the way colours are reported in the CSD, our results are certainly encouraging (Fig. 5). This is also reflected in the fact that the mean absolute error of our model is close to the mean variation of the colours in our survey.
In particular, we capture many interesting trends. For example, our model recognises that the addition of an amino group leads to a redshift for UiO-66. Likewise, we can analyse the influence of metal substitutions, e.g., the doping of UiO-66-NH2 with Nb leading to a redshift as described by Syzgantseva et al.47
What did the model learn?
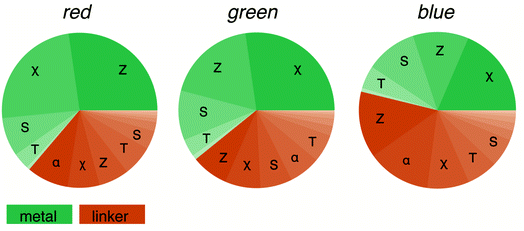
Machine learning is often seen as a black box, in which we replace our chemical knowledge and intuition by plain statistics.48 However, we can analyse the importance of the different features, and this feature analysis will tell us what the most important features are. Here, we are interested in which features make a MOF red (R), green (G), or blue (B) for our model.For this, we split the features into metal-centred and linker-centred contributions and evaluate their absolute importance as a function of the colour channel. Fig. 6 shows that for our model the characteristics of the metal is most important for the red colour channel. For the blue colour channel, being more relevant for absorption at longer wavelengths (the complementary colours are absorbed), the linker chemistry is more important for our model. But in no instance, the model relies solely on metal or linker and descriptors (for discussion of the interactions between the features see the ESI Section 7†). This supports the notion that for visible-light-driven applications of MOFs the interaction between metal and linker is important (linker-to-metal-cluster charge-transfer mechanism, LCCT).49
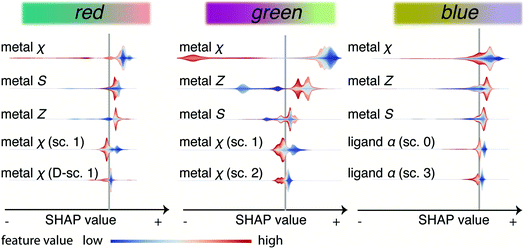
Our models indicate that linker modification is important to tune absorption in the visible regime, which triggers an important practical question. Can our model give us some insights about how we can tune the material to steer the optical response? We can get more insight into the direction in which the features influence the colours by analysing Fig. 7. This figure lists the five most important features (biggest slices in Fig. 6). This graph gives the SHAP value of each property, which is a measure for the impact on the output of the model, on the x-axis. A high absolute SHAP value means the feature has a large impact, which can be positive (increasing the R, G, or B values) or negative. For each material and feature, we get a SHAP value, and the colour coding indicates whether the feature value is high or low. For example, the violin plot for the red colour channel shows that metals with a low χ (blue), all have a positive SHAP value, indicating a low electronegativity leads to a higher output on the red colour channel.
Overall, we observe that the colourfulness primarily depends on the position of the metal (metal Z, χ, S) in the periodic table—broadly speaking, a high electronegativity tends to decrease the output on all colour channels (especially red and green). Similarly, we see increasing atomic number leading to increased output on all colour channels—but all those trends are not simple monotonic relationships. These observations can be thought of as a refined version of previous suggestions that an electron-rich metal centre (soft, i.e., low χ and large S) can be used to decrease the bandgap.50,51 But for our model, this does not happen equally for all colour channels. For example, for the blue colour channel we see strong interaction effects of the metal features with linker features. For instance, high and low values of χ and Z give rise to the same SHAP value and the impact on the output will depend on the value of linker features for a given value of the metal features. We suppose that this reflects that for an LCCT transition the energy levels of the metal cluster and the linker need to be properly aligned.
One interesting case to understand how the model learns is the case of HKUST-1 for which Müller et al. have shown that the green-blue colour that is typically observed for powders of this material is due to d–d transitions in defective paddlewheels (in perfect structures, the selection rules for the 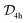 symmetry lead to only weak transitions).8 Not surprisingly, our model predicts a blue colour for this MOF as this is the colour reported in the CSD for HKUST-1 (CSD reference codes BODPAN, FIQCEN, but we excluded it from the training set). This is likely also one of the reasons why we predict blue for UMCM-152 (CSD reference code ANUGIA) which is reported as blue (dark-purple after drying) in the paper52 but as colourless in the CSD. Predicting colours that are due to defects would not be possible in a first-principles approach based on idealised crystal structures (for which transitions might be forbidden due to selection rules), but is, as we show, possible in a data-driven approach. Since the model learns chemical similarities in some descriptor space it will predict similar colours for similar MOFs that might have similar defects—which might not be directly clear from the crystal structure, as in the case of HKUST-1.
symmetry lead to only weak transitions).8 Not surprisingly, our model predicts a blue colour for this MOF as this is the colour reported in the CSD for HKUST-1 (CSD reference codes BODPAN, FIQCEN, but we excluded it from the training set). This is likely also one of the reasons why we predict blue for UMCM-152 (CSD reference code ANUGIA) which is reported as blue (dark-purple after drying) in the paper52 but as colourless in the CSD. Predicting colours that are due to defects would not be possible in a first-principles approach based on idealised crystal structures (for which transitions might be forbidden due to selection rules), but is, as we show, possible in a data-driven approach. Since the model learns chemical similarities in some descriptor space it will predict similar colours for similar MOFs that might have similar defects—which might not be directly clear from the crystal structure, as in the case of HKUST-1.
Conclusions
Predicting the colour of synthesised compounds was long deemed to be a “risky business”. In this work, we showed that it is possible to leverage a relatively small dataset of subjective and categorical assignments of colours to MOF structures to build a predictive model that outputs colours on a continuous scale. Furthermore, we show that the reasoning of our model is chemically meaningful, for example, recovering many aspects of an LCCT transition and recovering trends like colour changes for substitution of metal or ligand.In the process of building our model, we uncovered inadequacies in the way colours are reported. The common practice is to simply provide a name of the colour. Our simple survey shows, for example, that if one reports that the colour of a compound is beige, there is a large variation in colour different people associate with the name beige. In fact, that the variance of perception for most colours is above common tolerances for colour reproduction—ultimately limiting the learning our model. To remedy this, we propose a simple way to improve the reporting of colours. One can only imagine how much we could improve this model if we would have a large dataset of such high-quality data at disposal. Future work needs to focus on creating large scale, objective datasets mapping chemical structures to their colours.
Importantly, colours are only one example where chemical reporting can be improved. Generally, we envision that all reporting should happen in a digital, standardised and unique way. In this work, we provide an example of how this can be done using our electronic lab notebook (ELN). The idea is to take a picture with a smartphone of the sample together with a colour calibration card. This picture gets automatically uploaded into the ELN, and we have provided a tool that automatically recognises the calibration card and by clicking on the sample one can obtain the RGB value of the sample. In addition, from the ELN all data can be exported to a repository in a FAIR format, providing an alternative to the conventional SI in PDF.
Some of our ongoing work focuses on extending the set of characterisation techniques that are supported by our ELN to make this toolbox accessible to a wider group of chemists and materials scientists. Applying machine learning techniques to such standardised datasets might help us then to extract hidden, tactic, knowledge from this data.
Method
Online survey
We developed a custom tool (http://go.epfl.ch/colorjeopardy), based on the Plotly Dash53 Python framework, to conduct the online survey. Users were presented a random colour string (that was used to describe the colour of a MOF in the CSD) and then could use a colour picker to select the colour that most represents this colour string for them. We recorded the colour picked with the sRGB coordinates and the time the users took to select the colours. Note that our setup, similar to the one of the xkcd survey, did not ensure that the users see controlled colours (e.g., on a colour-calibrated monitor). The code is available under MIT License on GitHub (https://github.com/kjappelbaum/colorjeopardy, DOI: 10.5281/zenodo.3831841). The survey results are deposited on Zenodo (https://zenodo.org/record/3831845). Note that since the survey did not collect any personal information, no approval from the institutional review board was required.Colour calibration
More details can be found in Section 8 of the ESI.† The app is deployed at http://go.epfl.ch/colorcalibrator (code is available on GitHub at https://github.com/kjappelbaum/colorcalibrator).Featurisation
To numerically encode the MOF structures, we used RACs, as recently implemented for MOFs in the molSimplify code.46,54 Additionally, we used the SMILES strings of the linkers, as determined using the MOFid package,55 to calculate features that describe the chemistry of the linkers, focusing on aspects that we deem to be important for the colour of compounds—such as the size of the aromatic system, the number of double bonds or functional groups such as amides or carbonyls using Open Babel.56 We z-score standardised the features based on the mean and standard deviation of the training set. All preprocessing was performed using the scikit-learn Python library.57 The feature arrays are deposited on Materials Cloud archive (https://archive.materialscloud.org/record/2020.163).Machine learning
We used the LightGBM implementation of GBDTs, which implements techniques that greatly expedite the training for high feature dimensions and large datasets.58 To obtain prediction intervals, models were trained using the quantile loss function (0.5, i.e., the median prediction corresponding to the mean absolute error loss). For hyperparameter optimisation, we used a Bayesian approach with Gaussian processes as surrogate models (details like the parameter ranges are provided in the ESI Section 4†). For efficiency reasons, we used the same hyperparameters for every colour channel. To calculate the CIE2000 colour differences we used the implementation in the colormath Python package.59 Machine learning experiments were tracked using comet.ml (https://www.comet.ml/kjappelbaum/color-ml?shareable=jfE6okDmxlnYimYFFnsJcMCO6) and wandb (https://app.wandb.ai/kjappelbaum/colorml). The codes for the models (also for the failed attempts) and the analysis is available on GitHub (https://github.com/kjappelbaum/colorml). The numerical metrics that we report are calculated with respect to the median colour labels that we found by mapping the colour strings in the CSD to a distribution of colours through our survey. Confidence intervals (reported in parenthesis following the mean) are determined using the bootstrapping technique, typically with 5000 samples. To stabilise the model (reduce the variance), we employed bagging, i.e., the model was trained on 30 different bootstraps of the training set and the final prediction is the mean prediction of the sub-models.For the validation of our model, we dropped duplicates, structures that are similar to our case studies, and split the database in a training set (90%) and a test set (10%) using iterative stratification.60 For doing so, we binned each colour channel into three equally sized bins and then applied the iterative stratification algorithm to ensure that the train and test sets contain the same proportions of regions of the colour space.
The model is deployed as a web app with the name “MOFcolorizer” at https://go.epfl.ch/mofcolorizer (the code for this app is available on GitHub, https://github.com/kjappelbaum/mofcolorizer). In addition to the explicitly mentioned codes, our work made use of the following Python packages: colour-checker-detection,61 colour,62 crystal_toolkit,63 dokku,64 flask,65 gunicorn,66 iraspa,67 jupyter,68 matplotlib,69 numpy,70 OpenCV,71 pandas,72 Pillow,73 pymatgen,74 PyTelegramBotAPI,75 rdkit,76 scipy.77
Feature importance analysis
For feature importance analysis, we used the tree SHAP technique, marginalising over the training set.78 We averaged over the feature importance for each estimator of the bagged estimator.Export of characterisation data
The data is captured via an ELN,40 for which parsers are being developed for the relevant experimental data (all code is part of the cheminfo GitHub organisation, http://github.com/cheminfo, for this work, we, for example, used the parser for powder X-ray diffraction data79). The parsed data (which is stored in a CouchDB database) is then exported using the RESTful Application Programming Interface (REST-API) restoncouch,80 with other sample information to Zenodo. Spectra are typically stored in JCAMP-DX format,81,82 molecules in mol format, and sample information with metadata in JavaScript Object Notation (JSON). The characterisation data is available on Zenodo (DOI: 10.5281/zenodo.4044212) and can be visualised using a view developed with the Visualizer library (https://go.epfl.ch/zenodo_colorpaper).83,84 Large parts of the code for this view are also used in the ELN itself (eln.epfl.ch).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the EPFL community for participation in the survey. KMJ thanks Leopold Talirz for providing the Dokku instances for the apps, the cheminfo developer team for support, Fatma Pelin Kinik, Mish Ebrahim, Bardiya Valizadeh, and Mojtaba Rezaei for discussion. The research in this article was supported by the European Research Council (ERC) under the European Union's Horizon 2020 Research and Innovation Programme (grant agreement 666983, MaGic), by the NCCR-MARVEL, funded by the Swiss National Science Foundation, and by the Swiss National Science Foundation (SNSF) under Grant 200021_172759, and in part by the PrISMa Project (299659), funded through the ACT Programme (Accelerating CCS Technologies, Horizon 2020 Project 294766). Financial contributions from the Department for Business, Energy & Industrial Strategy (BEIS) together with extra funding from the NERC and EPSRC Research Councils, United Kingdom, the Research Council of Norway (RCN), the Swiss Federal Office of Energy (SFOE), and the U.S. Department of Energy are gratefully acknowledged. Additional financial support from TOTAL and Equinor is also gratefully acknowledged. MA acknowledges the Swiss Commission for Technology and Innovation (CTI) (the SCCER EIP-Efficiency of Industrial Processes) for financial support and the Swiss-Norwegian Beam Line BM01 at European Synchrotron Radiation Facility (ESRF) for the beamtime allocation.References
- M. V. Orna, Chemical Origins of Color, J. Chem. Educ., 1978, 55, 478 CrossRef CAS.
- Organic Spectroscopic Analysis, ed. R. J. Anderson, D. J. Bendell and P. W. Groundwater, Royal Society of Chemistry, 2004, vol. 7–23, https://pubs.rsc.org/en/content/chapter/bk9780854044764-00007/978-0-85404-476-4 Search PubMed.
- O. Laporte and W. F. Meggers, Some Rules of Spectral Structure, J. Opt. Soc. Am., 1925, 11, 459–463 CrossRef CAS.
- A. Rosen, S. Iyer, D. Ray, Z. Yao, A. Aspuru-Guzik, L. Gagliardi, J. Notestein and R. Q. Snurr, Machine Learning the Quantum-Chemical Properties of Metal–Organic Frameworks for Accelerated Materials Discovery with a New Electronic Structure Database, ChemRxiv preprint, 2020 DOI:10.26434/chemrxiv.13147616.v1.
- M. Fumanal, A. Ortega-Guerrero, K. M. Jablonka, B. Smit and I. Tavernelli, Charge Separation and Charge Carrier Mobility in Photocatalytic Metal-Organic Frameworks, Adv. Funct. Mater., 2020, 2003792 CrossRef CAS.
- G. Prandini, G.-M. Rignanese and N. Marzari, Photorealistic Modelling of Metals from First Principles, npj Comput. Mater., 2019, 5, 1–12 CrossRef.
- A. Ortega-Guerrero, M. Fumanal, G. Capano and B. Smit, From Isolated Porphyrin Ligands to Periodic Al-PMOF: A Comparative Study of the Optical Properties Using DFT/TDDFT, J. Phys. Chem. C, 2020, 124, 21751–21760 CrossRef CAS.
- K. Müller, K. Fink, L. Schöttner, M. Koenig, L. Heinke and C. Wöll, Defects as Color Centers: The Apparent Color of Metal–Organic Frameworks Containing Cu2+-Based Paddle-Wheel Units, ACS Appl. Mater. Interfaces, 2017, 9, 37463–37467 CrossRef.
- H. Furukawa, K. E. Cordova, M. O’Keeffe and O. M. Yaghi, The Chemistry and Applications of Metal-Organic Frameworks, Science, 2013, 341, 1230444 CrossRef.
- M. Ding, R. W. Flaig, H.-L. Jiang and O. M. Yaghi, Carbon Capture and Conversion Using Metal–Organic Frameworks and MOF-Based Materials, Chem. Soc. Rev., 2019, 48, 2783–2828 RSC.
- L. Jiao, Y. Wang, H.-L. Jiang and Q. Xu, Metal–Organic Frameworks as Platforms for Catalytic Applications, Adv. Mater., 2018, 30, 1703663 CrossRef.
- Y. Zhang, S. Yuan, G. Day, X. Wang, X. Yang and H.-C. Zhou, Luminescent sensors based on metal-organic frameworks, Coord. Chem. Rev., 2018, 354, 28–45 CrossRef CAS.
- M. D. Allendorf, C. A. Bauer, R. K. Bhakta and R. J. T. Houk, Luminescent Metal–Organic Frameworks, Chem. Soc. Rev., 2009, 38, 1330–1352 RSC.
- A. Fateeva, P. A. Chater, C. P. Ireland, A. A. Tahir, Y. Z. Khimyak, P. V. Wiper, J. R. Darwent and M. J. Rosseinsky, A Water-Stable Porphyrin-Based Metal-Organic Framework Active for Visible-Light Photocatalysis, Angew. Chem., Int. Ed., 2012, 51, 7440–7444 CrossRef CAS.
- M. A. Nasalevich, M. G. Goesten, T. J. Savenije, F. Kapteijn and J. Gascon, Enhancing optical absorption of metal–organic frameworks for improved visible light photocatalysis, Chem. Commun., 2013, 49, 10575–10577 RSC.
- S. Pu, L. Xu, L. Sun and H. Du, Tuning the optical properties of the zirconium–UiO-66 metal–organic framework for photocatalytic degradation of methyl orange, Inorg. Chem. Commun., 2015, 52, 50–52 CrossRef CAS.
- S. L. Anderson, D. Tiana, C. P. Ireland, G. Capano, M. Fumanal, A. Gładysiak, S. Kampouri, A. Rahmanudin, N. Guijarro, K. Sivula, K. C. Stylianou and B. Smit, Taking Lanthanides out of Isolation: Tuning the Optical Properties of Metal–Organic Frameworks, Chem. Sci., 2020, 11, 4164–4170 RSC.
- C. R. Groom, I. J. Bruno, M. P. Lightfoot and S. C. Ward, The Cambridge Structural Database, Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater., 2016, 72, 171–179 CrossRef CAS.
- D. Ongari, L. Talirz and B. Smit, Too Many Materials and Too Many Applications: An Experimental Problem Waiting for a Computational Solution, ACS Cent. Sci., 2020, 6, 1890–1900 CrossRef CAS.
- P. Z. Moghadam, A. Li, S. B. Wiggin, A. Tao, A. G. P. Maloney, P. A. Wood, S. C. Ward and D. Fairen-Jimenez, Development of a Cambridge Structural Database Subset: A Collection of Metal–Organic Frameworks for Past, Present, and Future, Chem. Mater., 2017, 29, 2618–2625 CrossRef CAS.
- K. M. Jablonka, D. Ongari, S. M. Moosavi and B. Smit, Big-Data Science in Porous Materials: Materials Genomics and Machine Learning, Chem. Rev., 2020, 120(16), 8066–8129 CrossRef CAS.
- H. Zollinger, Color Chemistry: Syntheses, Properties, and Applications of Organic Dyes and Pigments, Verlag Helvetica Chimica Acta, Wiley-VCH, Zürich, Weinheim, 3rd edn, 2003 Search PubMed.
- L. White, R. Togneri, W. Liu and M. Bennamoun, Learning of Colors from Color Names: Distribution and Point Estimation, 2020, arXiv:1709.09360 [cs].
- A. Mueller et al. , Amueller/Word_cloud: WordCloud 1.5.0, Zenodo, 2018, https://zenodo.org/record/1322068 Search PubMed.
- R. Munroe, Color Survey Results, 2010, https://blog.xkcd.com/2010/05/03/color-survey-results/ Search PubMed.
- J. Heer and M. Stone, Color Naming Models for Color Selection, Image Editing and Palette Design, Proceedings of the SIGCHI Conference on Human Factors in Computing Systems, New York, NY, USA, 2012, pp. 1007–1016 Search PubMed.
- P. Maheshwari, M. Ghuhan and V. Vinay, Learning Colour Representations of Search Queries, Proceedings of the 43rd International ACM SIGIR Conference on Research and Development in Information Retrieval, 2020 Search PubMed.
- L. Stearns, L. Findlater and J. E. Froehlich, Applying Transfer Learning to Recognize Clothing Patterns Using a Finger-Mounted Camera, Proceedings of the 20th International ACM SIGACCESS Conference on Computers and Accessibility, Galway Ireland, 2018, pp. 349–351 Search PubMed.
- C. I. Seresinhe, T. Preis and H. S. Moat, Quantifying the Impact of Scenic Environments on Health, Sci. Rep., 2015, 5, 16899 CrossRef CAS.
- W. Monroe, N. D. Goodman and C. Potts, Learning to Generate Compositional Color Descriptions, Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, 2016 Search PubMed.
- B. McMahan and M. Stone, A Bayesian Model of Grounded Color Semantics, Trans. Assoc. Comput. Linguist., 2015, 3, 103–115 CrossRef.
- G. Menegaz, A. L. Troter, J. Sequeira and J. M. Boi, A Discrete Model for Color Naming, EURASIP J. Adv. Signal Process., 2006, 2007, 029125 CrossRef.
- X. Han, P. Schulz and T. Cohn, Grounding Learning of Modifier Dynamics: An Application to Color Naming, 2019, arXiv:1909.07586 [cs].
- K. Kawakami, C. Dyer, B. R. Routledge and N. A. Smith, Character Sequence Models for ColorfulWords, 2016, arXiv:1609.08777 [cs].
- G. Sharma, W. Wu and E. N. Dalal, The CIEDE2000 Color-Difference Formula: Implementation Notes, Supplementary Test Data, and Mathematical Observations, Color Res. Appl., 2005, 30, 21–30 CrossRef.
- ISO, Proofing Processes Working Directly from Digital Data, Standard ISO 12647-7:2016, 2016 Search PubMed.
- R. G. Kuehni and R. T. Marcus, An Experiment in Visual Scaling of Small Color Differences, Color Res. Appl., 1979, 4, 83–91 Search PubMed.
- D. H. Brainard and A. C. Hurlbert, Colour Vision: Understanding #TheDress, Curr. Biol., 2015, 25, R551–R554 CrossRef CAS.
- V. Walsh, Perceptual constancy: why things look as they do, Cambridge University Press, Cambridge, UK, New York, NY, USA, 1998 Search PubMed.
- L. Patiny, M. Zasso, D. Kostro, A. Bernal, A. M. Castillo, A. Bolaños, M. A. Asencio, N. Pellet, M. Todd, N. Schloerer, S. Kuhn, E. Holmes, S. Javor and J. Wist, The C6H6 NMR Repository: An Integral Solution to Control the Flow of Your Data from the Magnet to the Public, Magn. Reson. Chem., 2018, 56, 520–528 CrossRef CAS.
- M. D. Wilkinson, M. Dumontier, I. J. Aalbersberg, G. Appleton, M. Axton, A. Baak, N. Blomberg, J.-W. Boiten, L. B. da Silva Santos, P. E. Bourne, J. Bouwman, A. J. Brookes, T. Clark, M. Crosas, I. Dillo, O. Dumon, S. Edmunds, C. T. Evelo, R. Finkers, A. Gonzalez-Beltran, A. J. G. Gray, P. Groth, C. Goble, J. S. Grethe, J. Heringa, P. A. C. ’t Hoen, R. Hooft, T. Kuhn, R. Kok, J. Kok, S. J. Lusher, M. E. Martone, A. Mons, A. L. Packer, B. Persson, P. Rocca-Serra, M. Roos, R. van Schaik, S.-A. Sansone, E. Schultes, T. Sengstag, T. Slater, G. Strawn, M. A. Swertz, M. Thompson, J. van der Lei, E. van Mulligen, J. Velterop, A. Waagmeester, P. Wittenburg, K. Wolstencroft, J. Zhao and B. Mons, The FAIR Guiding Principles for scientific data management and stewardship, Sci. Data, 2016, 3, 160018 CrossRef.
- J. C. Berry, N. Fahlgren, A. A. Pokorny, R. S. Bart and K. M. Veley, An Automated, High-Throughput Method for Standardizing Image Color Profiles to Improve Image-Based Plant Phenotyping, PeerJ, 2018, 6, e5727 CrossRef.
- J. P. Janet and H. J. Kulik, Resolving Transition Metal Chemical Space: Feature Selection for Machine Learning and Structure-Property Relationships, J. Phys. Chem. A, 2017, 121, 8939–8954 CrossRef CAS.
- J. P. Janet and H. J. Kulik, Predicting Electronic Structure Properties of Transition Metal Complexes with Neural Networks, Chem. Sci., 2017, 8, 5137–5152 RSC.
- A. Nandy, J. Zhu, J. P. Janet, C. Duan, R. B. Getman and H. J. Kulik, Machine Learning Accelerates the Discovery of Design Rules and Exceptions in Stable Metal–Oxo Intermediate Formation, ACS Catal., 2019, 9, 8243–8255 CrossRef CAS.
- S. M. Moosavi, A. Nandy, K. M. Jablonka, D. Ongari, J. P. Janet, P. G. Boyd, Y. Lee, B. Smit and H. J. Kulik, Understanding the Diversity of the Metal-Organic Framework Ecosystem, Nat. Commun., 2020, 11, 4068 CrossRef CAS.
- M. A. Syzgantseva, N. F. Stepanov and O. A. Syzgantseva, Band Alignment as the Method for Modifying Electronic Structure of Metal-Organic Frameworks, ACS Appl. Mater. Interfaces, 2020, 12, 17611–17619 CrossRef CAS.
- S. M. Moosavi, K. M. Jablonka and B. Smit, The Role of Machine Learning in the Understanding and Design of Materials, J. Am. Chem. Soc., 2020, 142(48), 20273–20287 CrossRef CAS.
- Y. Li, H. Xu, S. Ouyang and J. Ye, Metal–Organic Frameworks for Photocatalysis, Phys. Chem. Chem. Phys., 2016, 18, 7563–7572 RSC.
- P. Sippel, D. Denysenko, A. Loidl, P. Lunkenheimer, G. Sastre and D. Volkmer, Dielectric Relaxation Processes, Electronic Structure, and Band Gap Engineering of MFU-4-Type Metal-Organic Frameworks: Towards a Rational Design of Semiconducting Microporous Materials, Adv. Funct. Mater., 2014, 24, 3885–3896 CrossRef CAS.
- M. Usman, S. Mendiratta and K.-L. Lu, Semiconductor Metal–Organic Frameworks: Future Low-Bandgap Materials, Adv. Mater., 2017, 29, 1605071 CrossRef.
- J. K. Schnobrich, O. Lebel, K. A. Cychosz, A. Dailly, A. G. Wong-Foy and A. J. Matzger, Linker-Directed Vertex Desymmetrization for the Production of Coordination Polymers with High Porosity, J. Am. Chem. Soc., 2010, 132, 13941–13948 CrossRef CAS.
- C. Plotly/Dash Parmer, 2020, https://dash.plotly.com.
- E. I. Ioannidis, T. Z. H. Gani and H. J. Kulik, molSimplify: A Toolkit for Automating Discovery in Inorganic Chemistry, J. Comput. Chem., 2016, 37, 2106–2117 CrossRef CAS.
- B. J. Bucior, A. S. Rosen, M. Haranczyk, Z. Yao, M. E. Ziebel, O. K. Farha, J. T. Hupp, J. I. Siepmann, A. Aspuru-Guzik and R. Q. Snurr, Identification Schemes for Metal–Organic Frameworks To Enable Rapid Search and Cheminformatics Analysis, Cryst. Growth Des., 2019, 19, 6682–6697 CrossRef CAS.
- N. M. O'Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch and G. R. Hutchison, Open Babel: An Open Chemical Toolbox, J. Cheminf., 2011, 3, 33 Search PubMed.
- F. Pedregosa, et al., Scikit-Learn: Machine Learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- G. Ke; Q. Meng, T. Finley, T. Wang, W. Chen, W. Ma, Q. Ye and T.-Y. Liu, in Advances in Neural Information Processing Systems 30, ed. I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathan and R. Garnett, Curran Associates, Inc., 2017, pp. 3146–3154 Search PubMed.
- G. Taylor, Python-Colormath, 2020, https://github.com/gtaylor/python-colormath Search PubMed.
- K. Sechidis, G. Tsoumakas and I. Vlahavas, in Machine Learning and Knowledge Discovery in Databases, ed. D. Gunopulos, T. Hofmann, D. Malerba and M. Vazirgiannis, Springer Berlin Heidelberg, Berlin, Heidelberg, 2011, vol. 6913, pp. 145–158 Search PubMed.
- Colour Developers, Colour-Checker-Detection, colour-science, 2020, https://github.com/colour-science/colour-checker-detection/ Search PubMed.
- Colour Developers, Colour-Science/Colour, 2020, https://github.com/colour-science/colour Search PubMed.
- M. Horton, Materialsproject/Crystaltoolkit, 2020, https://github.com/materialsproject/crystaltoolkit Search PubMed.
- J. Lindsay, Dokku/Dokku Dokku, 2020, https://github.com/dokku/dokku.
- A. Ronacher, Pallets/Flask, 2020, https://palletsprojects.com/p/flask Search PubMed.
- B. Chesneau, Benoitc/Gunicorn, 2020, https://gunicorn.org/ Search PubMed.
- D. Dubbeldam, S. Calero and T. J. Vlugt, iRASPA: GPU-accelerated visualization software for materials scientists, Mol. Simul., 2018, 44, 653–676 CrossRef CAS.
- T. Kluyver, B. Ragan-Kelley, F. Pérez, B. Granger, M. Bussonnier, J. Frederic, K. Kelley, J. Hamrick, J. Grout, S. Corlay, P. Ivanov, D. Avila, S. Abdalla and C. Willing, Jupyter Notebooks – a Publishing Format for Reproducible Computational Workflows, Positioning and Power in Academic Publishing: Players, Agents and Agendas, 2016, pp. 87–90 Search PubMed.
- J. D. Hunter, Matplotlib: A 2D Graphics Environment, Comput. Sci. Eng., 2007, 9, 90–95 Search PubMed.
- C. R. Harris, et al., Array Programming with NumPy, Nature, 2020, 585, 357–362 CrossRef CAS.
- G. Bradski, The OpenCV Library, Dr Dobb's J. Software Tools, 2000 Search PubMed.
- W. McKinney, Data Structures for Statistical Computing in Python, Python in Science Conference, Austin, Texas, 2010, pp. 56–61 Search PubMed.
- H. V. Kemenade et al. , python-pillow/Pillow 8.0.1, 2020, https://zenodo.org/record/4118627 Search PubMed.
- S. P. Ong, W. D. Richards, A. Jain, G. Hautier, M. Kocher, S. Cholia, D. Gunter, V. L. Chevrier, K. A. Persson and G. Ceder, Python Materials Genomics (pymatgen): A robust, open-source python library for materials analysis, Comput. Mater. Sci., 2013, 68, 314–319 CrossRef CAS.
- GitHub user eternnoir, pyTelegramBotAPI, 2020, https://github.com/eternnoir/pyTelegramBotAPI.
- G. Landrum et al. , rdkit, 2020, https://zenodo.org/record/4288221 Search PubMed.
- SciPy 1.0 Contributors, et al., SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python, Nat. Methods, 2020, 17, 261–272 CrossRef.
- S. M. Lundberg, G. Erion, H. Chen, A. DeGrave, J. M. Prutkin, B. Nair, R. Katz, J. Himmelfarb, N. Bansal and S.-I. Lee, From Local Explanations to Global Understanding with Explainable AI for Trees, Nat. Mach. Intell., 2020, 2, 56–67 CrossRef.
- Cheminfo developers, Cheminfo/xrd-Analysis, 2020, https://github.com/cheminfo/xrd-analysis Search PubMed.
- M. Zasso, Cheminfo/Rest-on-Couch, 2020, https://github.com/cheminfo/rest-on-couch Search PubMed.
- R. S. McDonald and P. A. Wilks, JCAMP-DX: A Standard Form for Exchange of Infrared Spectra in Computer Readable Form, Appl. Spectrosc., 1988, 42, 151–162 CrossRef CAS.
- A. N. Davies and P. Lampen, JCAMP-DX for NMR, Appl. Spectrosc., 1993, 47, 1093–1099 CrossRef CAS.
- N. Pellet, NPellet/Visualizer, 2020, https://github.com/npellet/visualizer Search PubMed.
- N. Pellet, jsGraph and jsNMR—Advanced Scientific Charting, Challenges, 2014, 5, 294–295 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc05337f |
| This journal is © The Royal Society of Chemistry 2021 |