Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanobiotic formulations as promising advances for combating MRSA resistance: susceptibilities and post-antibiotic effects of clindamycin, doxycycline, and linezolid
Mennatallah A. Mohameda,
Maha Nasr b,
Walid F. Elkhatib
b,
Walid F. Elkhatib *cd,
Wafaa N. Eltayeba,
Aliaa A. Elshamye and
Gharieb S. El-Sayyad
*cd,
Wafaa N. Eltayeba,
Aliaa A. Elshamye and
Gharieb S. El-Sayyad *df
*df
aMicrobiology Department, Faculty of Pharmacy, Misr International University, Cairo 19648, Egypt
bPharmaceutics and Industrial Pharmacy Department, Faculty of Pharmacy, Ain Shams University, African Union Organization St., Abbassia, Cairo 11566, Egypt
cMicrobiology and Immunology Department, Faculty of Pharmacy, Ain Shams University, African Union Organization St., Abbassia, Cairo 11566, Egypt. E-mail: walid-elkhatib@pharma.asu.edu.eg; walid-elkhatib@gu.edu.eg; walid2005faisal@yahoo.com; Fax: +20-2-24051107; Tel: +20-2-24051120
dDepartment of Microbiology & Immunology, Faculty of Pharmacy, Galala University, New Galala City, Suez, Egypt
eMicrobiology and Public Health Department, Faculty of Pharmacy and Drug Technology, Heliopolis University for Sustainable Development, Cairo Belbes Road, Cairo 11788, Egypt
fDrug Radiation Research Department, National Center for Radiation Research and Technology (NCRRT), Egyptian Atomic Energy Authority (EAEA), Cairo, Egypt. E-mail: Gharieb.S.Elsayyad@eaea.org.eg; Gharieb.Elsayyad@gu.edu.eg; Fax: +20-2-22749298; Tel: +20-2-22727413
First published on 13th December 2021
Abstract
Antimicrobial activity and post-antibiotic effects (PAEs) are both important parameters in determination of the dosage regimen of antimicrobial agents. In the present study, antimicrobial activity and PAEs of clindamycin, doxycycline, linezolid, and their nanobiotic formulations were evaluated against two methicillin resistant Staphylococcus aureus clinical isolates (MRSA) encoded (MRSA-S1 and MRSA-S2). Nanobiotic formulations increased the susceptibility of MRSA isolates by 4–64 folds as compared to their conventional ones. The PAE values were determined after exposure of MRSA isolates for 1 h to 10× the MICs of the tested antibiotics. The duration of PAEs were recorded after bacterial growth in Mueller Hinton broth (MHB) free from antibiotic has been restored. The PAE values for MRSA-S1 were 2.5 h for the conventional antibiotics. However, the PAEs for nanobiotics were 4 h for both clindamycin and linezolid, while 3 h for doxycycline. For MRSA-S2, linezolid and linezolid nanobiotics PAEs were 3 h. PAEs of clindamycin and clindamycin nanobiotics were 3.75 h and 4 h, respectively. Doxycycline and doxycycline nanobiotics revealed the same PAEs patterns of 3.5 h. The findings of the current study may positively influence the pharmacodynamics of the antibiotics and consequently the dosage regimen of nanobiotics as well as on their clinical outcome.
1. Introduction
Sir Ogston mentioned Staphylococci and their part in formation of abscess and sepsis in a variety of clinical findings and experimental studies published in 1880 and 1882.1 After more than 100 years, Staphylococcus aureus (S. aureus) continues to be a major dynamic and harmful pathogen for humans. Both community and hospital acquired staphylococcal infections have exponentially increased.2 S. aureus is a prevalent pathogen related to severe skin and soft tissue infections, as well as pneumonia and bacteremia.3–5 Due to the rising incidence of antibiotic resistance and the emergence of multidrug-resistant strains, effective treatments provided for staphylococcal infections are becoming increasingly limited.6 Infections caused by resistant pathogens give rise to high morbidity and mortality rates, which leads to global rise in healthcare costs.7 This problem is even more complex when it comes to biofilm-associated infections.8 Bacteria in biofilm express different phenotypic characters from those expressed by their planktonic counterparts.9,10 Biofilms render the bacteria more resistant to the host defense mechanisms as well as to the action of antibiotics via many several mechanisms.11 Drug-resistant bacterial infections contributes to increased doses of drugs, combination of medications which elevates toxicity, long hospitalization, and higher mortality.12 Hence, different treatment strategies have become an essential requirement.6,13 However, there is no guarantee that the production of new antimicrobial drugs will timely keep up with the rapid and regular emergence of resistance by the microbial pathogens. However, nanoparticles (NPs) is one of the most promising approaches to tackle microbial resistance.14Antimicrobial NPs provide many remarkable characteristics as compared to traditional antibiotics in minimizing toxic effects, overcoming resistance and lowering costs.15 There are also numerous nanosized drug carriers offered for the efficient administration of antibiotics by enhancing their pharmacokinetic parameters and minimizing the side effects.16,17
In assumption, NPs are maintained in the body for extended period's thanlow molecular weight antibiotics, which may be useful for consistent therapeutic effects.18
Nanoemulsions are mostly oil-in-water (o/w) or water-in-oil (w/o) where stabilization of two dispersed immiscible liquids is carried out using a suitable surfactant.19 The average diameter of droplets obtained is usually less than 500 nm.20 Coarse emulsions are associated with milky white color while small droplet size emulsions show a transparent or hazy look.21
Post-antibiotic effect is a pharmacokinetic factor which lead to the slow regeneration of living bacteria even after removal of the antibiotics from the growth medium.22–25 The period of PAE is primarily influenced by the characteristics and the concentration of the antibiotic used as well as the bacterial species. Furthermore, PAE is affected by other factors such as growth media, temperature, pH, oxygen, and the body fluid. The clinical importance of the PAE is mainly related to the implementation of antibiotic dosing strategies in medical practice. The extended PAE could allow dose reductions without decreased effectiveness, and possibly a reduction incidence of adverse effects.23
Drugs without PAEs, for example, that require administration more frequently than those that slow PAEs.26,27
Tetracyclines are wide spectrum bacteriostatic agents which inhibit protein synthesis through binding to the 30S ribosomal subunit.28,29 A second generation of tetracycline antibiotics is the long-acting doxycycline.30 It is the most commonly used tetracycline with improved lipophilic characteristics, in contrast with earlier tetracyclines.30–33 Clindamycin is a derivative of lincomycin, which explicitly acts on the 50S subunit of the bacterial ribosome, probably by controlling peptide chain initiation, thus inhibiting protein synthesis in bacteria.34 For several years, clindamycin has been recommended for treatment of serious infections caused by S. aureus.35
Linezolid is a bacteriostatic agent and protein synthesis inhibitor demonstrating excellent activity against Staphylococcus biofilms.36–38 Although linezolid has a bacteriostatic effect in vitro, some authors have observed that it may act as a bactericidal antibiotic in vivo, by inhibiting the production of staphylococcal and streptococcal toxins.39 Linezolid was the first commercially available oxazolidinone licensed by Food and Drug Administration in 2000 at the United States.40 Oxazolidinones bind to 23S ribosomal RNA of the 50S ribosomal subunit, where they prevent the formation of 70S ribosomal unit and the initiation phase of translation, inhibiting protein synthesis of bacteria.39 In the present study, novel nanobiotic formulations of clindamycin, doxycycline, and linezolid were evaluated for the PAEs compared with their corresponding classical antibiotics against two selected biofilm forming MRSA isolates.
2. Materials and methods
2.1 Materials
The antimicrobial agents tested in the present analysis were clindamycin, doxycycline, and linezolid. The Egyptian Group for Pharmaceutical Industries (El Obour, Cairo, Egypt) supplied Clindamycin and linezolid were provided by EIPICO (10th of Ramadan City, Cairo, Egypt) provided doxycycline. From dry antibiotic powders, stock solutions were prepared at a concentration of 2560 μg mL−1 and stored at −20 °C according to the manufacturer's instructions. Tween 20, oleic acid, and ethanol used in nanoemulsion synthesis were purchased from Sigma Aldrich (Darmstadt, Germany).2.2 Bacterial isolates
The clinical isolates used in this research are biofilm forming (MRSA-S1 and MRSA-S2) and they have been obtained from blood specimens, from patients at Ain Shams University Specialized Hospital and El-Demerdash Hospital (Cairo, Egypt), respectively. No patients have been contacted by the investigators in this study and all the patient identifiers were carefully removed by the Microbiology labs of the above mentioned hospitals before obtaining the isolates. Accordingly, the need for consent was not required by the ethics committees. The two isolates were resistant to the three standard antimicrobial agents; clindamycin, doxycycline, and linezolid. The reference strain selected for this research was S. aureus ATCC 25923 and was kindly and formerly provided by the United States Naval Medical Research Unit (US-NAMRU), Cairo, Egypt.2.3 In vitro quantitative assessment of MRSA-S1 and MRSA-S2 biofilms
The two selected MRSA-S1 and MRSA-S2 were tested for formation of biofilm using 96-well flat-bottomed microtiter plates (Corning, New York, USA).39 Overnight S. aureus cultures in tryptic soy broth (TSB; Conda lab, Madrid, Spain) were diluted to the 0.5 McFarland turbidity standard in TSB (equivalent to 1.5 × 108 CFU mL−1). Next, the bacterial suspensions were diluted to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in TSB to which 2% w/v glucose and 2% w/v sodium chloride were added (EL-Nasr Pharmaceuticals Chemicals Co., Cairo, Egypt). A volume of 200 microliters of these suspensions were offloaded to each of three parallel wells of the microtiter plate. Negative control wells contained sterile TSB only and S. aureus ATCC 25923 was used as the positive control. Plates were then incubated at 37 °C for 24 h under static conditions, the absorbance was recorded at wavelength 562 nm using ASYS expert plus microplate reader (Biochrom, England, UK) as a measure of overall growth. In turn, the culture was aspirated and plates were rinsed 3 successive times with 200 μL of 0.1% tryptone water (Sigma-Aldrich, Darmstadt, Germany) to remove non-adherent cells and then were air dried at room temperature. The residual adhesive biofilms were fixed using 200 microlitres of 95% ethanol (Sigma-Aldrich, Darmstadt, Germany) for each well and after 15 min; the plates were drained and left to air dry. The biofilms formed were stained for 5 min with 100 μL per well of membrane filtered crystal violet solution (0.1% w/v, Sigma-Aldrich, Darmstadt, Germany) at room temperature. Excess solution of crystal violet was drained and the biofilms were treated two times with 200 mL phosphate buffered saline (Sigma-Aldrich, Darmstadt, Germany). The crystal violet dye bound to the biofilm was re-solubilized with a mixture of 80% ethanol and 20% acetone (100 μL per well) and the plate was then incubated at room temperature for 20 min. The re-solubilized crystal violet was diluted with ethanol/acetone mixture (1
100 in TSB to which 2% w/v glucose and 2% w/v sodium chloride were added (EL-Nasr Pharmaceuticals Chemicals Co., Cairo, Egypt). A volume of 200 microliters of these suspensions were offloaded to each of three parallel wells of the microtiter plate. Negative control wells contained sterile TSB only and S. aureus ATCC 25923 was used as the positive control. Plates were then incubated at 37 °C for 24 h under static conditions, the absorbance was recorded at wavelength 562 nm using ASYS expert plus microplate reader (Biochrom, England, UK) as a measure of overall growth. In turn, the culture was aspirated and plates were rinsed 3 successive times with 200 μL of 0.1% tryptone water (Sigma-Aldrich, Darmstadt, Germany) to remove non-adherent cells and then were air dried at room temperature. The residual adhesive biofilms were fixed using 200 microlitres of 95% ethanol (Sigma-Aldrich, Darmstadt, Germany) for each well and after 15 min; the plates were drained and left to air dry. The biofilms formed were stained for 5 min with 100 μL per well of membrane filtered crystal violet solution (0.1% w/v, Sigma-Aldrich, Darmstadt, Germany) at room temperature. Excess solution of crystal violet was drained and the biofilms were treated two times with 200 mL phosphate buffered saline (Sigma-Aldrich, Darmstadt, Germany). The crystal violet dye bound to the biofilm was re-solubilized with a mixture of 80% ethanol and 20% acetone (100 μL per well) and the plate was then incubated at room temperature for 20 min. The re-solubilized crystal violet was diluted with ethanol/acetone mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) in a new plate and the optical density was determined. Biofilm formation was categorized as strong (OD562 ≥ 1.11), weak (0.22 ≤ OD562 < 1.11), and negative (OD562 < 0.22) as previously reported.41
10) in a new plate and the optical density was determined. Biofilm formation was categorized as strong (OD562 ≥ 1.11), weak (0.22 ≤ OD562 < 1.11), and negative (OD562 < 0.22) as previously reported.41
2.4 Scanning electron microscopy (SEM) for the biofilms of MRSA isolates
Biofilms of the two MRSA isolates were examined and confirmed by SEM.42 Microscopy was performed at the Scanning Electron Microscopy Department, the Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo, Egypt. The images were captured using SEM JSM-5500LV (JEOL, Tokyo, Japan). Samples were examined using the secondary electron emission mode with accelerating voltages of 15 kV. The magnifications used in the examination were 5000×, 7000×, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00×, and 130
00×, and 130![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00×.43
00×.43
2.5 Preparation and characterization of nanobiotic formulations
The nanobiotics were prepared as previously mentioned.49–52 In short, clindamycin, doxycycline, and linezolid were solubilized in tween 20, oleic acid and ethanol mixture (Sigma-Aldrich, Darmstadt, Germany) using magnetic stirrer as stated in our previously published paper.53 The mixture was diluted with water dropwise till the formation of an oil in water nanoemulsion. The antibiotic concentration in the nano-formulation was 128 μg mL−1. All nanobiotic formulations were characterized for their size, charge, and homogeneity (Zetasizer ZS3600, Malvern Co., UK) as we described formerly.532.6 Determination of minimum inhibitory concentrations (MICs)
Minimum inhibitory concentrations were measured by broth microdilution method, referring to the Clinical and Laboratory Standards Institute (CLSI, 2017) for S. aureus.54 Minimum inhibitory concentrations were determined in MHB (Oxoid, England, UK) by using a two-fold serial dilution of antibiotics and nanobiotic with a 1–3 × 105 CFU mL−1 inoculum of the tested MRSA-S1 and MRSA-S2 isolates. After 24 hours incubation at 37 °C, the MIC values were estimated.55 The MIC is the lowest concentration of the antibiotic which prevents visible growth of the bacteria.56 Determination of susceptibility were made in triplicates, and the mean of those values were reported as MIC.2.7 Post-antibiotic effect determination by viable count technique
The viable count technique was used to detect the PAE as formerly mentioned.57 Clindamycin, doxycycline, and linezolid antibiotics as well as their nanobiotics were tested against MRSA isolates (MRSA-S1 and MRSA-S2). An overnight culture of S. aureus was diluted to 106 CFU mL−1 in MHB and then incubated at 37 °C until exponential growth phase was reached.In a shaker water bath, the cultures (106 CFU mL−1) were incubated at 37 °C together with the antibiotics at 10× MIC for 1 hour. Simultaneously, a suspension of each organism that was not subjected to antibiotics was used as a control and was exposed to the previous procedures. After the exposure time has ended, the supernatant was centrifuged at 1200g for 10 min, decanted and the pellet was re-suspended in new MHB. The bacterial counts, after ten-fold serial dilution, were performed at time zero for all cultures, before and after washing and every hour up to 6 h.58 The duration of PAEs were obtained following the recovery of bacterial growth in antibiotic-free MHB measured as colony forming units (CFU mL−1) on Mueller Hinton agar (Oxoid, England, UK). The relation between time and the counts of CFU mL−1 were plotted graphically on logarithmic scale.58 The PAE was determined by the following equation: PAE = T − C, where T is the time needed for the counts of CFU mL−1 in the test culture to rise 10 folds above the count detected instantly after removal of the antibiotic and C is the time needed for the count of CFU mL−1 in the control culture to increase by 10 folds above the count obtained directly after the same procedure used on the test culture following antibiotic removal has been completed.58
2.8 Statistical analysis
The statistical examination of the obtained outcomes had been conducted applying the ONE WAY ANOVA (at P < 0.05) and agreed to be Duncan's multiple series studies and the least significant difference summary (LSD).59 The outcomes and data were reviewed and examined through SPSS software version 15.3. Results
3.1 In vitro quantitative assessment of MRSA-S1 and MRSA-S2 biofilms
The biofilm activity of the two studied MRSA-S1 and MRSA-S2 showed optical densities of 1.806 and 1.893 indicating strong biofilm forming ability.413.2 Scanning electron microscopy of MRSA biofilms
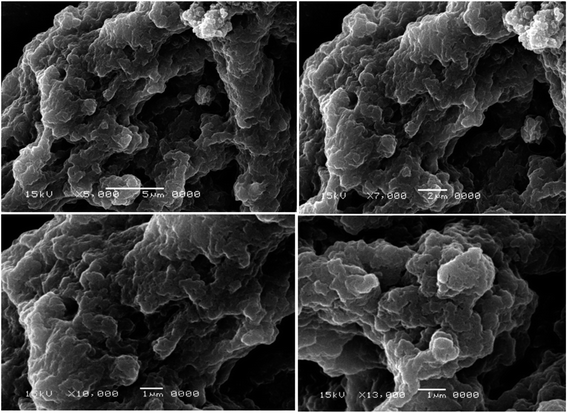
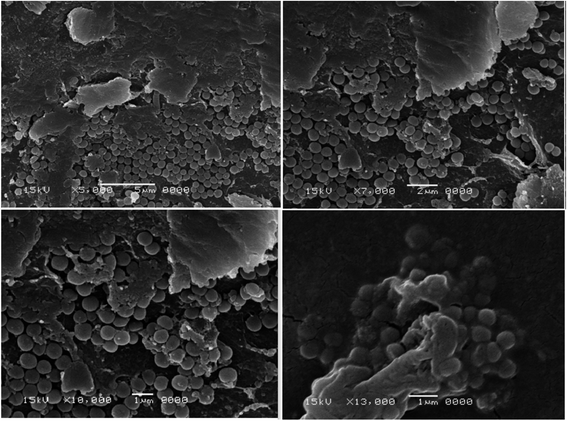
The two selected MRSA isolates were visually confirmed to have biofilm by SEM as shown in figures (Fig. 1 and 2). Scanning electron micrograph of MRSA-S1 shows a developed staphylococcal biofilm and attached coccoid staphylococcal cells were evident as shown in Fig. 1. In scanning electron micrograph of MRSA-S2 isolate, the biofilm is made of clustered cocci and it was possible to partly observe the fibriform extracellular matrix. The staphylococcal cells were mostly isolated from the biofilm in certain regions of the biofilm surface (Fig. 2).3.3 Characterization of the prepared nanobiotics
All nanobiotic formulations displayed a small particle size (11–14 nm), a negative charge (−10 to −13 mV), and moderate polydispersity of the formulation particles (data not shown).3.4 Minimum inhibitory concentrations of the tested antibiotics against MRSA clinical isolates
According to CLSI (2017), the MIC values for all tested antibiotics against MRSA-S1 and MRSA-S2 indicated their resistance or insensitivity (Table 1). On the other hand, the MIC values for clindamycin, doxycycline, and linezolid nanobiotics showed reduction by 8, 4, and 32 folds as compared to their conventional antibiotics, respectively against MRSA-S1.| Isolate code | MIC (μg mL−1) | |||||
|---|---|---|---|---|---|---|
| Clindamycin | Clindamycin nanobiotic | Doxycycline | Doxycycline nanobiotic | Linezolid | Linezolid nanobiotic | |
| a The cutoff values as proposed by the CLSI, (2017), MIC (μg mL−1) for linezolid S ≤ 4, R ≥ 8, for doxycycline S ≤ 4, I = 8, R ≥ 16, for clindamycin S ≤ 0.5, I from 1–2, R ≥ 4. | ||||||
| MRSA-S1 | 64 (R) | 8 (R) | 8 (I) | 2 (S) | 64 (R) | 2 (S) |
| MRSA-S2 | 64 (R) | 1 (I) | 64 (R) | 2 (S) | 64 (R) | 4 (S) |
Furthermore, the MIC values for clindamycin, doxycycline, and linezolid nanobiotics revealed reduction by 64, 32, and 16 folds as compared to their conventional antibiotics, respectively against MRSA-S2. Based on these results, nanoemulsion formulations of doxycycline and linezolid rendered both of MRSA-S1 and MRSA-S2 sensitive to such antibiotics. Moreover, nanoemulsion formulation of clindamycin could increase the susceptibility of the tested isolates to this antibiotic through lowering its MIC values as compared to the conventional one (Table 1).
3.5 Post-antibiotic effects (PAEs) determination
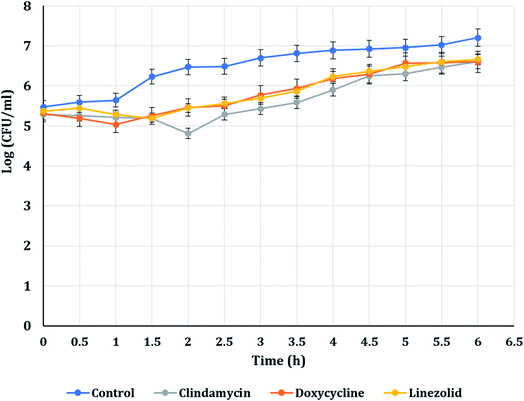
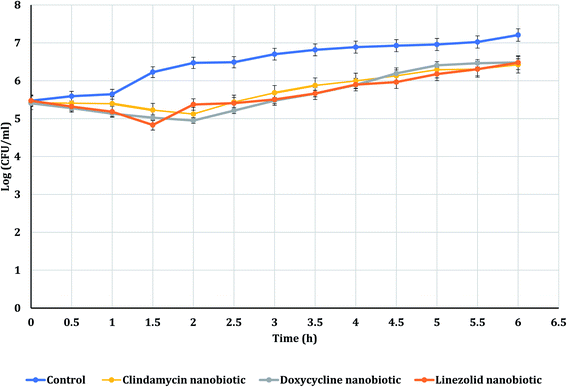
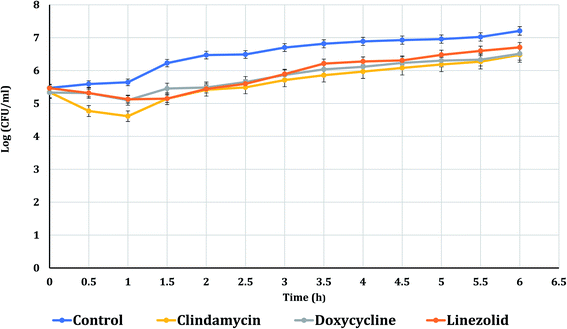
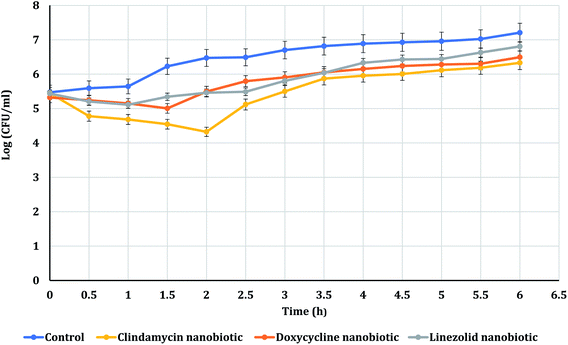
The PAE durations of clindamycin, doxycycline, and linezolid were determined at 10× MIC. The PAE values for MRSA-S1 obtained were 2.5 h for clindamycin, doxycycline, and linezolid while, PAEs were 4 h for nanobiotics of clindamycin and linezolid. On the other hand, the PAE was 3 h for doxycycline nanobiotic as shown in Fig. 3 and 4.As illustrated in Fig. 5 and 6, for the tested MRSA-S2, linezolid and linezolid-nanobiotic exhibited similar PAE patterns of 3 h. Clindamycin and clindamycin-nanobiotic exhibited PAEs of 3.75 h and 4 h, respectively. Doxycycline and doxycycline-nanobiotic combinations exhibited similar PAEs patterns of 3.5 h. Accordingly, the findings of the current analysis revealed that the formulation of antibiotics as nanoemulsions could produce either prolonged or similar PAEs as compared to their conventional antibiotics.
4. Discussion
Due to the prevalence of both community-acquired and hospital-acquired staphylococcal infections, methicillin resistant S. aureus was selected as a focus in the current study.60 Another reason is their ability to form biofilms where bacteria in biofilms are 1000 times more resistant to antibiotics relative to their planktonic forms.61–64 For these reasons, S. aureus nowadays is reported among ESKAPE organisms.13 The ESKAPE organisms, include Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, P. aeruginosa, and Enterobacter spp.65 These alert organisms are typical causes of hospital infections and are also multidrug resistant.66–68 As multidrug resistant bacteria are in a race with human to develop sustainable antimicrobial agents, biofilm forming Staphylococcus Spp. provoked our eagerness to challenge. The pillar of successful biofilm therapy, which is viewed as a global problem, is efficient biofilm assays.41 Although different methods are used to assess biofilm formation,69–71 in the present study crystal violet assay was chosen to assess S. aureus biofilm production.According to O'Toole (2011),72 a significant technique for studying the initial stages of biofilm formation is the microtiter plate assay. In addition, microtiter plate assay supports the development of a biofilm on the microtiter plate 's bottom and/or wall. Moreover, according to Xu et al. (2016),73 crystal violet can stain extracellular matrix, dead cells, and viable cells proving that crystal violet assay has a benefit in evaluating the overall biofilm production. Furthermore, according to Magana et al. (2018),74 one of the most widely used in vitro biofilm assessment techniques is crystal violet assay, enabling optical determination of biofilm thickness and total measurement of biomass, particularly in the early stages. However, in contrast to the previously mentioned advantages, owing to its high heterogeneity, crystal violet lacks reliability when the pigment binds un-specifically to negatively charged molecules or when it is unequally eluted by ethanol.73,74
Scanning Electron Microscope (SEM) continues to be unique in its power to analyze dimensional topography and allocation of visible structures despite the expanding range of specialized imaging instruments and the progress in types of microscopy.75,76 In the present study, biofilms of the two selected MRSA isolates were confirmed by SEM that has been efficiently used to capture biofilms due to its good resolution and magnification. Pattern and bulk of biofilm are essential characteristics that regulate the dynamics of substrate elimination by biofilms. SEM has been implemented on biofilms as it is an effective tool to show the fine details of living systems.77
Using the standard stages of fixation, post-fixation, dehydration, mounting, and sputter coating, all biofilm samples were prepared. To maintain the morphology of the biological sample, it was first fixed with aldehyde. Glutaraldehyde was used as a fixative in the current study because it permanently fixes the biofilm structure.78 Post-fixation with a fixative-containing heavy metal improved the strength of the cell structure and enhanced the contrast of the sample under the light source. In order to eliminate any reactive substance which could disrupt the performance of the electron microscope, the sample was dehydrated and left to dry.79 The dried sample was fixed to a conductive stub and covered with conductive material to minimize disruption of samples and improve topographical differentiation for enhanced imaging by SEM using secondary electron detection.80
Pharmacodynamics of antibiotics have been considered as vital parameters influencing the model of antibiotic intake.81 The PAE which is the slow bacterial regrowth after exposure to an antibiotic is a feature of pharmacodynamics studies.82 In the present study, viable count technique has been used to determine PAEs of different conventional antibiotics and their corresponding combinations with nanoemulsion. Methicillin resistant S. aureus was particularly chosen in the current study, because it is the primary cause of nosocomial infections in surgical wounds. Furthermore, biofilm forming MRSA can cause serious indwelling medical devices-associated infections.83,84
In the present study, the three antibiotics including linezolid, doxycycline, and clindamycin were encapsulated using nano-emulsion formulation.85 The nano-size gives the emulsion formulation a clear or blurry look in contrast to the coarse emulsion which appears milky white in color.86 Nanoemulsions were particularly designed in the present study as they are thermodynamically stable systems which are simple to make, and can dissolve hydrophobic medications, increasing the bioavailability of drugs or antibiotics.84,87 Compared to new antibiotics synthesis, production of antibiotic nano-systems could be cost-effective, and they are stable even with long-term storage. Antibiotic delivery using nanoparticles may afford multiple advantages including better solubility, controllable and consistent dissemination in specific tissues, sustained release, improved patient acceptance, reduced adverse effects, and increased cellular assimilation especially in biofilm forming isolates.88 Nanoemulsions used in this study are generally safe as previously described.53
The MIC values for all tested antibiotics against MRSA-S1 and MRSA-S2 isolates indicated their resistance. However, the MIC values for clindamycin, doxycycline, and linezolid nanobiotics showed reduction up to 32 folds as compared to their conventional antibiotics against MRSA-S1. Furthermore, the MIC values for clindamycin, doxycycline, and linezolid nanobiotics revealed reduction up to 64 folds as compared to their conventional antibiotics against MRSA-S2. In agreement with Allahverdiyev et al. (2011),89 the findings of the current study prove that combining antibiotics with NPs restores their inhibitory impact on different bacteria that have acquired resistance to them. As per Hussein-Al-Ali et al. (2014),90 NPs exert their antimicrobial effects either directly through their interactions with microbial cell targets, such as the cell envelope, or indirectly as potential transporters for antimicrobial agents, promoting their targeted transmission and enhanced diffusion into the bacterial cells. Compared to other antimicrobial agents, combination of bacterial protein synthesis inhibitors with nanoparticles showed higher antibacterial activity.91
In the present study, doxycycline exerted PAEs of 2.5 h and 3.5 h against MRSA-S1 and MRSA-S2, respectively. Doxycycline nanoemulsion exerted PAEs of 3 h and 3.5 h against MRSA-S1 and MRSA-S2 isolates, respectively. Earlier in a study performed by Cunha et al. (2000),58 it was confirmed that doxycycline exerted a dose-dependent PAE that varied between 2.5 h and 3.5 h. Recently, Sime and Roberts (2018),92 reported that tetracyclines exhibited a time-dependent bactericidal activity against various pathogens and could develop extended PAEs.
Clindamycin exerted PAEs of 2.5 h and 3.75 h against MRSA-S1 and MRSA-S2 isolates, respectively. Clindamycin nanoemulsion exerted PAE of 4 h against both MRSA-S1 and MRSA-S2 isolates. Results of the present study agreed with previous report, in which clindamycin exhibited an in vitro post antibiotic effect of 0.4–3.9 h against S. aureus isolates.93 Recently, Donaldson and Jason (2017),94 mentioned that clindamycin showed extended PAEs against some types of bacteria, which may be attributed to persistence of clindamycin at the ribosomal binding site. In addition, clindamycin has the ability to disrupt bacterial protein synthesis, causing changes in the bacterial cell wall, reducing bacterial adherence to host cells, and increasing intracellular destruction of susceptible organisms.94 Linezolid exhibited PAEs of 2.5 h and 3 h for MRSA-S1 and MRSA-S2 isolates, respectively. Linezolid nanoemulsion formulation exhibited PAEs of 4 h and 3 h, respectively. According to Sime and Roberts (2018),92 linezolid showed an antibacterial activity dependent on time and limited to small PAE.
Generally, nanoemulsions increase drug retention time in the desired area causing fewer adverse effects or toxicities where they act only on the wanted regions of the body.95–97 In nano-formulation, less drug quantity is needed due to the enhanced diffusion, improved bioavailability, increased retention time, and reduced drug loss.98,99 Although PAE is a crucial pharmacodynamic parameter of the antibiotic and could offer valuable clinical knowledge for a dose protocol, further studies are still needed to elucidate the mechanism. Many researches have documented that the growth kinetics, structure and biological activity of bacteria could be influenced by PAE.100,101 In addition, white blood cells could also exhibit stronger bactericidal activity in vivo. This can partly clarify why most antibiotics show longer PAE in vivo than in vitro.3
5. Conclusions
Formulations of the antibiotics as nanoemulsions could increase the susceptibility of MRSA isolates to these antibiotics through lowering the MIC values as compared to their conventional ones. Moreover, nanobiotic formulations revealed either prolonged or similar PAEs as compared to their conventional antibiotics. Consequently, this finding can influence the pharmacodynamic parameters of the antibiotic and may possess useful impacts on the dosage regimen of nanobiotics as well as on the clinical outcomes. In order to validate our findings and to determine the in vivo efficacy of the antibiotic formulations, more pharmacokinetic and pharmacodynamics studies are essential.Funding
This research has not received any specific grants from funding agencies in the public, commercial, or nonprofit sectors. The authors received no financial support for the research, authorship, and/or publication of this article.Compliance with ethics requirement
This article does not contain any studies involving human subjects or experimental animals.Consent to participate
No experimental investigation was performed on individuals within this research study.Consent for publication
All the co-authors are agreed for the research study publication.Availability of data and material
The authors stated and declare that all data is exist and available.Code availability
The authors stated and declare that all code is exist and available.Conflicts of interest
The authors stated and declare that no conflict or competing of interests.Acknowledgements
The authors would like to express their gratitude to Egyptian group for pharmaceutical industries, El Odour, Cairo, Egypt for supplying clindamycin and linezolid antibiotic powders as well as EIPICO, 10th of Ramadan City, Cairo, Egypt for providing doxycycline antibiotic powder.References
- A. Ogston, Rev. Infect. Dis., 1984, 6, 122–128 CrossRef.
- F. D. Lowy, N. Engl. J. Med., 1998, 339, 520–532 CrossRef CAS PubMed.
- H. Chen, L. Li, Y. Liu, M. Wu, S. Xu, G. Zhang, C. Qi, Y. Du, M. Wang and J. Li, Infect. Drug Resist., 2018, 11, 2107 CrossRef CAS PubMed.
- R. S. Daum, N. Engl. J. Med., 2007, 357, 380–390 CrossRef CAS PubMed.
- R. J. Gordon and F. D. Lowy, Clin. Infect. Dis., 2008, 46, S350–S359 CrossRef CAS PubMed.
- M. E. Falagas, A. P. Grammatikos and A. Michalopoulos, Expert Rev. Anti-Infect. Ther., 2008, 6, 593–600 CrossRef PubMed.
- A. El Kholy, H. Baseem, G. S. Hall, G. W. Procop and D. L. Longworth, J. Antimicrob. Chemother., 2003, 51, 625–630 CrossRef CAS PubMed.
- U. Römling and C. Balsalobre, J. Intern. Med., 2012, 272, 541–561 CrossRef PubMed.
- S. Miquel, R. Lagrafeuille, B. Souweine and C. Forestier, Front. Microb., 2016, 7, 592 Search PubMed.
- W. Elkhatib and A. Noreddin, Microb. Drug Resist., 2014, 20, 575–582 CrossRef CAS PubMed.
- A. Gupta, R. F. Landis, C.-H. Li, M. Schnurr, R. Das, Y.-W. Lee, M. Yazdani, Y. Liu, A. Kozlova and V. M. Rotello, J. Am. Chem. Soc., 2018, 140, 12137–12143 CrossRef CAS PubMed.
- M. A. Riley, S. M. Robinson, C. M. Roy, M. Dennis, V. Liu and R. L. Dorit, Biochem. Soc. Trans., 2012, 40, 1438–1442 CrossRef CAS PubMed.
- W. Elkhatib and A. Noreddin, J. Med. Devices, 2009, 3, 027543 CrossRef.
- R. Y. Pelgrift and A. J. Friedman, Adv. Drug Delivery Rev., 2013, 65, 1803–1815 CrossRef CAS PubMed.
- R. P. Allaker and G. Ren, Trans. R. Soc. Trop. Med. Hyg., 2008, 102, 1–2 CrossRef PubMed.
- V. Patravale, A. A. Date and R. Kulkarni, J. Pharm. Pharmacol., 2004, 56, 827–840 CrossRef CAS PubMed.
- S. Milewska, K. Niemirowicz-Laskowska, G. Siemiaszko, P. Nowicki, A. Z. Wilczewska and H. Car, Int. J. Nanomed., 2021, 16, 6593 CrossRef PubMed.
- A. J. Huh and Y. J. Kwon, J. Controlled Release, 2011, 156, 128–145 CrossRef CAS PubMed.
- R. P. Patel and J. R. Joshi, Int. J. Pharm. Sci. Res., 2012, 3, 4640 CAS.
- R. Najafi-Taher and A. Amani, Nanomed. Res. J., 2017, 2, 49–56 CAS.
- Y. Singh, J. G. Meher, K. Raval, F. A. Khan, M. Chaurasia, N. K. Jain and M. K. Chourasia, J. Controlled Release, 2017, 252, 28–49 CrossRef CAS PubMed.
- J. Bigger, Lancet, 1944, 497–500 CrossRef.
- A. Athamna, M. Athamna, B. Medlej, D. Bast and E. Rubinstein, J. Antimicrob. Chemother., 2004, 53, 609–615 CrossRef CAS PubMed.
- B. Bedenić, N. Beader, K. Godič-Torkar, E. Prahin, L. Mihaljević, M. Ćačić and J. Vraneš, J. Chemother., 2016, 28, 375–382 CrossRef PubMed.
- R. A. Sorg and J.-W. Veening, Nat. Commun., 2015, 6, 1–13 Search PubMed.
- I. Odenholt-Tornqvist, E. Löwdin and O. Cars, Antimicrob. Agents Chemother., 1992, 36, 1852–1858 CrossRef CAS PubMed.
- S. K. Spangler, G. Lin, M. R. Jacobs and P. C. Appelbaum, Antimicrob. Agents Chemother., 1998, 42, 1253–1255 CrossRef CAS PubMed.
- F. R. McSorley, J. W. Johnson and G. D. Wright, in Antimicrobial Resistance in the 21st Century, Springer, 2018, pp. 533–562 Search PubMed.
- H. Ullah and S. Ali, Antibact. Agents, 2017, 10, 1–15 Search PubMed.
- C. S. Monk, S. Y. Jeong, D. J. Gibson and C. E. Plummer, Vet. Ophthalmol., 2018, 21, 58–65 CrossRef CAS PubMed.
- A. D'Souza, K. Flynn, S. Chhabra, B. Dhakal, M. Hamadani, K. Jacobsen, M. Pasquini, D. Weihrauch and P. Hari, Contemporary Clinical Trials Communications, 2017, 8, 33–38 CrossRef PubMed.
- M. G. Maaland, M. G. Papich, J. Turnidge and L. Guardabassi, J. Clin. Microbiol., 2013, 51, 3547–3554 CrossRef CAS PubMed.
- D. Zhang, J. Zhao, Q. Wang, Y. Liu, C. Tian, Y. Zhao, L. Yu and M. Liu, Microb. Pathog., 2017, 105, 51–56 CrossRef CAS PubMed.
- J. Spížek and T. Řezanka, Appl. Microbiol. Biotechnol., 2004, 64, 455–464 CrossRef PubMed.
- G. Martínez-Aguilar, W. A. Hammerman, E. O. Mason Jr and S. L. Kaplan, Pediatr. Infect. Dis. J., 2003, 22, 593–599 Search PubMed.
- M. S. Butler, M. A. Blaskovich and M. A. Cooper, J. Antibiot., 2013, 66, 571–591 CrossRef CAS PubMed.
- S. R. Martinez, D. M. Rocca, V. Aiassa and M. C. Becerra, RSC Adv., 2016, 6, 101023–101028 RSC.
- R. Saginur, M. StDenis, W. Ferris, S. D. Aaron, F. Chan, C. Lee and K. Ramotar, Antimicrob. Agents Chemother., 2006, 50, 55–61 CrossRef CAS PubMed.
- M. De Rosa, M. Bonomo, A. Vassallo, G. Palma, L. Calabrone, S. Bimonte, N. Silvestris, N. Amruthraj, M. Sinicropi and G. Salzano, PharmacologyOnLine, 2018, 2, 134–148 CAS.
- F. A. K. Khan, R. N. Kaduskar, R. Patil, R. H. Patil, S. A. Ansari, H. M. Alkahtani, A. A. Almehizia, D. B. Shinde and J. N. Sangshetti, Bioorg. Med. Chem. Lett., 2019, 29, 623–630 CrossRef CAS PubMed.
- W. F. Elkhatib, A. S. Khairalla and H. M. Ashour, Future Microbiol., 2014, 9, 725–735 CrossRef CAS PubMed.
- A. I. El-Batal, H. G. Nada, R. R. El-Behery, M. Gobara and G. S. El-Sayyad, RSC Adv., 2020, 10, 9274–9289 RSC.
- G. S. El-Sayyad, M. Abd Elkodous, A. M. El-Khawaga, M. A. Elsayed, A. I. El-Batal and M. Gobara, RSC Adv., 2020, 10, 5241–5259 RSC.
- M. A. Maksoud, G. S. El-Sayyad, H. S. El-Bastawisy and R. M. Fathy, RSC Adv., 2021, 11, 28361–28374 RSC.
- S. Elbasuney, G. S. El-Sayyad, H. Tantawy and A. H. Hashem, RSC Adv., 2021, 11, 25961–25975 RSC.
- E. Fischer, B. Hansen, V. Nair, F. Hoyt and D. Dorward, Curr. Protoc. Microbiol., 2012, 25, 2B.2.1–2B.2.47 CrossRef PubMed.
- M. K. Abdel-Rafei, N. M. Thabet, M. Abdel Maksoud, M. Abd Elkodous, G. Kawamura, A. Matsuda, A. Ashour, A. I. El-Batal and G. S. El-Sayyad, Int. J. Mol. Sci., 2021, 22, 10171 CrossRef CAS.
- A. N. El-Shazly, G. S. El-Sayyad, A. H. Hegazy, M. A. Hamza, R. M. Fathy, E. El Shenawy and N. K. Allam, Sci. Rep., 2021, 11, 1–14 CrossRef PubMed.
- R. M. Hathout and M. Nasr, Colloids Surf., B, 2013, 110, 254–260 CrossRef CAS PubMed.
- M. Nasr and S. Abdel-Hamid, Drug Dev. Ind. Pharm., 2016, 42, 636–643 CrossRef CAS PubMed.
- M. Nasr, S. Abdel-Hamid, N. H Moftah, M. Fadel and A. A. Alyoussef, Curr. Drug Delivery, 2017, 14, 426–432 CrossRef CAS PubMed.
- S. A. Ramez, M. M. Soliman, M. Fadel, F. Nour El-Deen, M. Nasr, E. R. Youness and D. M. Aboel-Fadl, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 996–1002 CrossRef CAS PubMed.
- M. A. Mohamed, M. Nasr, W. F. Elkhatib and W. N. Eltayeb, BioMed Res. Int., 2018, 2018, 7658238 Search PubMed.
- T. C. Abbey and E. Deak, Clinical Microbiology Newsletter, 2019, 41, 203–209 CrossRef.
- M. Abd Elkodous, G. S. El-Sayyad, S. M. Youssry, H. G. Nada, M. Gobara, M. A. Elsayed, A. M. El-Khawaga, G. Kawamura, W. K. Tan and A. I. El-Batal, Sci. Rep., 2020, 10, 1–22 CrossRef PubMed.
- M. Abd Elkodous, G. S. El-Sayyad, I. Y. Abdelrahman, H. S. El-Bastawisy, F. M. Mosallam, H. A. Nasser, M. Gobara, A. Baraka, M. A. Elsayed and A. I. El-Batal, Colloids Surf., B, 2019, 180, 411–428 CrossRef CAS PubMed.
- W. Craig and S. Gudmundsson, Antibiotics in Laboratory Medicine, 1996, 296–329 Search PubMed.
- B. Cunha, P. Domenico and C. Cunha, Clin. Microbiol. Infect., 2000, 6, 270–273 CrossRef CAS PubMed.
- A. M. Brown, Computer Methods and Programs in Biomedicine, 2005, 79, 89–95 CrossRef PubMed.
- A. H. Salem, W. F. Elkhatib and A. M. Noreddin, J. Pharm. Pharmacol., 2011, 63, 73–79 CrossRef CAS PubMed.
- M. Chen, Q. Yu and H. Sun, Int. J. Mol. Sci., 2013, 14, 18488–18501 CrossRef PubMed.
- S. Darwish, A. Noreddin, R. Tiwari and W. F. Elkhatib, Int. J. Pept. Res. Ther., 2019, 25, 1075–1085 CrossRef CAS.
- W. Elkhatib and A. Noreddin, Antibiotics, 2014, 3, 64–84 CrossRef PubMed.
- W. Elkhatib, V. Haynes and A. Noreddin, J. Chemother., 2009, 21, 135–143 CrossRef CAS PubMed.
- L. B. Rice, J. Infect. Dis., 2008, 197(8), 1079–1081 CrossRef PubMed.
- J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417–433 CrossRef CAS PubMed.
- M. M. Sakr, K. M. Aboshanab, W. F. Elkhatib, M. A. Yassien and N. A. Hassouna, Appl. Microbiol. Biotechnol., 2018, 102, 10613–10622 CrossRef CAS PubMed.
- D. M. Osama, W. F. Elkhatib, A. M. Tawfeik, M. M. Aboulwafa and N. A.-H. Hassouna, Int. J. Biotechnol. Wellness Ind., 2017, 6, 12–21 CrossRef CAS.
- M. A. Maksoud, G. S. El-Sayyad, A. Ashour, A. I. El-Batal, M. A. Elsayed, M. Gobara, A. M. El-Khawaga, E. Abdel-Khalek and M. El-Okr, Microb. Pathog., 2019, 127, 144–158 CrossRef CAS PubMed.
- A. I. El-Batal, G. S. El-Sayyad, N. E. Al-Hazmi and M. Gobara, J. Cluster Sci., 2019, 30, 947–964 CrossRef CAS.
- A. I. El-Batal, N. M. Balabel, M. S. Attia and G. S. El-Sayyad, J. Cluster Sci., 2020, 31, 1021–1040 CrossRef CAS.
- G. A. O’Toole, J. Visualized Exp., 2011, 72(47), 2437 Search PubMed.
- Z. Xu, Y. Liang, S. Lin, D. Chen, B. Li, L. Li and Y. Deng, Curr. Microbiol., 2016, 73, 474–482 CrossRef CAS PubMed.
- M. Magana, C. Sereti, A. Ioannidis, C. A. Mitchell, A. R. Ball, E. Magiorkinis, S. Chatzipanagiotou, M. R. Hamblin, M. Hadjifrangiskou and G. P. Tegos, Clin. Microbiol. Rev., 2018, 31, 16–84 CrossRef PubMed.
- A. Eberle, S. Mikula, R. Schalek, J. Lichtman, M. K. Tate and D. Zeidler, J. Microsc., 2015, 259, 114–120 CrossRef CAS PubMed.
- A. Ashour, A. I. El-Batal, M. A. Maksoud, G. S. El-Sayyad, S. Labib, E. Abdeltwab and M. El-Okr, Particuology, 2018, 40, 141–151 CrossRef CAS.
- N. Raab and I. Bachelet, Journal of Biological Methods, 2017, 4, 70–75 CrossRef PubMed.
- M. A. Maksoud, G. S. El-Sayyad, A. M. El-Khawaga, M. Abd Elkodous, A. Abokhadra, M. A. Elsayed, M. Gobara, L. Soliman, H. El-Bahnasawy and A. Ashour, J. Hazard. Mater., 2020, 399, 123000 CrossRef PubMed.
- W. F. Khalil, G. S. El-Sayyad, W. M. El Rouby, M. Sadek, A. A. Farghali and A. I. El-Batal, Int. J. Biol. Macromol., 2020, 164, 1370–1383 CrossRef CAS PubMed.
- Y. Zhang, T. Huang, D. M. Jorgens, A. Nickerson, L.-J. Lin, J. Pelz, J. W. Gray, C. S. López and X. Nan, PLoS One, 2017, 12, e0176839 CrossRef PubMed.
- A. M. Noreddin and W. F. Elkhatib, Journal of Infection and Public Health, 2009, 2, 120–128 CrossRef PubMed.
- F. MacKenzie and I. Gould, J. Antimicrob. Chemother., 1993, 32, 519–537 CrossRef CAS PubMed.
- B. Cunha, Clin. Microbiol. Infect., 2005, 11, 33–42 CrossRef CAS PubMed.
- A. Salem, W. Elkhatib, G. Ahmed and A. Noreddin, J. Chemother., 2010, 22, 238–242 CrossRef CAS PubMed.
- C. Wu, L. Wang, Y. Hu, S. Chen, D. Liu and X. Ye, RSC Adv., 2016, 6, 20892–20900 RSC.
- N. Dasgupta and S. Ranjan, An Introduction to Food Grade Nanoemulsions, 2018, pp. 63–82 Search PubMed.
- M. Tariq, S. Mohurle, V. B. Patravale and K. Aruna, Int. J. Curr. Microbiol. Appl. Sci., 2016, 5, 190–201 CrossRef CAS.
- C. A. Omolo, R. S. Kalhapure, N. Agrawal, S. Rambharose, C. Mocktar and T. Govender, Mol. Pharmaceutics, 2018, 15, 3512–3526 CrossRef CAS PubMed.
- A. M. Allahverdiyev, K. V. Kon, E. S. Abamor, M. Bagirova and M. Rafailovich, Expert Rev. Anti-Infect. Ther., 2011, 9, 1035–1052 CrossRef CAS PubMed.
- S. H. Hussein-Al-Ali, M. E. El Zowalaty, M. Z. Hussein, B. M. Geilich and T. J. Webster, Int. J. Nanotechnol., 2014, 9, 3801 CAS.
- F. Shakeel, Chiang Mai J. Sci., 2017, 44, 1049–1055 CAS.
- F. B. Sime and J. A. Roberts, in Antibiotic Pharmacokinetic/Pharmacodynamic Considerations in the Critically Ill, Springer, 2018, pp. 17–29 Search PubMed.
- I. B. Xue, P. G. Davey and G. Phillips, Antimicrob. Agents Chemother., 1996, 40, 1403–1407 CrossRef CAS PubMed.
- B. Mark Donaldson and H. G. Jason, General dentistry, 2017 Search PubMed.
- M. Pandey, H. Choudhury, O. C. Yeun, H. M. Yin, T. W. Lynn, C. L. Tine, N. S. Wi, K. C. Yen, C. S. Phing and P. Kesharwani, Curr. Pharm. Biotechnol., 2018, 19, 276–292 CAS.
- G. S. R. Raju, L. Benton, E. Pavitra and J. S. Yu, Chem. Commun., 2015, 51, 13248–13259 RSC.
- H. Qin, H. Zhang, L. Li, X. Zhou, J. Li and C. Kan, RSC Adv., 2017, 7, 52684–52693 RSC.
- H. Yan, C. Bao, X. Chen, C. Yu, D. Kong, J. Shi and Q. Lin, RSC Adv., 2019, 9, 11649–11658 RSC.
- K. B. Sutradhar and M. L. Amin, Eur. J. Nanomed., 2013, 5, 97–110 Search PubMed.
- X. Meng, C. H. Nightingale, K. R. Sweeney and R. Quintiliani, J. Antimicrob. Chemother., 1994, 33, 721–728 CrossRef CAS PubMed.
- R. P. Singh, MedChemComm, 2015, 6, 259–272 RSC.
| This journal is © The Royal Society of Chemistry 2021 |