Open Access Article
Open Access ArticleMetabolome-based profiling of African baobab fruit (Adansonia digitata L.) using a multiplex approach of MS and NMR techniques in relation to its biological activity†
Mostafa H. Baky‡
 a,
Marwa T. Badawy‡b,
Alaa F. Bakrc,
Nesrine M. Hegazi
a,
Marwa T. Badawy‡b,
Alaa F. Bakrc,
Nesrine M. Hegazi d,
Ahmed Abdellatif
d,
Ahmed Abdellatif b and
Mohamed A. Farag
b and
Mohamed A. Farag *ef
*ef
aPharmacognosy Department, Faculty of Pharmacy, Egyptian Russian University, Badr City, Cairo, 11829, Egypt. E-mail: dr_mostafa1984@yahoo.com; mostafa-hasan@eru.edu.eg; Tel: +01007906443
bBiology Department, School of Sciences & Engineering, The American University in Cairo, New Cairo 11835, Egypt
cPathology Department, Faculty of Veterinary Medicine, Cairo University, Gamaa St., 12211 Giza, Egypt
dPhytochemistry and Plant Systematics Department, Division of Pharmaceutical Industries, National Research Centre, P. O. Box 12622, Cairo, Egypt
ePharmacognosy Department, College of Pharmacy, Cairo University, Kasr El Aini St., P.B. 11562 Cairo, Egypt. E-mail: mohamed.farag@pharma.cu.edu.eg; Fax: +011-202-25320005; Tel: +011-202-2362245
fChemistry Department, School of Sciences & Engineering, The American University in Cairo, New Cairo 11835, Egypt
First published on 13th December 2021
Abstract
Adansonia digitata L. also known as African baobab is one of the most important fruit-producing trees, widely distributed in the African continent. Baobab fruits are known to possess potential health benefits and nutritional value. This study aimed to holistically dissect the metabolome of A. digitata fruits using a novel comparative protocol using three different analytical platforms. Ultra high performance liquid chromatography coupled to high-resolution tandem mass spectrometry (UHPLC-HRMS/MS), and headspace solid-phase microextraction/gas chromatography coupled to mass spectrometry (HS-SPME/GC-MS) were respectively employed for phytonutrients and aroma profiling, whereas GC-MS post silylation provided an overview of nutrients i.e., sugars. UHPLC-HRMS/MS analysis allowed for the assignment of 77 metabolites, among which 50% are reported for the first time in the fruit. While GC-MS of silylated and aroma compounds led to the identification of 74 and 16 compounds, respectively. Finally, NMR-based metabolite fingerprinting permitted the quantification of the major metabolites for future standardization. In parallel, in vivo antidiabetic potential of the baobab fruit using a streptozotocin (STZ) induced diabetic rat model was assessed. Histopathological and immune-histochemical investigations revealed hepatoprotective and renoprotective effects of A. digitata fruit along with mitigation against diabetes complications. Moreover, the administration of A. digitata fruits (150 mg kg−1) twice a week lowered fasting blood glucose levels.
1. Introduction
African baobab (Adansonia digitata L.) belonging to the family Malvaceae is a large sub-tropical tree widely distributed in Southern, West, and East Africa.1 Baobab is considered an extremely important edible fruit with myriad traditional uses and is widely consumed in African countries owing to its medicinal importance in the treatment of fever, diarrhea, and dysentery. Additionally, it is used as an immuno-stimulant, analgesic, antimalarial, and insect repellent.1 Further, baobab fruit is recognized for its nutritional value and has been approved as a food ingredient in the European continent and the United States of America.1,2In West Africa, the fruit pulp and seeds are consumed as food and as an ingredient in other food-based products such as sauces, porridges, and beverages.3 Baobab fruit pulp is an essential source of vitamins, minerals, carbohydrates, amino acids, fatty acids, and sterols.3 In addition, baobab is highly rich in vitamin C as it supplies 150–500 mg/100 g fruit so it is useful as a potent antioxidant and about 40 g of baobab pulp consumption daily provides 100% of the daily required vitamin C.4 Baobab fruit is also rich in fibers, calcium, and vitamin B, and is low in proteins and fats.5 The fresh fruit pulp is acidic in taste and consumed in African countries as a snack or as a refreshing juice by adding milk or water. Baobab edible fruits were reported to be enriched in various phytochemical compounds belonging to different chemical classes such as flavonoids, triterpenoids, steroids, vitamins, amino acids, lipids, and carbohydrates.6 Reported biological properties of baobab include anti-hyperglycemic,7 analgesic and antipyretic, antibacterial, anti-inflammatory, antioxidant, and antiviral.5
Recently, modern analytical approaches especially metabolomics platforms have been used in the chemical analysis of food and spices.8 Among analytical tools, UHPLC-HRMS/MS, GC/MS and NMR spectroscopy have been extensively adopted to characterize the metabolite diversity, e.g., volatiles and non-volatiles in fruits and seeds.9,10 Lastly, SPME is considered a convenient solvent-free method that requires minimal sample preparation for the enhancement of volatiles recovery from food samples.11,12
To the best of our knowledge, this is the first report on holistic metabolites profiling of A. digitata L., metabolites using 3 different analytical platforms i.e., UHPLC-HRMS/MS, headspace SPME-GC/MS, GC/MS post silylation, and NMR simultaneously as a novel protocol for the determination of its quality characteristics. Additionally, molecular networks through the Global Natural Products Social Molecular Networking (GNPS) platform was exploited for the propagation of metabolites annotation from the UPLC-HRMS/MS dataset through the imaged structural similarities among the metabolites, with several novel metabolites reported for the first time. Moreover, the in vivo antidiabetic potential of A. digitata fruits was assessed in relation to the metabolites profile in streptozotocin (STZ)-induced diabetic animal model at multiple levels i.e., histopathological examination and biochemical markers.
2. Materials and methods
2.1. Plant material and extract preparation
Adansonia digitata L. fruits (700 g) were purchased from a local market in Khartoum state, Sudan in January 2019. The air-dried powdered fruits (100 g) were extracted by soaking in 500 mL methanol for 48 h. The extract was then filtered and evaporated under reduced pressure at 40 °C and kept in a freezer at −10 °C for further investigation (6 g). A voucher specimen of the fruit material was deposited at the College of Pharmacy Herbarium, Cairo University, Cairo, Egypt.2.2. Phytochemical investigation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 min to remove debris. 3 different samples for each A. digitata fruits accession were analyzed under the same conditions. Then, 100 μL of the methanol extract was aliquoted in screw-cap vials and left to evaporate under a nitrogen gas stream until complete dryness. For derivatization, 150 μL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) previously diluted 1
000 × g for 10 min to remove debris. 3 different samples for each A. digitata fruits accession were analyzed under the same conditions. Then, 100 μL of the methanol extract was aliquoted in screw-cap vials and left to evaporate under a nitrogen gas stream until complete dryness. For derivatization, 150 μL of N-methyl-N-(trimethylsilyl)-trifluoroacetamide (MSTFA) previously diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1% with anhydrous pyridine was mixed with the dried methanol extract and incubated for 45 min at 60 °C prior to analysis using GC-MS. Separation of silylated derivatives was achieved on a Rtx-5MS (30 m length, 0.25 mm inner diameter, and 0.25 mm film). The protocol to validate silylation was adopted as previously reported.18
1% with anhydrous pyridine was mixed with the dried methanol extract and incubated for 45 min at 60 °C prior to analysis using GC-MS. Separation of silylated derivatives was achieved on a Rtx-5MS (30 m length, 0.25 mm inner diameter, and 0.25 mm film). The protocol to validate silylation was adopted as previously reported.182.3. Antidiabetic activity
2.4. Statistical analysis
Data are presented as mean ± SD. One-way analysis of variance (ANOVA) followed by Tukey's comparison test was performed to compare parameters among groups. Data were analyzed by using R software (version 3.5.2) for biochemical analysis and parametric histological parameters. For non-parametric data (histological damage score), the statistics are presented as median ± SD using Kruskal–Wallis test, followed by Dunn's Multiple Comparison Test. A P-value of <0.05 was considered to be statically significant.3. Results and discussion
Functional foods have had always played a pivotal role in developing countries as a potential source of alternative medicine. In Africa, several plants have been used traditionally for the management of diabetes mellitus. The present study aimed to characterize metabolites in baobab fruits via UHPLC/PDA/ESI-MS, and GC-MS techniques. While the quantification of the major metabolites was accomplished through NMR metabolite fingerprinting for future standardization. Further, the antidiabetic potential of A. digitata L. fruit extract in streptozotocin (STZ) induced diabetic rats model was investigated.3.1. Phytochemical investigation
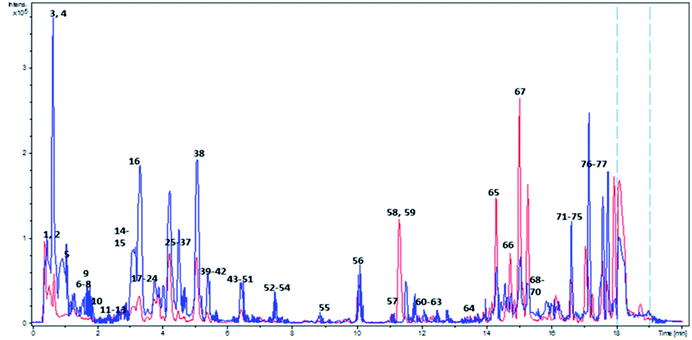 | ||
| Fig. 1 Overlaid BPC of Adansonia digitata L. fruit crude extract in both ionization modes. Red and blue colors correspond to positive and negative ionization modes respectively. Peaks numbering follow that listed in Table 2. | ||
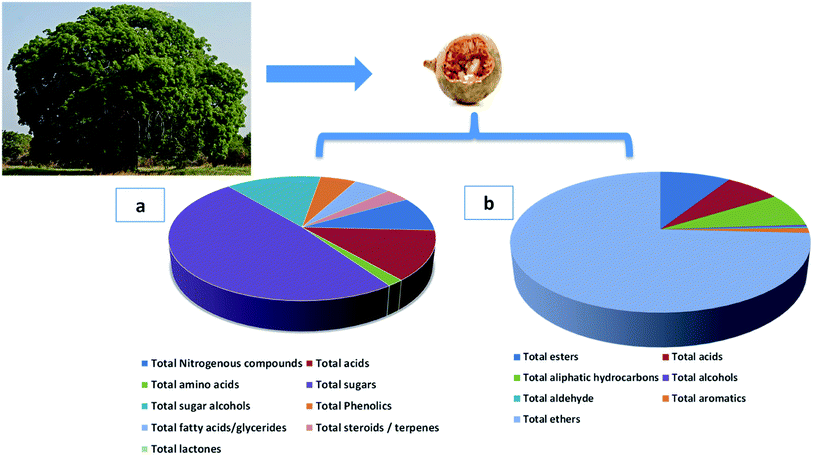
Metabolites dereplication was carried out based on their retention times, molecular formula, UV absorption maxima, and their fragmentation pattern in comparison to previously reported data. 77 compounds were annotated belonging to different classes including organic acids, sugars, alcohols, phenolics, coumarins, and fatty acids. Almost 50% of the identified metabolites (40 compounds) are reported for the first time in A. digitata fruits (Table 1).
| Peak | Rt | Metabolite class | M − H | MS2 | M + H | MS2 | Chemical formula | Error (ppm) | Name | References |
|---|---|---|---|---|---|---|---|---|---|---|
| a Metabolits detected for the first time from Adansonia digitata fruits. | ||||||||||
| 1 | 0.48 | Organic acid | 191.0198 | 129, 111 | C6H8O7 | −0.14 | Citric acid | 67 | ||
| 2 | 0.48 | Organic acid | 133.0142 | 115 | C4H6O5 | 0.3 | Malic acid | 25 | ||
| 3 | 0.62 | Saccharides | 341.1091 | 179, 133 | 343.1234 | 163 | C12H22O11 | 0.06 | Disaccharide (sucrose) | 25 |
| 4 | 0.66 | Saccharides | 371.1196 | 325, 193 | C13H24O12 | 0.16 | Unknown diasaccharidea | |||
| 5 | 0.75 | Organic acid | 191.0200 | 111 | C6H8O7 | −1.87 | Citric acid isomer | 67 | ||
| 6 | 1.0 | Organic acid | 205.0354 | 111 | C7H10O7 | −0.7 | Citric acid methyl estera | 23 | ||
| 7 | 1.3 | Saccharides | 385.1699 | 163, 145 | C15H28O11 | 1.36 | Butandiol pentoside hexosidea | 26 | ||
| 8 | 1.39 | Phenolic | 127.0393 | 109 | C6H6O3 | −4.05 | Pyrogallola | 28 | ||
| 9 | 1.53 | Benzoic acid | 167.0353 | 123 | C8H8O4 | 2.05 | Vanillic acida | 35 | ||
| 10 | 1.82 | Benzoic acid | 153.0193 | 109 | 155.0342 | 111 | C7H6O4 | −2.02 | Dihydroxy benzoic acida (protocatechuic acid) | 34 |
| 11 | 2.48 | Nitrogenous compound | 188.0701 | C11H9NO2 | 2.51 | Unknown | ||||
| 12 | 2.6 | Benzenoids | 137.0243 | 109 | C7H6O3 | −2.6 | Dihydroxybenzaldehydea | 29 | ||
| 13 | 2.84 | Organic acid | 175.0615 | 131, 157 | C7H12O5 | −1.8 | Hydroxyglutaric acida | 24 | ||
| 14 | 3.32 | Saccharides derivatives | 369.1753 | 295, 163, 145, 133, 115 | C15H28O10 | 0.38 | Butanol hexopyranosyl-deoxypyranosidea | 27 | ||
| 15 | 3.32 | Saccharides derivatives | 295.1012 | 163, 145, 133 | C11H18O9 | 2.55 | Dihydroxy—methylenebutanoate-O-hexosidea | Tuliposide B68 | ||
| 16 | 3.39 | Flavan-3-ol | 289.0717 | 245, 203 | 291.0861 | 207, 139 | C15H14O6 | 2.42 | Catechin/epicatechin | 34 and 67 |
| 17 | 3.5 | Cinnamic acid ester | 353.0891 | 191 | C16H18O9 | −2.1 | O-Caffeoyl quinic acid ester clorogenic acid | 34 | ||
| 18 | 3.53 | Nitrogenous compound | 352.1034 | 191 | C16H19NO8 | 0.85 | Unknowna | |||
| 19 | 3.72 | Cinnamic acid ester | 281.0660 | 193, 163, 145 | C13H14O7 | 4.39 | O-feruloyl lactic acid estera | 36 | ||
| 20 | 3.79 | Proanthocyanidin | 577.1345 | 407, 289 | C30H26O12 | −0.98 | Procyanidn dimer isomer | Xx34,67 | ||
| 21 | 3.8 | Cinnamic acid glycoside | 327.1082 | 165, 147 | C15H20O8 | 2.8 | Dihydrocoumaroyl O-hexoside (dihydromelilotoside)a | 39 | ||
| 22 | 3.9 | Proanthocyanidin | 577.0346 | 407, 289 | 579.1472 | 409, 301, 291 | C30H26O12 | 4.25 | Procyanidn dimer isomer I | 34 and 67 |
| 23 | 3.91 | Proanthocyanidin | 865.1987 | 289 | C45H38O18 | 0.2 | Procyanidin trimer isomer I | 34 and 67 | ||
| 24 | 3.98 | Cinnamic acids glycosides | 341.0893 | 179 [M-H-162], 161 | C15H18O9 | 4.5 | Caffeoyl–O-hexoside | 40 | ||
| 25 | 4.03 | Saccharides derivatives | 295.1018 | 163, 145, 133 | C11H18O9 | 1.88 | Dihydroxy–methylenebutanoate–O-hexoside isomer | 68 | ||
| 26 | 4.1 | Cinnamic acid glycosides | 487.1457 | 193[M − H − 132-162] | C21H28O13 | −1.3 | Feruloyl O-pentosyl hexosidea | 41 | ||
| 27 | 4.23 | Cinnamic acids | 165.0544 | 119, 107 | C9H8O3 | 3.67 | Coumaric acid | 34 | ||
| 28 | 4.24 | Flavan-3-ol | 289.0716 | 245, 203 | 291.0860 | C15H14O6 | −0.27 | Catechin/epicatechin | 34 and 67 | |
| 29 | 4.33 | Saccharides derivatives | 295.1018 | 163, 145, 133 | C11H18O9 | 1.9 | Dihydroxy-methylenebutanoic acid-O-hexoside | 68 | ||
| 30 | 4.47 | Cinnamic acid glycoside | 325.0925 | 163 | C15H18O8 | −0.29 | Coumaroyl-O-hexosidea | 41 | ||
| 31 | 4.52 | Saccharides derivatives | 383.1905 | 163, 145, 133, 155 | C16H30O10 | 1.7 | Methylbutyl-O-pentosyl-hexosidea | 68 | ||
| 32 | 4.52 | Saccharides derivatives | 295.1008 | 163, 145, 133 | C11H18O9 | 0.9 | Dihydroxy-methylenebutanoic acid-O-hexoside | 68 | ||
| 33 | 4.52 | Proanthocyanidin | 865.1992 | 577, 407, 289 | C45H38O18 | 1.03 | Procyanidin trimer isomer II | Xx34,67 | ||
| 34 | 4.7 | Proanthocyanidin | 865.1994 | 867.1867 | 579, 289 | C45H38O18 | 0.77 | Procyanidin trimer isomer | Xx34,67 | |
| 35 | 4.73 | Cinnamic acid ester selflooped | 367.1024 | 191, 173, 133 | C17H20O9 | −2.77 | O-feruloyl quinic acid | Xx67 | ||
| 36 | 4.8 | Phenolic | 461.1653 | 149 | C20H30O12 | 4.5 | Homovanillyl alcohol-O-pentosyl hexosidea | 30 | ||
| 37 | 4.8 | Cinnamic acid | 163.0401 | 119 | C9H8O3 | −0.1 | Coumaric acid isomer | 34 | ||
| 38 | 5.02 | Proanthocyanidin | 1153.2595 | 865, 577, 289 | C60H50O24 | −2.04 | Procyanidin tetramer | 34 | ||
| 39 | 5.1 | Cinnamate | 163.0391 | 135 | C9H8O3 | −0.5 | Unknowna | |||
| 40 | 5.49 | Cinnamic acid ester | 367.1031 | 191, 173, 133 | C17H20O9 | −0.52 | O-feruloyl quinic acid isomer | 67 | ||
| 41 | 5.59 | Furanochromones | 409.1111 | 247 | C19H20O10 | 4.37 | Khellol-O-hexosidea | 46 | ||
| 42 | 5.66 | Flavonoid | 609.1457 | 301 | C27H30O16 | −0.65 | Quercetin-O-deoxyhexosyl hexoside | 67 | ||
| 43 | 6.2 | Cinnamic acid glycoside | 325.0916 | 163 | C15H18O8 | −3.57 | Coumaroyl-O-hexoside | 40 | ||
| 44 | 6.27 | Flavonoid | 609.1467 | 301 | C27H30O16 | 1.04 | Quercetin-O-deoxyhexosyl hexoside | 67 | ||
| 45 | 6.43 | Proanthocyanidin | 577.1346 | 407, 289 | C30H26O12 | −0.93 | Procyanidn dimer isomer III | 34 and 67 | ||
| 46 | 6.46 | Flavonoid | 449.1073 | 287 | C21H20O11 | 1.17 | Kaempferol–O-hexoside | 67 | ||
| 47 | 6.44 | Flavonoid | 593.1505 | 447, 285 | 595.1617 | 449, 287 | C27H30O15 | 1.04 | Kaempferol–O-deoxyhexosyl hexosidea | 43 |
| 48 | 6.47 | Flavonoid | 287.0543 | 217, 151 | C15H10O6 | 3.07 | Kaempferol | 67 | ||
| 49 | 6.52 | Flavonoid | 593.1505 | 447, 285 | C27H30O15 | −2.46 | Kaempferol–O-deoxyhexosyl hexoside isomera | 43 | ||
| 50 | 6.62 | Cinnamic acid ester | 293.0671 | 163, 145, 119 | C14H14O7 | 1.28 | O-coumaroyl malic acid methyl estera | 37 | ||
| 51 | 6.88 | Cinnamic acid glycosides | 355.1022 | 193 | C16H20O9 | −3.48 | Feruloyl–O-hexoside | 40 | ||
| 52 | 7.3 | Cinnamic acid amide | 326.1032 | 206, 163, 145 | C18H17NO5 | −0.52 | Coumaroyl–N-tyrosinea | 38 | ||
| 53 | 7.5 | Cinnamic acid | 279.0870 | 163 | C14H16O6 | 1.6 | Unknowna | |||
| 54 | 7.63 | Flavonoid | 447.09314 | 285 | C21H20O11 | 0.34 | Kaempferol-O-hexoside | 67 | ||
| 55 | 8.9 | Megastigmane | 405.2123 | 225 | C19H34O9 | 3.8 | Megastigmane–O-hexosidea | 69 | ||
| 56 | 10.1 | Steroid | 507.2582 | 345 | C27H40O9 | 0.9 | Unknown | |||
| 57 | 11.2 | Phenolic | 149.0605 | 122 | C9H10O2 | −1.68 | Allyl catechola | 31 | ||
| 58 | 11.32 | Furanochromones | 261.0753 | 230 | C14H12O5 | 0.36 | Khellina | 70 | ||
| 59 | 11.38 | Furanochromones | 231.0648 | 203, 216 | C13H10O4 | 1.35 | Visnagina | 70 | ||
| 60 | 12.29 | Phenolic | 137.0600 | 122, 109 | C8H8O2 | −2.2 | Methoxy benzaldehyde | 32 | ||
| 61 | 12.31 | Phenolic | 133.0651 | 105 | C9H8O | 2.94 | Cinnamaldehyde | 33 | ||
| 62 | 12.32 | Phenolic | 165.0907 | 137, 124 | C10H12O2 | 1.63 | Eugenol | 32 | ||
| 63 | 13.6 | Cinnamic acid | 207.1017 | 165, 150 | C12H14O3 | −0.9 | Unknowna | |||
| 64 | 13.72 | Cinnamic acid | 211.0961 | 193, 181, 165 | C11H14O4 | 1.55 | Unknowna | |||
| 65 | 14.97 | Coumarins | 411.1410 [M + Na] | 329, 245, 227, 199 | C21H24O7 | 0.81 | Isovaleryl-acetyl khellactone (dihydrosamidin)a | 47 | ||
| 66 | 14.98 | Coumarins | 329.1377 | 245, 227 | C19H20O5 | 1.9 | Decursina | 51 | ||
| 67 | 15.1 | Fatty acid | 295.2274 | 277 (−H2O), 233 (−CO2) | C18H32O3 | 1.3 | Hydroxyoctadecadienoic acida | 71 | ||
| 68 | 15.1 | Fatty acid | 271.2278 | 253 | C16H32O3 | 1.2 | Hydroxyhexadecanoic acida | 72 | ||
| 69 | 15.4 | Fatty acid | 269.2108 | 251 | 225 | C16H30O3 | 5.1 | Oxohexadecanoic acida | 73 | |
| 70 | 16.1 | Fatty acid | 295.2271 | 277 (−H2O), 233 (−CO2) | C18H32O3 | −3.1 | Hydroxyoctadecadienoic acida | 74 | ||
| 71 | 16.1 | Fatty acid | 297.2423 | C18H34O3 | Hydroxy-octadecanoic acida | 71 | ||||
| 72 | 16.3 | Fatty acid | 311.2225 | 293 (−H2O), 275 (−H2O), 223 | C18H32O4 | 1.1 | Dihydroxy-octadecadienoic acida | 74 | ||
| 73 | 16.6 | Fatty acid | 277.2171 | 259 | C18H30O2 | 1.8 | Octadecatrienoic acida | 75 | ||
| 74 | 16.93 | Fatty acid | 355.3216 | 309 | C22H44O3 | 0.22 | Hydroxydocosanoic acida | 72 | ||
| 75 | 17.1 | Fatty acid | 279.2331 | 261 | C18H32O2 | −0.6 | Octadecadienoic acid (linoleic acid) | 75 | ||
| 76 | 17.5 | Fatty acid | 279.2331 | 261 | C18H32O2 | −1.8 | Octadecadienoic acid (linoleic acid) | 75 | ||
| 77 | 17.6 | Fatty acid | 255.2333 | C16H32O2 | −0.8 | Hexadecanoic acid (palmitic acid) | 76 | |||
3.1.2.1. Organic acids. Citric acid and its isomer were detected in peaks 1 & 5 with [M − H]− at m/z 191.0198, C6H8O7, and characteristic fragment at m/z 111. The negative molecular network (ESI Fig. S1,† cluster a) revealed a directly correlated ion at peak 6 [M − H]− 205.0354, C7H10O7, with a mass difference of 14 Da (CH2) and was annotated as citric acid methyl ester.23
Other organic acids included peak 2 with [M − H]− 133.0142, C4H6O5 annotated as malic acid, and peak 13 with [M − H]− 175.0615, C7H12O5.24
3.1.2.2. Saccharides and their derivatives. Cluster b of the negative spectral network (ESI Fig. S1†) compromised sugars and their derivatives from which peaks 3 & 4 corresponded to two disaccharides, where 3 had a molecular ion [M − H]− at m/z 341.1107, C12H22O11 and was assigned as sucrose previously identified in Adansonia fruits.25
Butanediol-O-pentosyl-hexoside, peak 7 in the positive mode, showed a molecular ion at m/z 385.1699 [M + H]+, C15H28O11.26 Similarly, peak 14 showed a molecular ion at m/z 369.1753 [M + H]+, C15H28O10 and a fragment at m/z 295 for the loss of C4H9O was assigned as butanol-pentosyl-hexoside.27
Additionally, the positive molecular network revealed the presence of 4 isomers of acylated hexoses; peaks 15, 25, 29 & 32 which shared a molecular ion at m/z 295.1018 [M + H]+ and corresponded to the elemental composition of C11H18O9.
3.1.2.3. Benzenoids & aromatics. Several phenolic metabolites were detected in both ionization modes and revealed themselves mostly as single nodes within both spectral networks. The first eluted at peak 8 was observed at [M + H]+ at m/z 127.0393, C6H6O3 with a fragment at m/z 109 for H2O loss and was annotated as pyrogallol.28 Following at peak 12, was dihydroxybenzaldehyde with [M − H]− at m/z 137.0243, C7H6O3 and a characteristic fragment at m/z 109 for the cleavage of CO.29 Lately eluted, peak 36 exhibited a molecular ion at m/z 461.1653 [M − H]− and elemental composition of C20H30O12 and showing a daughter ion at m/z 149 [M − H − 162 − 132]− corresponding to the loss of a hexoside and pentoyl moieties.30 Hence it was annotated as homovanillyl alcohol-O-pentosyl–O-hexoside. While peak 57 showed a molecular ion at m/z 149.0605 [M − H]−, C9H10O2 and a fragment at m/z 122 [M − H − C2H2]− was assigned as allylcatechol.31
Methoxybenzaldehyde, eluted in peak 60 with a molecular ion at m/z 137.0600 [M + H]+, C8H8O2 and fragment ion at m/z 122 and 109 for the consecutive cleavage of CH3 and CO.32 Similarly, peaks 61 & 62 with [M − H]+ at m/z 133.0651, C9H8O, & 165.0907, C10H12O2 and fragments at m/z 105 & 137 accounting for the loss of CO and were ascribed as cinnamaldehyde33 and eugenol,32 respectively.
The presence of benzoic acids in A. digitata fruits was previously reported34 and was firstly observed in peak 9 assigned as vanillic acid from its molecular ion [M − H]− at m/z 167.0353, C8H8O4 and fragment at m/z 123 [M − H − 44]− for the loss of CO2.35 Likewise, dihydroxybenzoic acid in peak 10 confirmed by its molecular ion at m/z 153.0193 [M − H]−, C7H6O4 and fragment at m/z 109 for the loss of CO2.
3.1.2.4. Hydroxy cinnamates. Peaks 27 & 37 were annotated as hydroxyl cinnamic acid, p-coumaric acid isomers as concluded from their molecular ions at m/z 163.0401 [M − H]−, 165.0544 [M + H]+ and formula of C9H8O3.34 Quinic acid esters were detected in peaks 17 annotated as O-caffeoyl quinic acid ester (chlorogenic acid) and 35 & 40 as two isomers of O-feruloyl quinic acid. Peak 17 [M − H]−at m/z 353.0891, C16H18O9 and a base peak at m/z 191 suggestive for O-caffeoyl quinic acid ester (chlorogenic acid).34 While peaks 35 & 40 showed same molecular ion at m/z 367.1024 [M − H]−, C17H20O9 and a base peak at m/z 191 confirming them to be isomers of O-feruloyl quinic acid esters.
Other identified cinnamate esters included peak 19 annotated as O-feruloyl lactic acid ester based on its [M − H]− at m/z 281.0660, C13H14O7, and daughter ions at m/z 193 for the loss of lactic acid moiety (C3H6O3).36 Peak 50 with a [M − H]−at m/z 293.0671 [M − H]−, C14H14O7 and fragment ions at m/z 163 [M − H − 130]− for the cleavage of C5H6O4 (methyl malic acid) and m/z 145 & 119 for dehydration and decarboxylation was annotated as O-p-coumaroyl malic acid methyl ester.37 Whereas, coumaroyl tyrosine conjugate was detected in peak 52 with [M − H]−at m/z 326.1032, and fragments at m/z 206 for tyrosine and subsequent fragments at m/z 163 and 145 for deamidation and dehydration.38
Cinnamic acid glycosides in peaks 21, 24, 30, 43, 51, all showed fragment ions [M − H − 162]− indicative of a hexoside moiety. Peak 21 was annotated as dihydrocoumaroyl O-hexoside (dihydromelilotoside) with [M − H]−at m/z 327.1082, C15H20O8,39 and peak 24 as caffeoyl acid O-hexoside showing a molecular ion at m/z 341.0893 [M − H]−, C15H18O9.40 Ferulic acid O-hexoside was observed in peak 51 with molecular ion at m/z 355.1022 [M − H]−, C16H20O9.41 Additionally, feruloyl O-pentosyl hexoside peak 26 assignment was based on [M − H]− at m/z 487.1457, C21H28O13 and fragment at m/z 193 corresponding to the loss of a hexoside and pentoside moieties.41
3.1.2.5. Proanthocyanidin and their monomers flavan-3-ol. Monomeric proanthocyanidins were detected in peaks 16 & 28 with molecular ions at m/z 289.0716 [M − H]−, 291.0860 [M + H]+, C15H14O6, assigned as catechin/epicatechin.34 In contrast, their dimeric procyanidins were observed in peaks 20, 22 & 45 with molecular ions at m/z 577.1346, C30H26O12 with fragments at m/z 407, and 289 which are characteristic of type B proanthocyanidin previously reported in A. digitata fruits.34 While, isomers of proanthocyanidin trimers were detected in peaks 23, 33 & 34 exhibiting molecular ions at m/z 865.1992, C45H38O18 and distinctive fragments at m/z 577, 407, 289.34 A procyanidin tetramer was recognized in peak 38 with a molecular ion at m/z 1153.2595 [M − H]−, following the same fragmentation pattern as the dimers and trimers.34 All detected proanthocyanidins were grouped in the negative molecular network as group c (ESI Fig. S1†) except for the tetramer which appeared as a self-looped node.
Proanthocyanidins are well known for their antidiabetic potential and to protect against diabetic complications to include oxidative stress, lipid peroxidation, diabetic encephalopathy, and retinopathy.42
3.1.2.6. Flavonoids. Aside from the flavan-3-ols, flavonols and their glycosides were depicted in peaks 42, 44, 46, 47, 48, 49 & 54. Quercetin-O-deoxyhexosyl hexoside isomers, 42 & 44 with a molecular ion at m/z 609.1457 [M − H]−, C27H30O16 showed the characteristic fragmentation for the loss of a hexoside and deoxyhexoside moieties to yield aglycone moiety at m/z 301.34 Similarly, isomers of kaempferol-O-deoxyhexosyl hexoside were detected in peaks 47 & 49 with m/z 593.1505 [M − H]−, C27H30O15 and fragments at m/z 447 and 285.43 While, peaks 46 & 54 were annotated as kaempferol-O-hexoside isomers at m/z 447.09314, C21H20O11 and fragment at m/z 285. Several studies have affirmed the potential of dietary flavonoids in diabetes management through improving glucose metabolism and the lipid profile, thus protecting against diabetes and its aggravations.44
3.1.2.7. Furano- & pyrano-coumarins. Furanochromones were observed for the first time in peaks 41, 58 & 59 as proved from their molecular ions and fragmentation pattern detected solely in the positive ionization mode.45
Peak 41 detected in the positive ionization mode exhibited [M + H]+ at m/z 409.1111, C19H20O10 and a distinct fragment at m/z 247 for the loss of a hexoside moiety,46 annotated as khellol-O-hexoside. While, peaks 58 & 59 with molecular ions at m/z 261.0753 [M + H]+, C14H12O5 and m/z 231.0648 [M + H]+, C13H10O4 were annotated as khellin and visnagin, respectively as concluded from their fragmentation pattern.
Likewise, pyranocoumarins were also detected for the first time in A. digitata fruits. An acylated derivative of the pyranocoumarin, khellactone was recognized at peak 65 as a Na adduct at m/z 411.1410 corresponding to C21H24O7 as previously detected by.47 Its MS2 spectrum showed fragments at m/z 329 [M + Na − C2H3O2]+ for the cleavage of an acetic acid moiety, m/z 245 for the consecutive loss of an isovaleryl moiety (C5H8O) followed by m/z 227 and 199 for the elimination of H2O and CO. Consequently, it was annotated as isovaleryl-acetyl khellactone.47 O-Substituted esters of khellactone are reported to exhibit numerous biological activities i.e., anti-obesity,48 anti-HIV,49 and anti-cancer50 activities.
Similarly, peak 66 found in the positive ionization mode, with molecular ion at m/z 329.1377, C19H20O5 was annotated as decursin following its fragmentation pattern with fragment ions at m/z 245 [M + H − C5H8O]+.51
| No | Class | RT | KI | Name | Relative percentile (n = 2) |
|---|---|---|---|---|---|
| 1 | Nitrogenous compounds | 8.203 | 1135.3 | Hydroxylamine (3TMS) | 0.32 ± 0.14 |
| 2 | 11.4365 | 1264.4 | Urea (2TMS) | 0.023 ± 0.01 | |
| 3 | 13.7775 | 1355.4 | Uracil (2TMS) | 0.001 ± 0.0 | |
| 4 | 18.5075 | 1545.7 | 4-Aminobutanoic acid (3TMS) | 0.35 ± 0.07 | |
| 5 | 25.5705 | 1864.6 | N-Acetyl-galactosamine (TMS) | 1.09 ± 0.46 | |
| 6 | 35.3 | 2357.7 | 9-Octadecenamide (TMS) | 2.55 ± 0.26 | |
| 7 | 35.7485 | 2380.5 | 5-Methyluridine, (3TMS) | 0.169 ± 0.12 | |
| 8 | 37.4285 | 2466 | (Z)-Docos-9-enenitrile (TMS) | 2.369 ± 0.01 | |
| 9 | 41.7965 | 2688.2 | 13-Docosenamide (TMS) | 1.97 ± 0.37 | |
| Total nitrogenous compounds | 8.842 | ||||
| 10 | Acids | 11.588 | 1270.3 | Benzoic acid (TMS) | 0.76 ± 0.34 |
| 11 | 12.9275 | 1322.4 | Maleic acid, (2TMS) | 0.17 ± 0.16 | |
| 12 | 13.5745 | 1347.5 | Glyceric acid, (3TMS) | 0.094 ± 0.06 | |
| 13 | 13.905 | 1360.3 | Itaconic acid, (2TMS) | 0.19 ± 0.07 | |
| 14 | 14.134 | 1369.2 | Fumaric acid, (2TMS) | 0.27 ± 0.21 | |
| 15 | 14.403 | 1379.6 | Nonanoic acid (TMS) | 0.114 ± 0.04 | |
| 16 | 15.5195 | 1423.7 | Pentanedioic acid (2TMS) | 0.0155 ± 0.01 | |
| 17 | 17.5605 | 1506 | Malic acid, (3TMS) | 4.80 ± 2.5 | |
| 18 | 21.061 | 1655.6 | Tartaric acid (4TMS) | 0.71 ± 0.22 | |
| 19 | 23.601 | 1770.6 | Aconitic acid (3TMS) | 0.90 ± 0.088 | |
| 20 | 24.961 | 1835 | Shikimic acid (4TMS) | 1.00 ± 0.23 | |
| 21 | 25.067 | 1840.2 | Citric acid (3TMS) | 2.51 ± 3.55 | |
| 22 | 25.818 | 1876.6 | Quininic acid (5TMS) | 0.994 ± 0.23 | |
| Total acids | 12.52 | ||||
| 23 | Amino acids | 11.775 | 1277.6 | L-Serine (3TMS) | 0.17 ± 0.02 |
| 24 | 12.677 | 1312.7 | L-Threonine (2TMS) | 0.054 ± 0.01 | |
| 25 | 15.9605 | 1441.4 | L-Aspartic acid (3TMS) | 0.68 ± 0.23 | |
| 26 | 18.2905 | 1536.6 | Pyroglutamic acid (3TMS) | 0.67 ± 0.04 | |
| 27 | 18.642 | 1551.4 | L-Glutamic acid (3TMS) | 0.11 ± 0.05 | |
| Total amino acids | 1.69 | ||||
| 28 | Sugars | 15.212 | 1411.4 | Ribonic acid (4TMS) | 0.097 ± 0.01 |
| 29 | 19.2745 | 1577.9 | Erythronic acid (4-TMS) | 0.64 ± 0.15 | |
| 30 | 20.551 | 1633.1 | Ribofuranose, (4TMS) | 0.12 ± 0.03 | |
| 31 | 20.801 | 1644.1 | Xylonic acid, (5TMS) | 0.60 ± 0.03 | |
| 32 | 21.6245 | 1680.4 | Arabinose, (4TMS) | 0.48 ± 0.19 | |
| 33 | 22.9725 | 1741.7 | Rhamnose (4TMS) | 0.30 ± 0.12 | |
| 34 | 23.2415 | 1754.1 | Ribono-1,4-lactone (3TMS) | 0.21 ± 0.01 | |
| 35 | 24.4295 | 1809.3 | Fructofuranoside (5TMS) | 2.52 ± 1.85 | |
| 36 | 25.9845 | 1884.7 | Galactopyranose (5TMS) | 0.40 ± 0.07 | |
| 37 | 26.118 | 1891.2 | Fructose (5TMS) | 5.19 ± 0.83 | |
| 38 | 26.398 | 1904.9 | Arabinose (4TMS) isomer | 6.47 ± 2.69 | |
| 39 | 26.554 | 1912.9 | Galactopyranose (5TMS) isomer | 0.14 ± 0.03 | |
| 40 | 26.730 | 1921.8 | Glucose (5TMS) | 12.16 ± 0.08 | |
| 41 | 27.030 | 1937.1 | Glucose (5TMS) | 3.18 ± 4.5 | |
| 42 | 27.161 | 1943.8 | Glucono-1,4-lactone (4TMS) | 0.14 ± 0.19 | |
| 43 | 27.399 | 1955.9 | Allose (5TMS) | 0.43 ± 0.02 | |
| 44 | 27.532 | 1962.6 | Galactaric acid (6TMS) | 1.43 ± 0.16 | |
| 45 | 28.3115 | 2002.3 | Talose (5TMS) | 1.86 ± 0.49 | |
| 46 | 28.566 | 2015.2 | Gluconic acid (6TMS) | 0.05 ± 0.06 | |
| 47 | 37.7745 | 2483.6 | Sucrose (8TMS) | 0.66 ± 0.43 | |
| 48 | 42.444 | 2721.1 | Sedoheptulose anhydride (4TMS) | 0.25 ± 0.18 | |
| 49 | 43.6855 | 2784.3 | Lactose (8TMS) | 0.51 ± 0.38 | |
| 50 | 44.253 | 2813.1 | Melibiose (8TMS) | 2.88 ± 0.15 | |
| 51 | 44.4365 | 2822.5 | Galactinol (9TMS) | 4.07 ± 1.54 | |
| 52 | 44.5525 | 2828.4 | Melibiose (8TMS) isomer | 2.22 ± 2.60 | |
| Total sugars | 47.00 | ||||
| 53 | Sugar alcohol | 17.834 | 1517.5 | Threitol (4TMS) | 0.11 ± 0.04 |
| 18.017 | 1525.2 | Threitol (4TMS) isomer 1 | 0.21 ± 0.08 | ||
| 54 | 21.624 | 1680.4 | Threitol (4TMS) isomer 2 | 0.17 ± 0.23 | |
| 55 | 27.262 | 1948.9 | Mannitol (6TMS) | 6.17 ± 8.39 | |
| 56 | 29.185 | 2046.7 | Myo-inositol (6TMS) | 1.09 ± 1.55 | |
| 57 | 30.361 | 2106.5 | Myo-inositol (6TMS) isomer | 4.60 ± 0.62 | |
| 58 | 36.0015 | 2393.4 | Myo-inositol, (5TMS) phosphate | 0.87 ± 0.25 | |
| Total sugar alcohols | 13.22 | ||||
| 59 | Phenolics | 18.6975 | 1553.7 | Pyrogallol (3TMS) | 0.032 ± 0.0 |
| 60 | 20.194 | 1617.3 | Pyrogallol (3TMS) (isomer) | 0.07 ± 0.07 | |
| 61 | 42.7805 | 2738.2 | Catechin (5TMS) | 4.90 ± 0.06 | |
| Total phenolics | 5.0 | ||||
| 62 | Fatty acids/glycerides | 29.6455 | 2070.1 | Palmitic acid (TMS) | 0.65 ± 0.91 |
| 63 | 32.77 | 2229 | Oleic acid (TMS) | 0.92 ± 0.04 | |
| 64 | 33.248 | 2253.4 | Stearic acid (TMS) | 0.45 ± 0.1 | |
| 65 | 36.0975 | 2398.3 | Oleamide (TMS) | 1.06 ± 1.09 | |
| 66 | 36.505 | 2419 | Stearamide (TMS) | 0.80 ± 0.34 | |
| 67 | 38.8645 | 2539 | Monopalmitin (2TMS) | 1.24 ± 0.11 | |
| 68 | 42.5505 | 2726.5 | Lignoceric acid (TMS) | 0.08 ± 0.05 | |
| Total fatty acids/glycerides | 5.2 | ||||
| 69 | Steroids/terpenes | 44.6665 | 2834.2 | α-Tocopherol (TMS) | 0.83 ± 0.22 |
| 70 | 48.153 | 3011.5 | Campesterol (TMS) | 0.25 ± 0.02 | |
| 71 | 48.557 | 3032 | Stigmasterol (TMS) | 0.15 ± 0.03 | |
| 72 | 49.603 | 3085.2 | Stigmast-5-ene (TMS) | 2.13 ± 0.22 | |
| Total steroids/terpenes | 3.36 | ||||
| 73 | Inorganics | 12.1065 | 1290.5 | Phosphoric acid (3TMS) | 2.94 ± 0.83 |
| Total inorganics | 2.94 | ||||
| 74 | Lactones | Dihydro-2(3H)-furanone (2TMS) | 0.01 ± 0.02 | ||
| Total lactones | 0.01 | ||||
3.1.3.1. Sugars/sugar alcohols. Sugars, mostly mono-saccharides represented the most dominant primary metabolite class at ca. 47% of total detected metabolites, comprising 25 different sugars. Glucose (peaks 40 and 41) was the main sugar (15%) detected in baobab fruit followed by arabinose (peak 38) and fructose (peak 37) detected at 6.5 and 5.2%, respectively. Such high level of glucose and other monosaccharides accounts for baobab fruits sweet taste. In contrast, major disaccharides in baobab were found to be melibiose (peak 50 and 52) and galactinol (peak 51) to amount for ca. 5% of the total sugars. Galactinol is one of the raffinose family oligosaccharides that was identified in baobab fruit, with potential antioxidant action.52
Sugar alcohols amounted for 13.2% of baobab fruit total metabolites detected using GC/MS. Mannitol (6.2%) and myo-inositol amounted for the major sugar alcohols in the fruit. Myo-inositol (peaks 56 and 57), the most abundant inositol isomer in cereals, and fruits, improves insulin sensitivity.53 Consequently, baobab fruit could represent a potential functional food for diabetic patients based on these results.
3.1.3.2. Organic acids. Baobab fruit showed a considerable level of organic acids to account for the fruits acidic taste (12.5%) represented by malic acid (4.8%) and citric acid (2.5%). Malic acid is a common acid in unripe fruit, is used to enhance beverages flavor and to act as a food acidulant.54
3.1.3.3. Nitrogenous compounds/amino acids. Nitrogenous compounds were detected in A. digitata fruit at 8.8% represented mostly by 9-octadecenamide (peak 6) and docos-9-enenitrile (peak 8) at 2.55 and 2.37%, respectively. N-Acetyl-D-galactosamine (peak 5), an amino sugar derivative of galactose was detected in baobab fruit which plays a role in mucin formation, aside for its effect on immune system and cancer.55 The low amino acids level in baobab fruit (1.7%) suggests for its low pool in free amino acids, represented by pyroglutamic (peak 26), aspartic (peak 25), and glutamic (peak 27) acids.
3.1.3.4. Fatty acids/acyl esters/sterols. Fatty acids were detected in baobab fruit reaching 5.2% represented mostly by palmitic and oleic acids. Concerning fatty acyl esters, 1-monopalmitin (peak 67) was detected at 1.2%. Minor steroids were detected in baobab fruit to reach 3.36%, with stigmast-5-ene (peak 72, 2.13%) was found to be the dominant steroid.
3.1.3.5. Inorganic acids. Phosphoric acid (peak 73) was the only inorganic detected in A. digitata fruits at (2.9%). Phosphoric acid was reported to improve the antioxidant activity of tocopherols, aside from functioning as food preservative.56
3.1.3.6. Phenolics. Another considerable metabolite class detected in A. digitata fruits were phenolics detected at 5%. The major identified phenolics included catechin (peak 61), also detected using LC/MS. Catechin, a flavan-3-ol derivative common with antioxidant, antihyperlipidemic, and antidiabetic activities, and likely to account for fruit effects.57
| No | Class | RT | KI | Name | Relative percentile |
|---|---|---|---|---|---|
| 1 | Ester | 2.095 | 398 | Acetic acid, methyl ester | 0.45 ± 0.0 |
| 2 | 11.8654 | 1468 | Terpinene 4-acetate | 0.57 ± 0.0 | |
| 3 | 16.1491 | 1916 | Hexadecanoic acid methyl ester | 8.05 ± 0.0 | |
| Total esters | 9.07 | ||||
| 4 | Acids | 2.3652 | 430 | Acetic acid | 6.80 ± 0.0 |
| 5 | 8.9189 | 1166 | Octanoic acid | 0.34 ± 0.0 | |
| Total acids | 7.14 | ||||
| 6 | Aliphatic hydrocarbon | 5.3152 | 784 | 1,3-Butadiene, 2,3-dimethyl | 3.17 ± 0.0 |
| 7 | 7.1919 | 1007 | 2,4-Hexadiene | 1.81 ± 0.0 | |
| 8 | 9.283 | 1200 | Dodecane | 0.57 ± 0.0 | |
| 9 | 11.163 | 1399 | Tetradecane | 0.68 ± 0.0 | |
| 10 | 12.0777 | 1489 | Pentadecane | 0.79 ± 0.0 | |
| 11 | 13.2017 | 1598 | Hexadecane | 0.79 ± 0.0 | |
| Total aliphatic hydrocarbons | 7.82 | ||||
| 12 | Alcohol | 8.2609 | 1106 | β-Linalool | 0.45 ± 0.0 |
| Total alcohols | 0.45 | ||||
| 13 | Aldehyde | 8.304 | 1110 | Nonanal | 0.34 ± 0.0 |
| Total aldehyde | 0.34 | ||||
| 14 | Aromatic | 8.5455 | 1132 | o-Cymene | 0.03 ± 0.0 |
| 15 | 12.0261 | 1484 | p-Cymene | 1.10 ± 0.0 | |
| Total aromatics | 1.13 | ||||
| 16 | Ether | 10.8708 | 1368 | Isoeugenol | 74.49 ± 0.0 |
| Total ethers | 74.49 | ||||
Ethers were detected as the major class in A. digitata fruit (74.5%), represented mostly by isoeugenol (peak 18). Isoeugenol is a common flavoring agent used as a food preservative owing to its antimicrobial activity.58 The relative amounts of both organic acids and esters in A. digitata were at ca. 9.1 and 7.1% of total detected aroma compounds, respectively. Hexadecanoic acid methyl ester (peak 3, 8.1%) was the most abundant ester in A. digitata fruit to mediate for its antimicrobial properties.59
In contrast, alcohols were detected at trace levels of 0.5% represented by linalool (peak 14) a monoterpene alcohol used as flavoring and antibacterial agent.60 Aliphatic hydrocarbons accounted for 7.8% of the volatile composition in baobab fruit. Major forms included 1,3-butadiene, 2,3-dimethyl (peak 6), and 2,4-hexadiene (peak 7) at 3.2 and 1.8%, respectively. Aromatic compounds were also detected at a low level at 1.13% of the total volatiles represented by o-cymene (peak 16) and p-cymene (peak 17).
Typically, 1H-NMR spectrum of A. digitata fruits compromised two main regions, a low field region (δ 0.5–5.5 ppm) with more abundant signals of the primary metabolites (ESI Fig. 2a†), and a high field region of much lower abundance corresponding to secondary metabolites to account more for fruits health effects (ESI Fig. 2b†). Primary metabolites identified included fatty acids (N1 & N2), amino acids (N3–N8), organic acids (N9 & N10), and sugars (N11–N13). While secondary metabolites belonged to flavonoids (N14–N15), ESI Table S1.†
The first recognized metabolites in spectrum belonged to ω-6 fatty acids (N1) as evident from the broad signal at δ 0.87–0.91 ppm for the terminal methyl, δ 2.77 ppm for the allylic methylene and δ 5.36 for the olefinic protons.13 A fatty acid ester (N2) was recognized from the upfield shifts of H-2 and H-3 at δ 2.26 and 1.53 ppm, respectively. Several amino acids were detected including (N5) leucine (δ 0.96 ppm, d, 5.4), (N6) alanine (δ 1.41 ppm, d, 7.2), and (N8) proline (δ 4.17 ppm, dd, 2.4, 7.2). Further, downfield region revealed the presence of aromatic amino acid i.e.,(N7) phenylalanine via its characteristic signals at δ 7.55 ppm (d, 1.8), δ 7.57 ppm (d, 2.4), and δ 7.58 ppm (d, 2.4).
The presence of other nitrogenous compounds i.e., γ-aminobutyric acid (N3) was also evident from its signals at δ 1.973 ppm (m), 2.209 ppm (t, 7.2), and δ 2.72 ppm along with choline (N4) through the signal at δ 3.202 ppm (s). Besides the presence of organic acids were evident from signals ascribed to (N9) citric acid detected at δ 2.540 ppm (d, 15.6) and δ 2.567 ppm (d, 15.6), and (N10) malic acid with its signal at δ 4.091 ppm (dd, 12, 6). Citric and malic acids were detected also in GC-MS which indicate the efficiency of identification process, Table 2.
Sugars abundance observed from GC/MS Table 2, was also confirmed using 1H-NMR spectrum, and in agreement with previous reports.25 Sugars were assigned based on their characteristic anomeric signals at δ 4.401 ppm (d, 7.8) for β-glucose (N11), δ 5.033 ppm (d, 3.6) for α-glucose (N12), and δ 5.319 ppm (d, 3.6) for sucrose (N13).
Comparably, the aromatic region revealed much less abundant signals, mostly for flavonoids. As previously described, catechin and its conjugates (N11), were the major flavonoids assigned from signals at δ 5.841 ppm (d, 2.4), δ 5.86 ppm (d, 2.4) for H-6 & H-8, and characteristic multiplet at δ 3.965 ppm for H-3. Quercetin, a common flavonol in planta was also detected based on signals at δ 6.924 ppm (d, 2.4, H-6), δ 6.936 ppm (d, 2.4, H-8), and δ 7.148 ppm (d, 2.4). The enrichment of flavonoids is likely to account for fruits reported antioxidant and antidiabetic effects.61
3.2. Antidiabetic activity
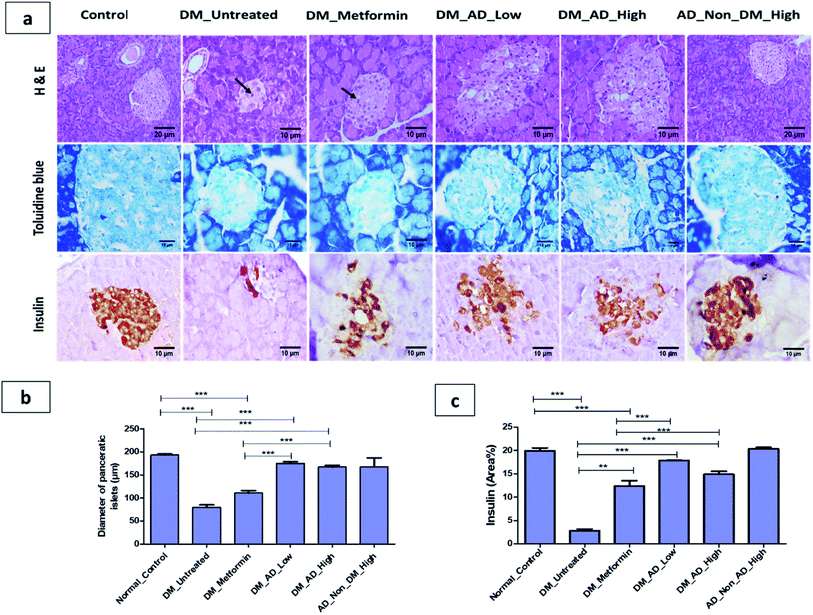
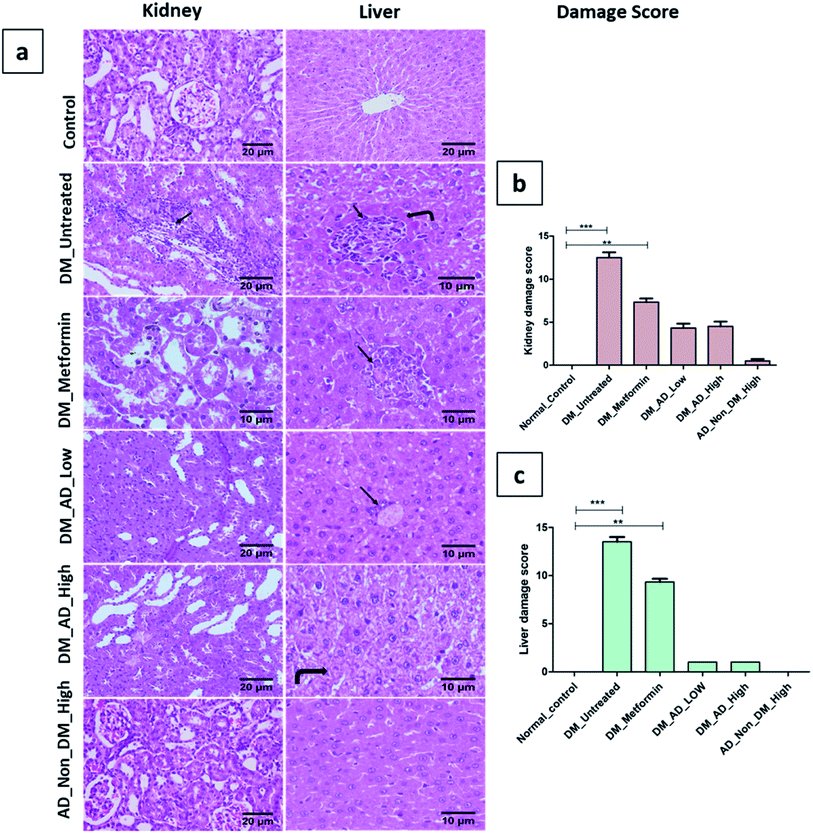
A. digitata fruits' antidiabetic and its effect on tissues histopathology and different biochemical parameters related to diabetes were investigated in vivo.Similarly, vacuolar degeneration, necrobiotic changes, and interstitial nephritis were observed in diabetic untreated and metformin-treated groups. Treatment of diabetic rats with either low or high doses of A. digitata upgraded the histological structure of the kidney to normal (Fig. 4a), while the scoring description of kidney damage is depicted in (Fig. 4b).
In the same context, administration of A. digitata at both low and high doses improved the histological structure of the liver tissue of diabetic rats. Rats receiving 150 mg kg−1 of A. digitata showed restoration of cellular structure with minimal infiltration of inflammatory cells (Fig. 4a). The normal structure was observed in the group treated with only a high dose of A. digitata extract (Fig. 4a).
4. Conclusion
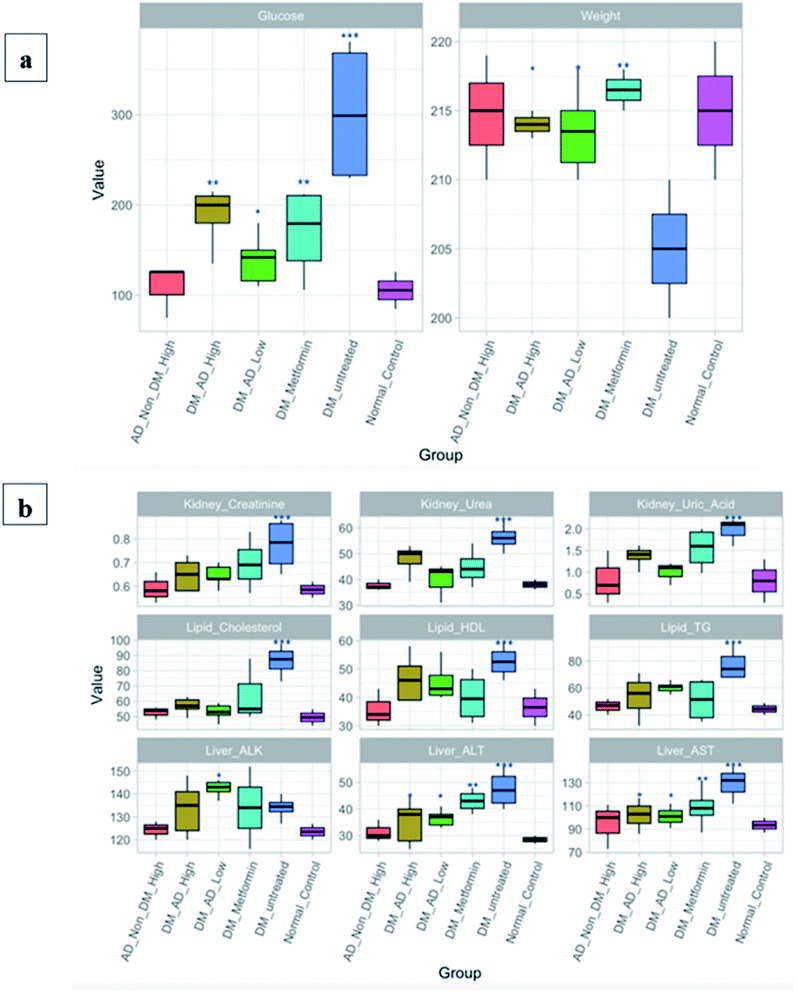
In the present study, holistic metabolites profiling of A. digitata fruits was achieved via different metabolomics platforms. UPLC-MS/MS permitted for the dereplication of 77 compounds belonging to different classes, including organic acids, sugars, alcohols, phenolics, coumarins, and fatty acids. Almost 50% of the identified metabolites (40 compounds) are reported for the first time in A. digitata fruits. GC/MS investigation for both non-volatile and volatile metabolites revealed the identification of 74 non-volatile and 16 volatile compounds. Lastly, identification and quantification of the major metabolites was possible through the employment of 1D-NMR and 2D-NMR spectroscopy. While the investigated fruit is from one origin, same metabolomics platform can be used to assess fruits from other origins for comparative analysis. Simultaneously, the antidiabetic potential of A. digitata fruits extract was also investigated revealing potent activity owing to the abundance of bioactive phytochemicals in the fruits. Histopathological and immunohistochemical studies demonstrated the hepatoprotective and renoprotective effects of A. digitata fruits in STZ induced diabetic rats. Biochemical assays revealed its capacity to lower the fasting glucose, lipid profile, hepatic and renal markers. Further investigations are required for deeper pharmacological, physiological, and biochemical insights and the determination of the optimum therapeutic dose.Funding
The work was funded by AUC financial support.Author contributions
Mostafa H. Baky: data curation; writing – review. Marwa T. Badawy: data curation, writing – review. Alaa F. Bakr: writing – review. Nesrine M. Hegazi: data curation; writing – review. Ahmed Abdellatif: writing – review. Mohamed A. Farag: supervision, writing – review & editing.Conflicts of interest
There is no conflict of interest.Acknowledgements
Dr Mohamed Farag would like to thank the Alexander Von Humboldt foundation, Germany for funding. The authors would like to acknowledge the contribution of Ms. Eman A. Khalil in facilitating some procedures in histopathology investigation.References
- G. Kamatou, I. Vermaak and A. Viljoen, S. Afr. J. Bot., 2011, 77, 908–919 CrossRef.
- A. Lamien-Meda, C. E. Lamien, M. M. Compaoré, R. N. Meda, M. Kiendrebeogo, B. Zeba, J. F. Millogo and O. G. Nacoulma, Molecules, 2008, 13, 581–594 CrossRef CAS PubMed.
- F. Chadare, A. Linnemann, J. Hounhouigan, M. Nout and M. Van Boekel, Crit. Rev. Food Sci. Nutr., 2008, 49, 254–274 CrossRef PubMed.
- J. Scheuring, M. Sidibé and M. Frigg, Sight and Life Newsletter, 1999, 1, 21–24 Search PubMed.
- C. Buchmann, S. Prehsler, A. Hartl and C. R. Vogl, Ecol. Food Nutr., 2010, 49, 145–172 CrossRef PubMed.
- Y. Shukla, S. Dubey, S. Jain and S. Kumar, J. Med. Aromat. Plant Sci., 2001, 23, 429–434 CAS.
- M. Y. Gwarzo and H. u. Y. Bako, Int. J. Anim. Vet. Adv., 2013, 5, 108–113 CrossRef.
- M. A. Farag, A. R. Khattab, A. A. Maamoun, M. Kropf and A. G. Heiss, Food Res. Int., 2019, 115, 379–392 CrossRef CAS PubMed.
- S. M. Afifi, A. El-Mahis, A. G. Heiss and M. A. Farag, ACS Omega, 2021, 6, 5775–5785 CrossRef CAS PubMed.
- M. A. Farag, S. M. Afifi, D. M. Rasheed and A. R. Khattab, J. Food Compos. Anal., 2021, 104073 CrossRef CAS.
- M. A. Farag, D. Fathi, S. Shamma, M. S. A. Shawkat, S. M. Shalabi, H. R. El Seedi and S. M. Afifi, LWT, 2021, 142, 111046 CrossRef CAS.
- M. H. Baky, M. A. Farag and D. M. Rasheed, ACS omega, 2020, 5, 31370–31380 CrossRef CAS PubMed.
- M. A. Farag, A. Otify, A. Porzel, C. G. Michel, A. Elsayed and L. A. Wessjohann, Anal. Bioanal. Chem., 2016, 408, 3125–3143 CrossRef CAS PubMed.
- M. Farag, E. Mahrous, T. Lübken, A. Porzel and L. Wessjohann, Metabolomics, 2013, 10, 21–32 CrossRef.
- M. A. Farag, A. Otify, A. Porzel, C. G. Michel, A. Elsayed and L. A. Wessjohann, Anal. Bioanal. Chem., 2016, 408, 3125–3143 CrossRef CAS PubMed.
- L. Olmo-García, K. Wendt, N. Kessler, A. Bajoub, A. Fernández-Gutiérrez, C. Baessmann and A. Carrasco-Pancorbo, Eur. J. Lipid Sci. Technol., 2019, 121, 1800336 CrossRef.
- M. E. Smoot, K. Ono, J. Ruscheinski, P. L. Wang and T. Ideker, Bioinformatics, 2011, 27, 431–432 CrossRef CAS PubMed.
- M. A. Farag, A. R. Khattab, S. Shamma and S. M. Afifi, Foods, 2021, 10, 728 CrossRef CAS PubMed.
- N. A. Qinna and A. A. Badwan, Drug Des., Dev. Ther., 2015, 9, 2515–2525 CrossRef CAS PubMed.
- D. Ahmed, V. Kumar, A. Verma, G. S. Shukla and M. Sharma, Springerplus, 2015, 4, 1–17 CrossRef CAS PubMed.
- R. Klopfleisch, BMC Vet. Res., 2013, 9(1), 1–15 CrossRef PubMed.
- G. B. Ha, E. J. Park, J. E. Shin and H. Y. Shon, PloS one, 2014, 9(7), 1–12 CrossRef PubMed.
- L. Makila, O. Laaksonen, A.-L. Alanne, M. Kortesniemi, H. Kallio and B. Yang, J. Agric. Food Chem., 2016, 64, 4584–4598 CrossRef CAS PubMed.
- P. Lorenz, J. Conrad, J. Bertrams, M. Berger, S. Duckstein, U. Meyer and F. C. Stintzing, Phytochem. Anal., 2012, 23, 60–71 CrossRef CAS PubMed.
- A. J. T. Sokeng, A. P. Sobolev, A. Di Lorenzo, J. Xiao, L. Mannina, D. Capitani and M. Daglia, Food Chem., 2019, 272, 93–108 CrossRef PubMed.
- H. Sun, J. Liu, A. Zhang, Y. Zhang, X. Meng, Y. Han, Y. Zhang and X. Wang, J. Sep. Sci., 2016, 39, 496–502 CrossRef CAS PubMed.
- E. Fujimatu, T. Ishikawa and J. Kitajima, Phytochemistry, 2003, 63, 609–616 CrossRef CAS PubMed.
- H. Tohma, İ. Gülçin, E. Bursal, A. C. Gören, S. H. Alwasel and E. Köksal, J. Food Meas. Charact., 2017, 11, 556–566 CrossRef.
- M. Lutter, A. C. Clark, P. D. Prenzler and G. R. Scollary, Food Chem., 2007, 105, 968–975 CrossRef CAS.
- Y. Zhang, L.-J. Yang, K. Jiang, C.-H. Tan, J.-J. Tan, P.-M. Yang and D.-Y. Zhu, Carbohydr. Res., 2012, 361, 114–119 CrossRef CAS PubMed.
- N. F. Abdullah and R. Mohamad Hussain, J. Liq. Chromatogr. Relat. Technol., 2015, 38, 289–293 CrossRef CAS.
- O. Razanamaro, E. Rasoamanana, B. Rakouth, J. R. Randriamalala, E. Rabakonadrianina, A. Clément-Vidal, J.-M. L. P. Tsy, C. Menut and P. Danthu, Biochem. Syst. Ecol., 2015, 60, 238–248 CrossRef CAS.
- J. A. Seukep, A. G. Fankam, D. E. Djeussi, I. K. Voukeng, S. B. Tankeo, J. A. Noumdem, A. H. Kuete and V. Kuete, Springerplus, 2013, 2, 363 CrossRef PubMed.
- B. B. Ismail, Y. Pu, M. Guo, X. Ma and D. Liu, Food Chem., 2019, 277, 279–288 CrossRef CAS PubMed.
- J. Gruz, O. Novák and M. Strnad, Food Chem., 2008, 111, 789–794 CrossRef CAS.
- S. Wu, A. E. Wilson, L. Chang and L. Tian, Molecules, 2019, 24, 3814 CrossRef CAS PubMed.
- M. K. Lee, H. Y. Jeon, K. Y. Lee, S. H. Kim, C. J. Ma, S. H. Sung, H.-S. Lee, M. J. Park and Y. C. Kim, Planta Med., 2007, 73, 782–786 CrossRef CAS PubMed.
- M. N. Clifford and S. Knight, Food Chem., 2004, 87, 457–463 CrossRef CAS.
- L. Yang, N. Nakamura, M. Hattori, Z. Wang, S. A. Bligh and L. Xu, Rapid Commun. Mass Spectrom., 2007, 21, 1833–1840 CrossRef CAS PubMed.
- X.-N. Li, J. Sun, H. Shi, L. L. Yu, C. D. Ridge, E. P. Mazzola, C. Okunji, M. M. Iwu, T. K. Michel and P. Chen, Food Res. Int., 2017, 99, 755–761 CrossRef CAS PubMed.
- I. M. Abu-Reidah, D. Arráez-Román, A. Segura-Carretero and A. Fernández-Gutiérrez, Food Res. Int., 2013, 51, 354–362 CrossRef CAS.
- A. Rauf, M. Imran, T. Abu-Izneid, S. Patel, X. Pan, S. Naz, A. S. Silva, F. Saeed and H. A. R. Suleria, Biomed. Pharmacother., 2019, 116, 108999 CrossRef CAS PubMed.
- F. Ferreres, S. Magalhães, A. Gil-Izquierdo, P. Valentão, A. R. Cabrita, A. J. Fonseca and P. B. Andrade, Food Chem., 2017, 214, 678–685 CrossRef CAS PubMed.
- R. Vinayagam and B. Xu, Nutr. Metab., 2015, 12, 1–20 CrossRef PubMed.
- M. Ganzera, S. Sturm and H. Stuppner, Chromatographia, 1997, 46, 197–203 CrossRef CAS.
- R. Schuster, Chromatographia, 1980, 13, 379–385 CrossRef CAS.
- D. N. Olennikov, I. A. Fedorov, N. I. Kashchenko, N. K. Chirikova and C. Vennos, Molecules, 2019, 24, 2286 CrossRef CAS PubMed.
- N. Taira, R. N. Nugara, M. Inafuku, K. Takara, T. Ogi, T. Ichiba, H. Iwasaki, T. Okabe and H. Oku, J. Funct. Foods, 2017, 29, 19–28 CrossRef CAS.
- L. Xie, Y. Takeuchi, L. M. Cosentino and K.-H. Lee, J. Med. Chem., 1999, 42, 2662–2672 CrossRef CAS PubMed.
- J. Y.-C. Wu, W.-F. Fong, J.-X. Zhang, C.-H. Leung, H.-L. Kwong, M.-S. Yang, D. Li and H.-Y. Cheung, Eur. J. Pharmacol., 2003, 473, 9–17 CrossRef CAS PubMed.
- H. J. Lee, H. J. Lee, E. O. Lee, J. H. Lee, K. S. Lee, K. H. Kim, S.-H. Kim and J. Lü, Am. J. Chin. Med., 2009, 37, 127–142 CrossRef CAS PubMed.
- A. Nishizawa, Y. Yabuta and S. Shigeoka, Plant Physiol., 2008, 147, 1251–1263 CrossRef CAS PubMed.
- M. L. Croze and C. O. Soulage, Biochimie, 2013, 95, 1811–1827 CrossRef CAS PubMed.
- C. Pereira, L. Barros, A. M. Carvalho and I. C. Ferreira, Food Anal. Methods, 2013, 6, 1337–1344 CrossRef.
- L. Wells, in Handbook of Glycomics, Elsevier, 2010, pp. 45–57 Search PubMed.
- A. Saba, O. Benini and A. Cupisti, Clinical Aspects of Natural and Added Phosphorus in Foods, 2017, pp. 133–141 Search PubMed.
- T. Chanadiri, T. Sanikidze, M. Esaishvili, I. Chkhikvishvili and I. Datunashvili, Georgian Med. News, 2005, 61–63 CAS.
- T. Koeduka, E. Fridman, D. R. Gang, D. G. Vassão, B. L. Jackson, C. M. Kish, I. Orlova, S. M. Spassova, N. G. Lewis and J. P. Noel, Proc. Natl. Acad. Sci., 2006, 103, 10128–10133 CrossRef CAS PubMed.
- M. T. Shaaban, M. F. Ghaly and S. M. Fahmi, J. Basic Microbiol., 2021, 61, 557–568 CrossRef CAS PubMed.
- M. Y. Issa, E. Mohsen, I. Y. Younis, E. S. Nofal and M. A. Farag, Ind. Crops Prod., 2020, 144, 112002 CrossRef CAS.
- R. G. C. Barros, U. C. Pereira, J. K. S. Andrade, C. S. de Oliveira, S. V. Vasconcelos and N. Narain, Food Res. Int., 2020, 136, 109614 CrossRef CAS PubMed.
- A. M. Otify, A. M. El-Sayed, C. G. Michel and M. A. Farag, Metabolomics, 2019, 15, 119 CrossRef PubMed.
- M. A. Farag, M. G. Sharaf El-Din, M. A. Selim, A. I. Owis, S. F. Abouzid, A. Porzel, L. A. Wessjohann and A. Otify, Molecules, 2021, 26, 761 CrossRef CAS PubMed.
- T. C. Wallace, J. K. Blusztajn, M. A. Caudill, K. C. Klatt, E. Natker, S. H. Zeisel and K. M. Zelman, Nutr. Today, 2018, 53, 240 CrossRef PubMed.
- Y. Jaiswal, P. Tatke, S. Gabhe and A. Vaidya, J. Tradit. Complement. Med., 2017, 7, 421–427 CrossRef CAS PubMed.
- M. Y. Gwarzo, Int. J. Anim. Vet. Adv., 2013, 5, 108–113 CrossRef.
- A. Braca, C. Sinisgalli, M. De Leo, B. Muscatello, P. L. Cioni, L. Milella, A. Ostuni, S. Giani and R. Sanogo, Molecules, 2018, 23, 3104 CrossRef PubMed.
- I. M. Abu-Reidah, D. Arráez-Román, M. Al-Nuri, I. Warad and A. Segura-Carretero, Food Chem., 2019, 279, 128–143 CrossRef CAS PubMed.
- K. Singh, B. K. Dubey, A. C. Tripathi, A. P. Singh and S. K. Saraf, Nat. Prod. Res., 2014, 28, 1313–1317 CrossRef CAS PubMed.
- P. Vanachayangkul, V. Butterweck and R. F. Frye, J. Chromatogr. B, 2009, 877, 653–656 CrossRef CAS PubMed.
- A. Napolitano, A. Cerulli, C. Pizza and S. Piacente, Food Chem., 2018, 269, 125–135 CrossRef CAS PubMed.
- K. Feussner and I. Feussner, in High-Throughput Metabolomics, Springer, 2019, pp. 167–185 Search PubMed.
- K.-H. Nam, H. J. Kim, I.-S. Pack, H. J. Kim, Y. S. Chung, S. Y. Kim and C.-G. Kim, Appl. Biol. Chem., 2019, 62, 15 CrossRef.
- Y. Liang, G. Y. Yan, J. L. Wu, X. Zong, Z. Liu, H. Zhou, L. Liu and N. Li, Phytochem. Anal., 2018, 29, 398–405 CrossRef CAS PubMed.
- M. A. Osman, Plant Foods Hum. Nutr., 2004, 59, 29–33 CrossRef CAS PubMed.
- G. Razafimamonjison, J. M. Leong Pock Tsy, M. Randriamiarinarivo, P. Ramanoelina, J. Rasoarahona, F. Fawbush and P. Danthu, Chem. Biodiversity, 2017, 14, e1600441 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08277a |
| ‡ Equally contributed to the work. |
| This journal is © The Royal Society of Chemistry 2021 |