Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
New triterpenes from Cimicifuga yunnanensis down-regulating the mRNA expression of CD147, MMP-2, and MMP-9†
Ni-Hong Lu,
Jie Li,
Yong-Rui Yang,
Hong-Lu Liu and
Ying-Rong Du *
*
Department of Respiratory Medicine, The Third People's Hospital of Kunming, Yunnan, 650041, People's Republic of China. E-mail: 602157606@qq.com
First published on 17th November 2021
Abstract
Eleven new 9,19-cycloartane triterpenes (1–9, 11–12) and one undescribed lanostane-type aglycone (10) were identified from the aerial parts of Cimicifuga yunnanensis. The new structures were elucidated by analysis of spectroscopic data. Compounds 3–5, 7–9, and 11, without obvious cytotoxicity at 50 μM, were evaluated for inhibiting the mRNA expressions of atherosclerosis-related factors of CD147 (extracellular matrix metalloproteinase inducer, EMMPRIN), matrix metalloproteinase 2 (MMP-2) and MMP-9 in phorbol-12-myristate-13-acetate (PMA) induced Human monocytic THP-1 cells by using a quantitative real-time PCR method (q-PCR). Among them, aglycones 7 and 8 showed potent activities, whereas all tested glycosides were inactive. Compounds 7 and 8 suppressed the mRNA expression of CD147 in a dose-dependent manner, with an IC50 value of 3.38 ± 0.27 μM and 8.25 ± 0.33 μM, respectively. Besides, 7 dose-related down-regulated the mRNA expression of MMP-2, and MMP-9, having an IC50 value of 6.32 ± 0.31 μM and 11.57 ± 0.23 μM, respectively. Meanwhile, 8 at 10 μM reduced the mRNA expression of MMP-2 and MMP-9 by 35% and 25%, respectively. Significantly, the migration ability of the induced THP-1 cells was potently and dose-dependently inhibited by 7, with an IC50 value of 5.87 ± 0.27 μM.
1. Introduction
Cardiovascular diseases (CVDs) are leading causes of death globally, by which more than 17.0 million people die every year.1,2 Pathologically, atherosclerosis, a chronic inflammatory disease, is a critical causing factor of CVDs.1,3 Patients who can maintain stability of atherosclerotic plaques will adapt to stable angina pectoris. Otherwise, a life-threatening acute coronary syndrome, including acute myocardial infarction (AMI) and unstable angina pectoris (UA) would happen to them.4–6 CD147, extracellular matrix metalloproteinase inducer, and metalloproteinases (MMPs), such as MMP-2 and MMP-9 (a novel marker of AMI), are overexpressed in advanced atherosclerotic plaques by monocytes/macrophages and have been shown to contribute to AMI and UA through degradation of the extracellular matrix.7–10 Of note, CD147 stimulates macrophages to produce MMP-2 and MMP-9 in a paracrine or autocrine way.4,11 Therefore, to suppress the expression of CD147 and MMPs represents a promising strategy for anti-atherosclerosis.Plants of Cimicifuga genus (C. racemosa, C. foetida, and C. simplex) are famous herb medicines in Europe, the United States, and East Asia.12,13 These herbs mainly contain 9,19-cycloartane triterpenes (CTs) with diverse bioactivities, such as cytotoxicity,14,15 anti-angiogenic,16 anti-inflammatory,17,18 and neuro-protective.19,20 Recently, we identified two CTs, yunnanterpene G (YG) and 12β-hydroxycimiacerol (HC), with anti-atherosclerosis potentials by potently suppressing the mRNA expressions of CD147 and MMPs.21,22 As a part of our successive program to explore bioactive CTs from Cimicifuga spp, nine unreported CTs glycosides (1–6, 9, and 11–12) and two new aglycones (7 and 8), together with one undescribed lanostane-type triterpene (10) were identified from the aerial parts of C. yunnanensis (Fig. 1), an indigenous species distributed in the southwest region of China.16 Significantly, the q-PCR experiments showed that aglycones 7 and 8 dose-dependently attenuated the mRNA expression of CD147, with an IC50 value of 3.38 ± 0.27 μM and 8.25 ± 0.33 μM, respectively, in PMA-induced THP-1 cells. Of note, the CD147 mRNA inhibitory effect of 7 at 10 μM is more potent than that of YG (positive control). While, 8 has comparable activity as YG at this concentration. Moreover, 7 dose-dependently down-regulated the mRNA expression of MMP-2, and MMP-9 and suppressed the migration ability of the induced THP-1 cells, having an IC50 value of 6.32 ± 0.31 μM, 11.57 ± 0.23 μM, and 5.87 ± 0.27 μM, respectively. Conversely, all tested glycosides (3–5, 9 and 11) were inactive at 10 μM. Described herein are the isolation, structure elucidation, and biological activities of compounds 1–12.
2. Results and discussion
2.1. Structural elucidation of compounds 1–12
The same molecular formula C37H54O10 for compounds 1 and 2 were determined by the HRESIMS ([M]+ m/z 658.3703, calcd 658.3717 for 1, and [M]+ m/z 658.3694, calcd 658.3717 for 2, respectively). The IR spectra showed absorptions for OH (3443 cm−1 for 1 and 3441 for 2), and carbonyl (1733 cm−1 both for 1 and 2) groups. In the 1H NMR (Table 1) spectrum of 1, downfield shifted cyclopropane methylene signals at δH 0.57 (1H, d, J = 3.9 Hz) and 1.07 (1H, overlapped), a secondary methyl group at δH 0.95 (d, J = 6.0 Hz), six tertiary methyl groups at δH 0.95–1.47, and an anomeric proton at δH 4.79 (1H, d, J = 7.2 Hz) were observed. The 13C spectrum of 1 (Table 2) showed the existence of an ester carbonyl group at δC 171.1 (s), two olefinic carbons at δC 114.5 (C-7, d) and 148.2 (C-7, s), and six oxygenated carbon atoms at δC 88.3 (C-3, d), 77.1 (C-12, d), 73.5 (C-16, d), 63.6 (C-24, d), 63.7 (C-25, s), and 69.2 (C-26, t), respectively. These data suggested that 1 was a highly oxygen-bearing CTs glycoside with a seven-ring skeleton.| Position | 1a | 2a | 3b | 4b | 5b | 6b | 7b | 8a | 9b | 10a | 11b | 12a |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a Recorded at 500 MHz in pyridine-d5.b Recorded at 600 MHz in pyridine-d5.c Signals overlapped. | ||||||||||||
| 1 | 1.57 m | 1.54 m | 1.53 m | 1.50 m | 1.55 m | 1.46 m | 1.81 m | 1.55m | 2.77 m | 1.72 m | 1.61 m | 1.64 m |
| 1.16 m | 1.11 m | 1.08 m | 1.10 m | 1.12 m | 1.12c | 1.53c | 1.21 m | 1.71 m | 1.26 m | 1.21c | 1.28c | |
| 2 | 2.30 m | 2.24 m | 2.37 m | 2.33 m | 2.27 m | 2.27 m | 2.70 ddd (27.8, 13.9, 6.2) | 2.00 m | 2.46 m | 1.92 m (2H) | 2.37 m | 2.35 m |
| 1.90 m | 1.85 m | 1.90 m | 1.89 m | 1.81a | 1.89 m | 2.24 m | 1.90 m | 2.11 m | 1.95 m | 2.23c | ||
| 3 | 3.45 dd (11.7, 4.2) | 3.39 m | 3.47 dd (11.5,4.1) | 3.48 dd (11.7, 4.2) | 3.43 dd (11.7, 4.2) | 3.45 dd (11.5, 3.9) | 3.55 m | 3.59 dd (11.4, 3.9) | 3.48 m | 3.46 dd (11.5, 3.9) | 3.48 m | |
| 4 | ||||||||||||
| 5 | 1.19 m | 1.11 m | 1.27c | 1.24 m | 1.18 m | 1.19 m | 1.53c | 1.30 dd (12.6, 4.1) | 1.36c | 1.21c | 1.25a | 1.24 m |
| 6 | 1.82 m | 1.73 m | 2.50 m | 1.46 m | 1.81c | 1.97 m | 1.83 m | 1.56 m | 1.92 m | 1.76 m | 1.89 m | 1.77 m |
| 1.54 m | 1.44c | 1.05 m | 0.71 m | 1.54 m | 1.61c | 1.75 m | 0.75 m | 1.76 m | 1.55 m | 1.00c | 1.44 m | |
| 7 | 5.15c | 5.08c | 1.57 m | 1.25 m | 5.13 d (7.2) | 6.73 brd (6.3) | 6.75 d (7.1) | 1.57 m | 5.21 brd (6.3) | 2.66 m | 5.25 brd (5.8) | 5.31 d (6.1) |
| 0.73 dd (25.0,12.5) | 0.94 m | 1.03 m | 2.46 m | |||||||||
| 8 | 2.00 dd (12.5,4.0) | 1.55 m | ||||||||||
| 9 | ||||||||||||
| 10 | ||||||||||||
| 11 | 2.96 dd (16.1, 9.1) | 2.91 dd (15.7, 9.3) | 2.97 dd (16.2, 9.4) | 2.66 dd (15.9, 9.4) | 2.93 dd (16.0, 9.3) | 2.91 dd (16.2, 8.9) | 2.82 dd (15.3, 8.6) | 1.98 m | 4.60 brd (6.6) | 2.08 m | 2.06 m | 2.09 m |
| 1.27 m | 1.23c | 1.16 d (15.3) | 1.16 m | 1.25 m | 1.36 m | 1.57c | 1.10c | 2.02 m | 1.55 m | 1.14 m | ||
| 12 | 5.23 d (8.5) | 5.20 d (8.4) | 5.74 d (7.8) | 5.34 dd (9.3, 4.0) | 5.23 d (8.9) | 5.49 d (8.5) | 4.53 d (8.5) | 1.59 m (2H) | 2.85 dd (13.6, 9.5) | 1.73 m | 1.92 m | 1.85 m (2H) |
| 2.08 m | 1.26 m | 1.76 m | ||||||||||
| 13 | ||||||||||||
| 14 | ||||||||||||
| 15 | 2.16 m | 2.09 m (2H) | 1.97 m | 2.17c | 1.94 m | 2.56 m | 9.82 s | 5.88 s | ||||
| 2.03 m | 1.88 m | 2.05 dd (13.6, 5.9) | 1.66 m | 2.27 d (7.2) | ||||||||
| 16 | 4.63 dd (14.1, 7.4) | 4.31c | 4.23 m | 4.89 m | 5.01 dd (16.2, 7.9) | 4.54 d (8.7) | ||||||
| 17 | 1.79 m | 1.76 m | 2.10 m | 1.81c | 2.38 d (7.0) | 2.47 d (7.4) | 1.61 m | 2.25 d (4.8) | 1.50c | 2.72 d (4.4) | 2.35 m | |
| 18 | 1.41 s | 1.46 s | 1.67 | 1.35 s | 1.42 s | 1.00 s | 1.64 s | 1.24 s | 1.26 s | 0.92 s | 1.54 s | 1.22 s |
| 19 | 1.07c | 0.99c | 0.58 d (4.0) | 0.59 d (3.7) | 1.02c | 1.07c | 1.32c | 0.51 d (3.9) | 2.01 d (3.4) | 1.07 s | 0.89 d (3.8) | 0.97c |
| 0.57 d (3.9) | 0.47 brs | 0.26 d (4.0) | 0.20 d (4.1) | 0.52 d (3.8) | 0.57 d (4.0) | 0.92 d (4.3) | 0.24 d (4.1) | 1.02 d (3.4) | 0.49 d (3.8) | 0.50 d (3.9) | ||
| 20 | 1.82 m | 2.22 m | 3.37 dd (13.8,7.0) | 2.48 m | 1.95 m | 2.02 m | 1.58c | 2.31 m | 2.17 m | 1.68 m | 2.29 m | 2.59 m |
| 21 | 0.95 d (6.0) | 0.98 d (6.2) | 1.61 d (6.9) | 1.15 d (7.2) | 0.99 brd (4.0) | 1.11 d (6.2) | 1.53 d (6.2) | 1.26 d (6.4) | 0.90 d (5.9) | 0.91 d (6.9) | 1.00c | 1.06 d (6.6) |
| 22 | 2.19 m | 1.55 m | 2.71 2H m | 5.22 d (3.2) | 2.85 brd (11.7) | 2.53 d (13.0) | 2.61 d (12.8) | 3.94 d (10.7) | 2.51 m | 2.29 m | 2.09 dd (13.5, 6.9) | 3.57 dd (15.0, 3.2) |
| 1.65 m | 1.42 m | 1.58 m | 1.68 d (15.5) | 1.72 m | 2.43 m | 1.05 m | 1.89 m | 2.74 dd (18.0, 8.2) | ||||
| 23 | 4.64 m | 4.64 brd (10.4) | 4.71 brd (9.9) | 4.80 d (9.0) | 5.13 brd (11.5) | |||||||
| 24 | 3.73 s | 3.66 s | 3.64 s | 4.35 brs | 3.78 brs | 3.59 brs | 3.59 brs | 4.22 s | 4.51 s | 3.82 s | 3.74 d (4.6) | 3.72 s |
| 25 | ||||||||||||
| 26 | 3.97 d (10.0) | 4.03 d (10.0) | 1.60 s | 1.63 s | 1.84 s | 1.58 s | 1.59c | 1.81 s | 1.50 s | 1.66 s | 1.33 s | |
| 3.91 d (10.0) | 3.60 d (10.0) | |||||||||||
| 27 | 1.47 s | 1.46 s | 1.67 s | 1.68 s | 1.78 s | 1.59 s | 1.61c | 1.72 s | 1.52 s | 1.71 s | 1.33 s | |
| 28 | 1.07 s | 1.02 s | 1.31 s | 0.88 s | 1.05 s | 1.54 s | 1.60c | 0.87 s | 1.60 s | 1.29 s | 1.59 s | 1.29 s |
| 29 | 0.99 s | 0.93 s | 1.27 s | 1.01 s | 0.99c | 1.29 s | 1.11 s | 1.09 s | 1.38 s | 1.08 s | 1.25 s | 1.02 s |
| 30 | 1.32 s | 1.26 s | 0.97 s | 1.32 s | 1.29 s | 1.50 s | 1.02 s | 1.24 s | 1.12 s | 1.21 s | 0.99 s | 1.30 s |
| 1′ | 4.79 d (7.2) | 4.74 d (7.1) | 4.79 d (7.0) | 4.81 d (7.1) | 4.76 d (7.2) | 4.76 d (7.1) | 4.82 d (7.0) | 4.77 d (7.0) | 4.78 d (7.2) | |||
| 2′ | 4.49 m | 4.44 t (7.8) | 4.47 m | 4.49 t (8.8) | 4.45 t (8.0) | 4.45 t (8.0) | 4.47 m | 4.45 t (5.9) | 4.47 m | |||
| 3′ | 4.19 m | 4.14 brd (7.3) | 4.16 dd (8.8, 3.0) | 4.19 dd (9.0, 3.4) | 4.14 dd (8.8, 3.2) | 4.15 dd (8.8,3.1) | 4.17 dd (8.7,3.0) | 4.17 brd (8.7) | 4.16 dd (8.9, 3.2) | |||
| 4′ | 4.34 brs | 4.30 brs | 4.31 m | 4.34 brs | 4.30 brs | 4.31 m | 4.32 m | 4.33 m | 4.32 brs | |||
| 5′ | 4.32 m | 4.29c | 4.28 m | 4.32 m | 4.29 brd (10.9) | 4.28 m | 4.28 m | 4.30 m | 4.30 m | |||
| 3.80 m | 3.76 d (11.2) | 3.79 d (10.6) | 3.82 m | 3.77 brd (9.6) | 3.79 m | 3.79 m | 3.80 brd (10.8) | 3.79 m | ||||
| 12-OCOCH3 | 2.22 s | 2.17 s | 2.28 s | 2.14 s | 2.16 s | 2.24 s | ||||||
| 15-OCOCH3 | 3.36 s | 2.23 s | ||||||||||
| Position | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a 13C Recorded at 150 MHz in pyridine-d5. | ||||||||||||
| 1 | 30.6 t | 30.2 t | 32.1 t | 32.4 t | 30.3 t | 30.1 t | 31.8 t | 32.8 t | 27.3 t | 36.5 t | 31.0 t | 30.2 t |
| 2 | 29.9 t | 29.5 t | 29.8 t | 30.3 t | 29.4 t | 29.3 t | 36.7 t | 31.7 t | 29.7 t | 29.2 t | 29.3 t | 29.4 t |
| 3 | 88.3 d | 87.9 d | 87.9 d | 87.9 d | 87.9 d | 87.7 d | 214.6 s | 78.3 d | 88.3 d | 78.5 d | 87.7 d | 88.1 d |
| 4 | 40.8 s | 40.4 s | 41.1 s | 41.7 s | 40.4 s | 40.2 s | 48.7 s | 41.5 s | 40.6 s | 39.9 s | 40.2 s | 40.4 s |
| 5 | 42.8 d | 42.4 d | 46.8 d | 47.5 d | 42.5 d | 41.4 d | 43.1 d | 47.8 d | 44.0 d | 51.3 d | 40.4 d | 42.5 d |
| 6 | 22.2 t | 21.8 t | 25.6 t | 20.8 t | 21.9 t | 21.5 t | 21.7 t | 21.7 t | 21.9 t | 19.1 t | 22.2 t | 21.9 t |
| 7 | 114.5 d | 114.1 d | 20.4 t | 26.1 t | 114.0 d | 117.2 d | 116.7 d | 27.2 t | 113.6 d | 28.5 t | 122.0 d | 115.2 d |
| 8 | 148.2 s | 147.7 s | 40.6 d | 47.1 d | 147.9 s | 140.3 s | 141.3 s | 48.1 d | 147.4 s | 134.9 s | 140.2 s | 146.1 s |
| 9 | 21.7 s | 21.2 s | 19.9 s | 20.9 s | 21.3 s | 21.2 s | 29.8 s | 20.1 s | 27.3 s | 136.0 s | 18.8 s | 21.4 s |
| 10 | 28.7 s | 28.2 s | 26.8 s | 27.6 s | 28.3 s | 28.3 s | 22.6 s | 26.7 s | 29.0 s | 38.0 s | 28.4 s | 28.7 s |
| 11 | 36.9 t | 36.6 t | 36.8 t | 36.9 t | 36.7 t | 35.6 t | 39.2 t | 26.9 t | 63.3 d | 21.2 t | 24.6 t | 24.9 t |
| 12 | 77.1 d | 76.8 d | 71.0 d | 77.4 d | 76.9 d | 76.1 d | 71.3 d | 33.9 t | 48.8 t | 32.8 t | 30.8 t | 33.3 t |
| 13 | 48.5 s | 48.1 s | 49.4 s | 49.4 s | 48.1 s | 43.6 s | 45.1 s | 45.7 s | 46.2 s | 41.6 s | 42.9 s | 41.4 s |
| 14 | 51.1 s | 50.5 s | 54.5 s | 48.3 s | 50.7 s | 54.6 s | 57.8 s | 47.3 s | 50.7 s | 49.5 s | 59.3 s | 48.5 s |
| 15 | 42.9 t | 43.0 t | 207.3 s | 46.2 t | 42.7 t | 211.6 s | 212.2 s | 43.8 t | 48.4 t | 76.3 d | 200.3 d | 81.6 d |
| 16 | 73.5 d | 74.5 d | 148.6 s | 75.2 d | 71.8 d | 95.9 s | 96.1 s | 72.8 d | 81.9 s | 112.7 s | 173.7 s | 214.0 s |
| 17 | 57.2 d | 56.6 d | 151.0 s | 52.8 d | 57.4 d | 55.6 d | 56.9 d | 52.8 d | 63.5 d | 58.7 d | 55.3 d | 58.9 d |
| 18 | 14.3 q | 14.2 q | 25.7 q | 13.6 q | 14.9 q | 13.9 q | 15.1 q | 21.2 q | 21.0 q | 18.2 q | 21.9 q | 21.7 q |
| 19 | 29.1 t | 28.8 t | 31.3 t | 30.3 t | 28.8 t | 28.0 t | 27.6 t | 30.9 t | 18.5 t | 19.6 q | 28.5 t | 27.9 t |
| 20 | 26.2 d | 23.1 d | 28.0 d | 25.1 d | 25.9 d | 25.4 d | 26.1 d | 35.2 d | 25.7 d | 24.9 d | 27.9 d | 27.9 d |
| 21 | 22.1 q | 21.4 q | 17.3 q | 26.1 q | 21.2 q | 23.4 q | 23.7 q | 17.9 q | 20.5 q | 20.5 d | 24.7 q | 20.8 q |
| 22 | 37.1 t | 37.2 t | 40.7 t | 106.2 d | 42.1 t | 32.4 t | 32.5 t | 87.4 d | 44.7 t | 38.6 t | 36.2 t | 46.5 t |
| 23 | 106.4 s | 105.9 s | 68.3 d | 154.2 s | 102.5 s | 75.7 d | 76.1 d | 106.5 s | 211.2 s | 72.5 d | 78.0 d | 205.5 s |
| 24 | 63.6 d | 62.3 d | 78.7 d | 79.9 d | 77.5 d | 78.7 d | 78.5 d | 83.7 d | 82.3 d | 90.7 d | 79.7 d | 65.8 d |
| 25 | 63.7 s | 62.5 s | 73.6 s | 73.5 s | 75.1 s | 72.6 s | 72.8 s | 84.0 s | 71.4 s | 72.2 s | 60.8 s | |
| 26 | 69.2 t | 68.1 t | 27.7 q | 27.9 q | 29.4 q | 26.1 q | 28.5 q | 28.2 q | 27.6 q | 25.8 q | 24.6 q | |
| 27 | 15.2 q | 14.8 q | 27.1 q | 26.4 q | 28.9 q | 28.5 q | 26.2 q | 25.4 q | 25.8 q | 29.0 q | 18.4 q | |
| 28 | 27.2 q | 26.9 q | 23.6 q | 21.3 q | 26.8 q | 24.2 q | 24.2 q | 20.2 q | 28.0 q | 18.3 q | 18.6 q | 19.5 q |
| 29 | 14.6 q | 14.3 q | 25.5 q | 15.7 q | 14.2 q | 25.4 q | 22.3 q | 14.9 q | 25.8 q | 16.9 q | 25.3 q | 14.2 q |
| 30 | 26.2 q | 25.8 q | 15.2 q | 26.3 q | 25.7 q | 15.7 q | 19.8 q | 26.2 q | 14.4 q | 29.0 q | 13.7 q | 25.8 q |
| 1′ | 108.0 d | 107.3 d | 107.4 d | 108.1 d | 107.3 d | 107.4 d | 107.4 d | 107.3 d | 107.4 d | |||
| 2′ | 73.5 d | 72.9 d | 72.9 d | 73.4 d | 72.9 d | 72.8 d | 72.8 d | 72.8 d | 72.9 d | |||
| 3′ | 75.2 d | 74.6 d | 74.6 d | 75.1 d | 74.6 d | 74.5 d | 74.5 d | 74.5 d | 74.7 d | |||
| 4′ | 70.2 d | 69.4 d | 69.5 d | 70.1 d | 69.5 d | 69.5 d | 69.5 d | 69.4 d | 69.5 d | |||
| 5′ | 67.5 t | 66.6 t | 66.8 t | 67.4 t | 66.7 t | 66.8 d | 66.8 t | 66.7 t | 66.8 t | |||
12-O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OCH3 OCH3 |
171.2 s | 170.8 s | 170.8 s | 171.2 s | 170.6 s | 170.7 s | ||||||
12-OCO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
21.4 q | 21.6 q | 21.4 q | 21.7 q | 21.6 q | 21.2 q | ||||||
15-O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) OCH3 OCH3 |
170.2 s | |||||||||||
15-OCO![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3 H3 |
20.9 q | |||||||||||
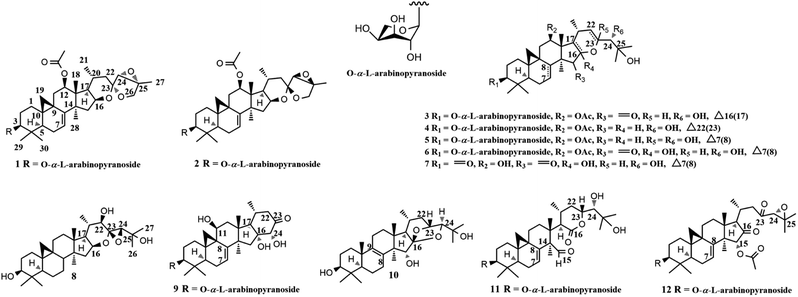
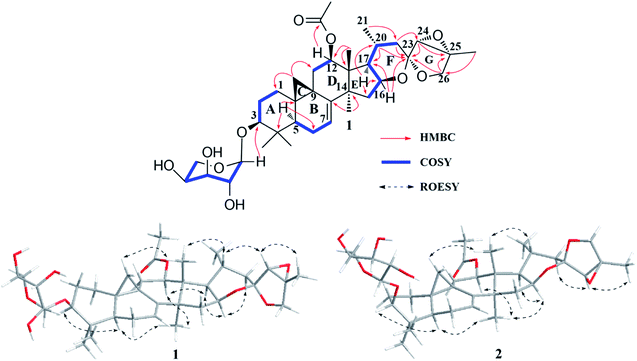
The 1H–1H COSY (Fig. 2) spin system of –CH2CHCHCH(CH3)CH2– (for C-15 to C-17, C-20 to C-22), together with the diagnostic ketal signal at δC 106.4 (C-23, s), as well as the pair of geminal signals for CH2-26 at δH 3.91 and 3.97 (each 1H, d, J = 10.0 Hz), indicated that 1 was a acteol-type CTs.12,23 HMBC couplings from H-16 (δH 4.63), H-22 (δH 1.65 and 2.19) to C-23 (δC 106.4), H-22 (δH 1.65 and 2.19) to C-24 (δC 63.6), and H-26 (δH 3.91 and 3.97) and H-24 (δH 3.73) to C-23 (δC 109.5) and C-25 (δC 83.4), further supported this deduction. The location of sugar unit at C-3 was inferred from HMBC correlation between the anomeric proton at δH 4.85 (1H, d, J = 8.6 Hz) and the methine signal at δC 88.5 (C-3). In addition, the sugar was determined as L-arabinose by comparing its TLC and specific rotation with a standard after acid hydrolysis. Structurally, 1 resembles to 26-deoxyactein,23 with main differences as that an acetoxy group substituted at C-12 and the presence of a double bond at C-7 and C-8. These elucidations were confirmed by HMBC association from H-12 (δH 5.23) to the ester carbonyl group (δC 171.1), and the 1H–1H COSY correlation of the olefinic proton resonance (δH 5.15) and H-6 (δH 1.54 and 1.82).
In the ROESY spectrum (Fig. 2), cross-peaks of H-3 with H-5 (biogenetically α-oriented), H-16 and H-17 with CH3-28 (biogenetically α-oriented), and H-20 with CH3-18 (biogenetically β-oriented) were observed, which helped to establish the relative configuration of the core structure of 1. Moreover, the characteristic ROESY correlations of H-21/H-24 and H-24/CH3-27 further decided the configuration of ring F and G, as well as the ternary epoxy ring and ring G as shown (Fig. 2). Intensive analysis of 1 D and 2D NMR spectra demonstrated that compound 2 had the same core structure as that of 1, with the major differences being at C-24, C-25 and C26. Diagnostically, the ROESY correlation of H-21/H-24 was absence in 2, instead, the association of H-22α/H-24 was observed. Thus, the configuration of ring F and G of 2 was determined as same to 23-epi-26-deoxyactein (Fig. 2).23 Finally, the structure of 1 and 2 were determined as 7(8)-en-acteol-3-O-α-L-arabinopyranoside and 7(8)-en-23-epi-acteol-3-O-α-L-arabinopyranoside, respectively.
Compound 3, white powder, had molecular formula C37H58O12 based on the HRTOF-ESIMS at m/z 717.3823 [M + Na + H2O]+ (calcd 717.3826). The 1H NMR spectrum (Table 1) showed characteristic cyclopropane methylene signals at δH 0.26 (1H, d, J = 4.0 Hz) and 0.58 (1H, d, J = 4.0 Hz), and an anomeric proton at δH 4.79 (1H, d, J = 7.2 Hz). The 13C NMR spectrum (Table 2) indicated that 3 had resonances corresponding to an ester carbonyl group at δC 171.1 (s), and an α,β-unsaturated ketone unit at δC 207.3 (C-15, s), 148.6 (C-16, s) and 151.0 (C-17, s). Aforementioned data indicated that 3 was a CTs glycoside. The sugar unite was determined as L-arabinose by the same way as that of 1. The NMR data of aglycone part of 3 resembled that of 24-O-acetyl-16(17)-en-hydroshengmanol-3-O-β-D-xylopyranoside,20 except that the acetoxy group was changed to C-12 and the OH-16 was replaced by a carbonyl group. HMBC correlations of H-12 (δH 5.74) with the ester carbonyl group (δC 170.8), and CH3-28 (δH 1.31) with the carbonyl carbon (δC 207.3) further confirmed these elucidations. ROESY cross-peaks of H-3/H5, H-12/CH3-28, and H-20/CH3-18 in 3 suggested the α-orientation of H-3, H-12, and CH3-21. The β-orientation of H-24 was deduced by the ROESY correlation of H-24/H-20. In addition, identical to isodahurinyl-type molecules, H-24 of 3 was a singlet in 1H NMR spectrum, suggesting S configuration of C-24 (the coupling constant of H-24 and H-23 of dahurinyl-type compounds, with R configuration of C-24, is around 6–9 Hz).15 Therefore, the structure of 3 was determined as 12β-acetoxy-16(17)-en-isodahurinyl-3-O-α-L-arabinopyranoside.
Compound 4 possessed the molecular formula of C37H58O10 based on the HREIMS at m/z 662.4058 [M]+ (calcd 662.4030). The NMR spectroscopic data for 4 resembled those of (16S,20S,24R)-12β-acetoxy-16,23-epoxy-24,25-dihydroxy-3β-(β-D-xylopyranosyloxy)-9,19-cyclolanost-22(23)-ene (AC),24 with major differences in sugar unit. In addition, the sugar moiety was attached to C-3 and determined as L-arabinose by the same way as that of 1. A α-orientation of H-3, H-12, H-16, H-17, and CH3-21 were determined by ROESY couplings of H-3 with H-5, H-12, H-16, and H-17 with CH3-28, and H-20 with H-17, respectively. Whereas, a β-orientation of H-8 was deduced by the correlation of H-8/CH3-18. In addition, identical to that of AC (16S,20S,24R)-12β-acetoxy-16,23-epoxy-24,25-dihydroxy-3β-(β-D-xylopyranosylo-xy)-9,19-cyclolanost-22(23)-ene, the characteristic ROESY association of CH3-18/CH3-26 was observed in 4, indicating it shares the same configuration at C-24 as R in AC (As shown in Fig S100,† when configuration of C-24 is S, it is impossible to see the cross-peak of CH3-18/CH3-26). Thus, the structure of 4 was determined as 12β-acetoxy-22(23)-en-15-deoxy-isodahurinyl-3-O-α-L-arabinopyranoside.
Compound 5 was purified as white powder, with the molecular formula C37H58O11, given by the HREIMS ([M]+ m/z 678.3990, calcd 678.3979). The IR spectrum showed the presence of hydroxyl (3431 cm−1), carbonyl (1730 cm−1) and olefinic (1629 cm−1) groups. The NMR data of aglycone part for 5 (Tables 1 and 2) were similar to those of actaeaepoxide-3-O-α-D-xylopyranoside.25 The main differences were that a methine (C-22) at δC 86.6 was absent, instead, there's another methylene (δC 42.1), and the upfield shifts of C-23, C-24, and C-25 by 3.1 ppm, 5.4 ppm and 8.6 ppm, respectively. These changes could be explained as that, in 5, a methylene replaced a methine at C-22, and two hydroxy groups instead of the ternary epoxy ring at C-23 and C-24. These deductions were further confirmed by the HMBC coupling of H-20 (δH 4.81)/C-22 (δC 42.1). The sugar unit was connected to C-3 and identified as L-arabinose using same approaches as those of 1. In addition, the orientations of core structure and the configuration of C-24 of 5 were determined on the basis of the ROESY associations as those of 4. Therefore, the structure of 5 was determined as 12β-acetoxy-23,24-dihydroxy-7(8)-en-15-deoxy-isodahurinyl-3-O-α-L-arabinopyranoside.
On the basis of the HRTOF-ESIMS peak at m/z 715.3666 [M + Na]+ (calcd 715.3670), the molecular formula of 6 was determined as C37H56O12. 1H NMR resonances due to a downfield shifted cyclopropane methylene at δH 0.57 (1H, d, J = 4.0 Hz) and 1.07 (1H, overlapped), an olefinic proton at δH 6.73 (brd, J = 6.3 Hz), an acetyl methyl group at δH 2.24, a secondary methyl signal at δH 1.11 (d, J = 6.2 Hz), six singlet methyl groups at δH 1.00–1.59, as well as an anomeric proton at δH 4.76 (1H, d, J = 7.1 Hz) were observed (Table 1), indicating 6 is a CTs glycoside with an acetoxy group and a double bond. The sugar unit was connected to C-3 and deduced as L-arabinose by using same approaches as those of 1. Comparison of NMR data of 6 and hydroxyshengmanol-7(8)-en-15-one-3-O-β-D-xylopyranoside26 revealed the aglycone part of the two compounds were identical, except for an acetoxy group substituted at C-12 in 6, which further supported by the HMBC correlation of H-12 (δH 5.49) and the ester carbonyl group (δC 170.7). A α-orientation of the substituents at C-3, and C-12 were determined by ROESY correlations of H-3/H-5 and H-12/CH3-28. Whereas, correlations of H-20/CH3-18 and H-23/H-20 indicated the β-orientation of H-23. The stereochemistry of C-24 was elucidated as S by comparison of coupling constant of H-24 with known compounds (S, J ≤ 2 Hz; R, J ≈ 6).15,26 The molecular formula C30H44O7 of 7 was deduce from its HRTOF-ESIMS at m/z 539.2988 [M + Na]+ (calcd 539.2985). The spectroscopic features of 7 were identical to 6 except for a carbonyl group and a hydroxy group at C-3 and C-12, respectively. HMBC correlation of CH3-29 (δH 1.11) with the carbonyl carbon (δC 214.6) and the upfield shift of C-12 by 4.8 ppm further confirmed these elucidations. Same orientations of H-8, H-12, H-17, and H-23, as well as the configuration of C-24 between 7 and 6 were determined on the basis of the ROESY associations and comparison of coupling constant of H-24 with known compounds. Thus, the structure of 6 and 7 were determined as 12β-acetoxy-7(8)-en-15-one-hydroxyshengmanol-3-O-α-L-arabinopyranoside and 12β-hydroxy-7(8)-en-3,15-dione-hydroxyshengmanol, respectively.
The molecular composition of compound 8, C30H48O5, was established by HREIMS ([M]+ m/z 488.3499, calcd 488.3502), indicating 7 degrees of unsaturation. The 30 carbon signals of 8 were similar to the aglycone resonances of actaeaepoxide-3-O-β-D-xylopyranoside.25 The main differences were that no double bond at C-7 and C-8, and the absence of an acetoxy group at C-12 in 8. Moreover, 1H–1H COSY correlations of H-6 (each 1H, δH 0.75 and 1.56) with H-7 (each 1H, δH 1.03 and 1.57), H-11 (each 1H, δH 1.10 and 1.98) with H-12 (2H, δH 1.59) confirmed these deductions. The orientations of H-16, H-17, and H-22 were assigned as α by analysis of ROESY spectrum. Besides, the characteristic ROESY correlation of H-22/H-24 further suggested the configuration between ring E and the ternary epoxy ring as shown (Fig S100†). Therefore, the structure of 8 was determined as actaeaepol.
The molecular formula of compound 9 was determined as C32H48O9 from HRTOF-ESIMS at m/z 599.3188 [M + Na]+ (calcd 599.3196). In the 1H NMR spectrum (Table 1), signals due to an extremely downfield shifted cyclopropane methylene at δH 1.02 (1H, d, J = 3.4 Hz) and 2.01 (1H, d, J = 3.9 Hz), an anomeric proton at δH 4.82 (d, J = 7.0 Hz), an olefinic hydrogen atom at δH 5.21 (brd, J = 6.3 Hz), four tertiary methyl groups at δH 1.12–1.60, and a secondary methyl signal at δH 0.90 (d, J = 5.9 Hz), were observed. The 1H–1H COSY spectrum indicated that 9 had part structure of –CHCH(CH3)CH2– (for C-17, C-20 to C-22). Aforementioned data together with HMBC associations of H-22 (2H, δH 2.43 and 2.51) with the carbonyl carbon (δC 211.2) and the oxygenated carbon at δC 82.3 (C-24, d), and H-17 (δH 2.25) with the oxygenated carbon at δC 81.9 (C-16, s) and C-24, exhibited that 9 was a foetidonol-type CTs glycoside. The sugar unit was connected to C-3 and identified as L-arabinose using same ways as those of 1. Compound 9 had a similar structure as that of foetidonol-3-O-β-D-xylopyranoside,27 except for a double bond at C-7 and C-8, and a hydroxy group at C-11. The 1H–1H COSY coupling of H-6 and the olefinic proton at δH 5.21, and the HMBC correlation of H-11 (δH 4.60) and C-9, as well as the molecule weight further confirmed these elucidations. Therefore, the structure of 9 was determined as 11β-hydroxy-7(8)-en-foetidonol-3-O-α-L-arabinopyranoside.
The HREIMS of 10 exhibited a molecular ion at m/z 488.3508 [M]+ (calcd 488.3502) for the molecular formula of C30H48NO5. The 1D NMR spectroscopic data (Table 1) of 10 showed seven tertiary methyl groups at δH 0.92–1.52, and a secondary methyl signal at δH 0.91 (d, J = 6.9 Hz). These data indicated that 10 possessed one more tertiary methyl group in the skeleton than a usual CTs. In 13C NMR spectrum, a pair of tetrasubstituted olefinic carbons at δC 134.9 (C-8, s) and 136.0 (C-9, s), and the characteristic ketal signal for C-16 of cimigenol-type CTs at δC 112.7 (s) were observed. HMBC associations of H-7/C-8 (δC 134.9, s), H-11/C-9 (δC 136.0, s), H-1/CH3-19 (δC 19.6, q), and CH3-19/C-9 and C-10, located the double bond at C-8 and C-9, and the CH3-19 at C-10, respectively. The rest of NMR resonances of 10 were identical to those of cimigenol.28 Therefore, the structure of 10 was determined as 19β-methyl-8(9)-en-cimigenol.
Compound 11 gave a pseudo-molecular ion at m/z 657.3613 [M + Na]+ (calcd 657.3615) in the positive ion HRTOF-ESIMS, corresponding to the molecular formula C35H54NaO10, which is 2 Da less than that of 15,16-seco-14-formyl-16-oxo-hydroshengmanol-3-O-α-L-arabinopyranoside.14 When its spectroscopic data (Table 1 and 2) were compared with 15,16-seco-14-formyl-16-oxo-hydroshengmanol-3-O-α-L-arabinopyranoside, the resonances of a methylene and a methine were absent in 11, showing instead a pair of double bond at C-7 and C-8. The 1H–1H COSY coupling of H-6 and the olefinic proton at δH 5.25 further supported this deduction. Therefore, the structure of 11 was determined as 15,16-seco-14-formyl-16-oxo-7(8)-en-hydroshengmanol-3-O-α-L-arabinopyranoside.
Compound 12 was assigned a molecular formula C37H54O10 from its HREIMS at m/z 658.3707 [M]+ (calcd 658.3717). The spectroscopic features of 12 resembled to those of 15,23-O-diacetyl-7(8)-en-shengmanol-3-O-α-L-arabinopyranoside29 except for the substituent group at C-23. For 12, the oxygenated methine of C-23 (δC 72.3, d) was absent, showing instead a carbonyl carbon at δC 205.5. This difference was due to a carbonyl group at C-23 in 12, which confirmed by the HMBC correlations of H-22 and H-24 with carbonyl signal at δC 205.5. The sugar unit and the relative configuration of 12 were elucidated by the same ways as those of aforementioned compounds. Therefore, the structure of 12 was determined as 15α-acetoxy-23-oxo-7(8)-en-cimicidanol-3-O-α-L-arabinopyranoside.
2.2. Alteration of morphology and phenotype on PMA-induced THP-1 cells
The normal THP-1 cells are ball-shaped without adhering to the surfaces of the plastic culture plates.11 Cultured by adding 100 nM PMA for 24 h, the cells became flat and amoeboid in shape, and adhered to the dish bottom (Fig. S97A and B†). Moreover, the differentiation of monocyte to macrophage was determined on the basis of 97% of the PMA-induced THP-1 cells were CD68 positive and 33% of these cells were CD11b positive with flow cytometry analysis (Fig. S97C and D†).2.3. Cytotoxic activities of compounds 1–12 on PMA-induced THP-1 cells
Before conducting further bioassays, the cytotoxicities of compounds 1–12 on PMA-induced macrophages were tested by MTT assay. As shown in Fig S98,† compounds 1, 2, 6, 10, and 12 indicated notable cytotoxic effect (25% to 75% inhibition) on the cell viability from the concentration of 25 μM. Thus, these molecules were discarded for further studies. Conversely, compounds 3–5, 7–9 and 11 were chose for successive investigations due to their negligible cytotoxicity even at 50 μM (about 10–15% reduction on the cell viability) (Fig. S98†). In addition, the experimental concentration range was set as10 to 50 μM in the present study.2.4. Downregulation of mRNA expression of CD147 and MMPs in PMA-induced THP-1 cells by compounds 3–5, 7–9 and 11
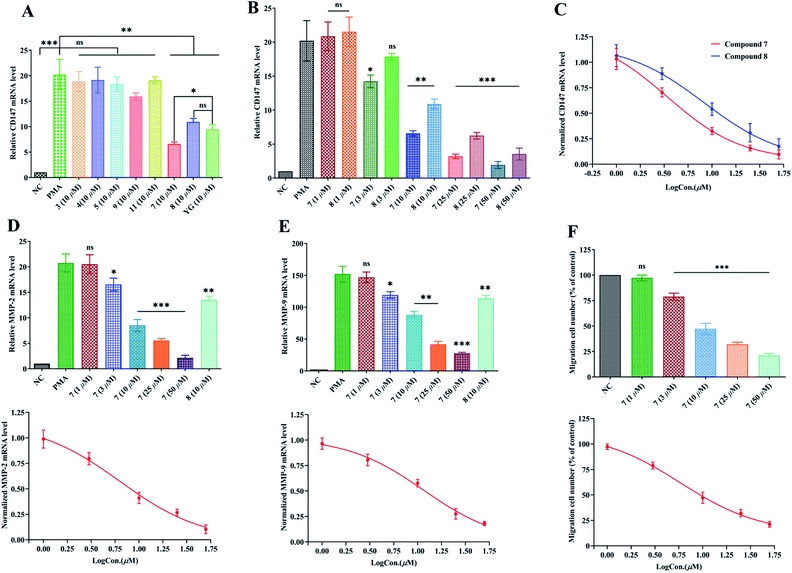
Fig. 3A revealed that, the expression of CD147 was significantly augmented in PMA-induced THP-1 cells as compared with the NC group (P < 0.001) with q-PCR 2−ΔΔCt method. Compounds 3–5, 7–9 and 11, and Yunnanterpene G (YG), the previously identified active compound (positive control), were firstly tested at 10 μM in the differentiated THP-1 cells for 24 hours. The aglycones (7 and 8) noticeably suppressed the mRNA expression of CD147 (P < 0.001). Conversely, the tested glycosides, 3–5, 7 and 11, were inactive. Of note, 7 is more potent than YG (P < 0.05), while, 8 showed same level of activity as YG. Dose–response studies further revealed that 7 and 8 down-regulated the mRNA expression with an IC50 value of 3.38 ± 0.27 μM and 8.25 ± 0.33 μM, respectively (Fig. 3B and C). Importantly, CD147 could regulate the expression of MMP-2 and MMP-9 in the activated macrophage.4,11 Thus, the effect of 7 and 8 on the mRNA production of these metalloproteinases were further investigated. As shown in Fig. 3D and E, 8 (10 μM) potently decreased the MMP-2 and MMP-9 mRNA expression by 35% (P < 0.01) and 25% (P < 0.01), respectively. In addition, the mRNA expression of MMP-2, and MMP-9 were significantly and dose-dependently down-regulated by 7, having an IC50 value of 6.32 ± 0.31 μM and 11.57 ± 0.23 μM, respectively.2.5. Inhibition of PMA-induced THP-1 cells migration by compound 7
The enhanced migration or invasion of peripheral macrophages is a characteristic feature in pathological process of atherosclerosis.4,5 Because of compound 7 showed significant inhibition on the mRNA expression of CD147 and MMPs, the regulatory effect of this molecule on the migration of PMA-induced THP-1 cells was further studied by scratch wound assay. As a result, 7 (after 24 h incubation) potently and dose-related decreased the number of migrated cells with an IC50 value of 5.87 ± 0.27 μM (Fig. 3F and S99†).3. Conclusion
As mentioned in the introduction, substances, which inhibit CD147 and MMPs expression may hold great potentials to prevent the development of atherosclerosis. Indeed, anti-atherogenic drugs, such as fluvastatin, attenuating the MMPs and EMMPRIN productions partially contribute to their clinical effects.30–32Natural products (NPs) are important resources of active molecules for modern drug development.33 Previously, two undescribed CTs (YG and HC) from C. foetida, with notable inhibitions on CD147 and MMPs mRNA expression, were identified. Successive investigations on the aerial parts of C. yunnanensis led to characterize eleven new CTs, including nine glycosides (1–6, 9, and 11–12) and two aglycones (7 and 8), along with one undescribed lanostane-type triterpene (10). Compounds 7 and 8, two aglycones, showed noticeable inhibitory effects on the mRNA expression of CD147 and MMPs, as well as migration ability of the induced THP-1 cells. By contrast, all tested glycosides were inactive. It is worth noting that 7 is the most potent molecule among these four active CTs. Given the critical roles of CD147 and MMPs in stabilizing atherosclerotic plaques, 7 may have a promising effect in retarding the development of the vulnerability of the plaque, deserve to conduct more sophisticated animal studies in future.
In summary, our studies show that CTs, specially aglycones, are potential resources of anti-atherosclerosis bioactive agents and deserve further extensive exploration.
4. Experimental section
4.1. General experimental procedures
Column chromatography (CC) were conducted with Silica gel (200–300 mesh, Qingdao Marine Chemical, Inc.) and Lichroprep RP-18 (40–63 μm, Merck). Waters 2695 liquid chromatography system was applied to run Semipreparative HPLC with a YMC-Pack 10 mm × 250 mm column (Pro C18 RS). Thin-layer chromatography was carried out on precoated TLC plates (200–250 μm thickness, silica gel 60 F254, Qingdao Marine Chemical, Inc.). Bruker DRX-500 and Avance III-600 MHz spectrometers (Bruker, Zűrich, Switzerland) were used to record 1D and 2D NMR data with solvent signal as internal reference. ESIMS, HREIMS and HRESIMS were obtained from a Shimadzu LCMS-IT-TOF MS (Shimadzu, Kyoto, Japan), a Waters AutoSpec Premier P776 MS (Waters Corporation, Milford, USA) or an Agilent G6230 TOF MS (Agilent Technologies, Palo Alto, USA). Shimadzu IR-450 instrument was used to evaluate infrared spectra with KBr pellets. A JASCO P-1020 digital polarimeter was applied to test optical rotations, using MeOH as solvent. Quantitative-PCR (q-PCR) was conducted on ProFlex™ PCR system (Thermo Fisher, Shanghai, China).4.2. Materials
4.3. Extraction and isolation
The aerial parts of Cimicifuga yunnanensis (4.7 kg) was extracted at room temperature by MeOH (20 L, 3 times, 7 days each). The extract (489.7 g) was obtained after evaporation of MeOH under vacuum at 50 °C. The extract was divided by silica gel CC (12.0 kg, 30 × 200 cm) eluted with CHCl3–MeOH [100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 (35 L), 50
0 (35 L), 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (30 L), 10
1 (30 L), 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (20 L), 5
1 (20 L), 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (10 L), 0
1 (10 L), 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (15 L)] to yield fractions A (52.6 g), B (67.4 g), C (55.7 g), D (29.2 g) and E (23.3 g). Sub-fractions (B.1–B.7) were further obtained by silica CC (4 kg, 10 × 150 cm), eluted with CHCl3–Me2CO from 40
100 (15 L)] to yield fractions A (52.6 g), B (67.4 g), C (55.7 g), D (29.2 g) and E (23.3 g). Sub-fractions (B.1–B.7) were further obtained by silica CC (4 kg, 10 × 150 cm), eluted with CHCl3–Me2CO from 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 gradient to 5
1 gradient to 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Compounds 7 (3.2 mg), 8 (2.4 mg) and 10 (2.2 mg) were purified from fraction B.4 (7.9 g) by RP-18 CC (300 g, 5 × 50 cm) eluting with MeOH–H2O from 60
1. Compounds 7 (3.2 mg), 8 (2.4 mg) and 10 (2.2 mg) were purified from fraction B.4 (7.9 g) by RP-18 CC (300 g, 5 × 50 cm) eluting with MeOH–H2O from 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 to 100
40 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 and semipreparative HPLC (eluted with CH3CN–H2O, gradient from 60
0 and semipreparative HPLC (eluted with CH3CN–H2O, gradient from 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 to 80
40 to 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20). Subsequently, further silica CC (2 kg, 10 × 150 cm) on fraction C, eluting with CHCl3–Me2CO from 20
20). Subsequently, further silica CC (2 kg, 10 × 150 cm) on fraction C, eluting with CHCl3–Me2CO from 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 gradient to 1
1 gradient to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, gave six sub-fractions (C.1–C.6). Another five sub-fractions (C.3.1–C.3.5) were obtained by RP-18 CC (1 kg, 10 × 50 cm), eluting with MeOH–H2O from 50
1, gave six sub-fractions (C.1–C.6). Another five sub-fractions (C.3.1–C.3.5) were obtained by RP-18 CC (1 kg, 10 × 50 cm), eluting with MeOH–H2O from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 to 100
50 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0. Fraction C.3.3 (7.7 g) gave compounds 1 (2.7 mg), 2 (3.2 mg), 3 (2.1 mg), 4 (2.2 mg), 11 (2.1 mg), and 12 (3.1 mg) by RP-18 CC (250 g, 5 × 50 cm) eluting with MeOH–H2O from 60
0. Fraction C.3.3 (7.7 g) gave compounds 1 (2.7 mg), 2 (3.2 mg), 3 (2.1 mg), 4 (2.2 mg), 11 (2.1 mg), and 12 (3.1 mg) by RP-18 CC (250 g, 5 × 50 cm) eluting with MeOH–H2O from 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 to 75
40 to 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 and semipreparative HPLC (eluted with CH3CN–H2O, gradient from 50
25 and semipreparative HPLC (eluted with CH3CN–H2O, gradient from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 to 65
50 to 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35). Compound 5 (2.5 mg), 6 (2.3 mg), and 9 (3.7 mg) were purified from fraction C.3.4 (6.9 g) by successively silica gel CC (120 g, 5 × 40 cm, eluted with CHCl3–Me2CO from 10
35). Compound 5 (2.5 mg), 6 (2.3 mg), and 9 (3.7 mg) were purified from fraction C.3.4 (6.9 g) by successively silica gel CC (120 g, 5 × 40 cm, eluted with CHCl3–Me2CO from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 2
1 to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and semipreparative HPLC (eluted with CH3CN–H2O, 65
1), and semipreparative HPLC (eluted with CH3CN–H2O, 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35).
35).
4.4. Cell culture and differentiation
Human monocytic THP-1 cells were cultured at 37 °C by DMEM/F12 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin in a humidified atmosphere of 5% CO2. Cells (5 × 105 to 106 per mL) were subjected to differentiation by 100 nM PMA for 24 h with DMEM/F12 serum medium. After incubation, the adherent cells were washed with DMEM/F12. Nonattached cells were removed by aspiration.4.5. Flow cytometry analysis
The expression of CD68 and CD11b on surface of the differentiated THP-1 cells were tested by suing Flow cytometry. In a dark condition, cells (5 × 105) were washed 3 times by phosphate-buffered saline (PBS) and then respectively treated with fluorescein isothiocyanate (FITC)-conjugated anti-CD68 antibody, and FITC-conjugated anti-CD11b antibody, for 20 minutes. Cells were washed by PBS and then tested on a PARTEC CyFlow® Cube flow cytometer. Data were processed by the CytExpert software.4.6. MTT assay
The differentiated THP-1 cells were seeded onto gelatinized 96-well culture plates (5 × 105 cells per mL, 0.1 mL per well), and incubated at 37 °C with 5% CO2 for 24 h. Then, 0.1 mL of DMEM/F12 with different concentrations of compounds 1–12 (0, 5, 10, 25, 50, 75, and 100 μM) were added to instead of the original medium, and cultured for another 48 h. Cell viability was evaluated by MTT assay: each well was added with 20 μL of MTT to a final concentration of 0.5 g L−1 for 4 h before using 150 μL DMSO to solubilize the reactive dye. Each well was recorded by a Bio-Rad microplate reader to absorbance value of 570 nm. All the experiments were repeated in triplicate.4.7. Isolation of total RNA and RT-PCR
RevertAid™ First Strand cDNA Synthesis Kit was applied to extract total RNA of differentiated THP-1 cells treated with compounds 3–5, 9 and 11 (10 μM), along with 7 and 8 (1, 3, 10, 25, and 50 μM) for 24 h (5 × 105 cells per mL) following the manufacturer's instructions. cDNA was synthesized from the isolated total RNA based on the instruction of the PrimeScript RT reagent Kit. Briefly, PrimeScript RT Enzyme Mix 1 (0.5 μL), 5 × PrimeScriptTM Buffer (2 μL), oligo dt Primer (0.5 μL) and Random 6 mers (0.5 μL) were gently mixed with 1 μg RNA from each sample to a reaction volume of 10 μL with RNase free water, then incubated for 15 minutes at 37 °C to activate the reverse transcriptase enzyme. Finally, the reaction was stopped by 85 °C for 5 seconds.After reverse transcription, cDNA was used to carry out real-time quantitative RT-PCR on ProFlex™ PCR system by SYBR Premix Ex Taq (Takara). The final volume of TR-PCR reaction is 25 μL, containing 12.5 μL SYBR green master mix, 1 μL cDNA, 0.5 μL each forward and reverse primer, and 10.5 μL nuclease-free water. For information of primers see Table S1.† Thermal cycling conditions for all genes were as follows: template pre-denaturation (10 min at 95 °C), denaturation (15 seconds at 95 °C), annealing and extension (30 seconds at 60 °C) for 40 cycles. Internal reference is GAPDH mRNA, and fold changes of mRNA expression for each target relative to GAPDH were calculated by the 2−ΔΔCt method. Expression of mRNA is determined as the change in mRNA copy numbers relative to negative control cells (undifferentiated THP-1 cells). All the experiments were carried out in triplicate.
4.8. Wound-healing migration assay
The differentiated THP-1 cells were seeded and grown into full confluence in 6 well plates. 2% FBS DMEM/F12 media was used to inactivated cell proliferation for 12 h, then wounded with pipette tips. Fresh DMEM/F12 medium with or without 1, 3, 10, 25, and 50 μM of 7 was added to the scratched monolayers. Images were took after 24 hours using a Nickon inverted microscope (magnification, 10×). The migration cell number of PMA-induced THP-1 cell group was defined as control. All the experiments were performed in triplicate.4.9. Statistical analysis
Data are presented as mean ± SD. Statistical analysis of data was performed with Student's t-test. (*) P < 0.05, (**) P < 0.01, and (***) P < 0.001 are considered significant.Annotations for abbreviations
| CD147 | Extracellular matrix metalloproteinase inducer, EMMPRIN |
| MMP-2 | Matrix metalloproteinase 2 |
| MMP-9 | Matrix metalloproteinase 9 |
| PMA | Phorbol-12-myristate-13-acetate |
| q-PCR | Quantitative real-time PCR method |
| CVDs | Cardiovascular diseases |
| AMI | Acute myocardial infarction |
| UA | Unstable angina pectoris |
| CTs | 9,19-Cycloartane triterpenes |
| YG | Yunnanterpene |
| HC | 12β-Hydroxycimiacerol |
| NPs | Natural products |
Author contributions
Ni-Hong Lu: investigation; methodology; writing-original draft. Jie Li and Yong-Rui Yang: formal analysis; writing-original draft. Hong-Lu Liu: data curation. Ying-Rong Du: supervision; writing – review & editing.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This project was financially supported by Regional Project of National Natural Science Foundation of China (81960096) and Joint Special key projects of Local Colleges and Universities in Yunnan Province (201901N070046).Notes and references
- A. M. Ruiz-León, M. Lapuente, R. Estruch and R. Casas, Front Immunol, 2019, 10, 837 CrossRef PubMed.
- G. A. Roth, M. H. Forouzanfar, A. E. Moran, R. Barber, G. Nguyen, V. L. Feigin, M. Naghavi, G. A. Mensah and C. J. L. Murray, N. Engl. J. Med., 2015, 372(14), 1333–1341 CrossRef CAS PubMed.
- R. Ross, N. Engl. J. Med., 1999, 340(2), 115–126 CrossRef CAS PubMed.
- R. Schmidt, A. Bultmann, M. Ungerer, N. Joghetaei, O. Bülbül, S. Thieme, T. Chavakis, B. P. Toole, M. Gawaz, A. Schömig and A. E. May, Circulation, 2006, 113(6), 834–841 CrossRef CAS PubMed.
- D. L. Brown, M. S. Hibbs, M. Kearney, C. Loushin and J. M. Isner, Circulation, 1995, 91(8), 2125–2131 CrossRef CAS PubMed.
- R. P. Choudhury, J. M. Lee and D. R. Greaves, Nat. Clin. Pract. Cardiovasc. Med., 2005, 2(6), 309–315 CrossRef CAS PubMed.
- C. P. Wang, R. Jin, X. L. Zhu, J. C. Yan and G. H. Li, J. Cardiovasc. Trans. Res., 2015, 8(1), 59–66 CrossRef PubMed.
- Y. W. Yoon, H. M. Kwon, K. C. Hwang, E. Y. Choi, B. K. Hong, D. Kim, H. S. Kim, S. H. Cho, K. S. Song and G. Sangiorgid, Atherosclerosis, 2005, 180(1), 37–44 CrossRef CAS PubMed.
- T. C. Major, L. Liang, X. Lu, W. Rosebury and T. M. Bocan, Arterioscler. Thromb. Vasc. Biol., 2002, 22(7), 1200–1207 CrossRef CAS PubMed.
- Z. Q. Huang, L. S. Wang, S. Meng, Y. Wang, T. Chen and C. Q. Wang, Int. J. Cardiol., 2011, 146(2), 153–158 CrossRef PubMed.
- J. Zhou, P. Zhu, J. L. Jiang, Q. Zhang, Z. B. Wu, X. Y. Yao, H. Tang, N. Lu, Y. Yang and Z. N. Chen, BMC Cell Biol., 2005, 6(1), 837 CrossRef PubMed.
- J. X. Li and Z. Y. Yu, Curr. Med. Chem., 2006, 13(24), 2927–2951 CrossRef CAS PubMed.
- Y. Q. Guo, T. Yin, X. M. Wang, F. Zhang, G. X. Pan, H. Lv, X. R. Wang, J. O. Orgah, Y. Zhu and H. H. Wu, J. Ethnopharmacol., 2017, 209, 264–282 CrossRef CAS PubMed.
- Y. Nian, H. Zhu, W. R. Tang, Y. Luo, J. Du and M. H. Qiu, J. Nat. Prod., 2013, 76(5), 896–902 CrossRef CAS PubMed.
- Y. Nian, H. Yan, X. N. Li, L. Zhou and M. H. Qiu, RSC Adv., 2017, 7, 38557–38564 RSC.
- Y. Nian, J. Yang, T. Y. Liu, Y. Luo, J. H. Zhang and M. H. Qiu, Sci. Rep., 2015, 5, 9026–9031 CrossRef CAS PubMed.
- Y. Su, L. Wu, Q. H. Wang, B. Y. Yang and H. X. Kuang, Bioorg. Med. Chem. Lett., 2014, 24(24), 5688–5691 CrossRef CAS PubMed.
- Y. Su, L. Wu, G. R. Mu, Q. H. Wang, B. Y. Yang, G. H. Cheng and H. X. Kuang, Bioorg. Med. Chem., 2017, 25(17), 4917–4923 CrossRef CAS PubMed.
- C. N. Lv, F. Yang, R. L. Qin, Z. Y. Qi, W. R. Zhou and J. C. Lu, Bioorg. Med. Chem. Lett., 2017, 27(15), 3305–3309 CrossRef CAS PubMed.
- M. A. Findeis, F. Schroeder, T. D. McKee, D. Yager, P. C. Fraering, S. P. Creaser, W. F. Austin, J. Clardy, R. Wang, D. Selkoe and C. B. Eckman, ACS Chem. Neurosci., 2012, 3(11), 941–951 CrossRef CAS PubMed.
- N. H. Lu, Z. W. Zhang, R. W. Guo, L. X. Yang, Y. X. Song, J. S. Ye and Y. K. Shi, RSC Adv., 2018, 8, 15036–15043 RSC.
- N. H. Lu, Y. R. Yang, X. F. Li, H. L. Liu, Z. R. Zhao and Y. R. Du, Phytochem. Lett., 2021, 42, 109–116 CrossRef CAS.
- S. N. Chen, W. K. Li, D. S. Fabricant, B. D. Santarsiero, A. Mesecar, J. F. Fitzloff, H. H. S. Fong and N. R. Farnsworth, J. Nat. Prod., 2002, 65(4), 601–605 CrossRef CAS PubMed.
- Z. J. Fang, T. Zhang, S. X. Chen, Y. L. Wang, C. X. Zhou, J. X. Mo, Y. J. Wu, Y. K. Xu, L. G. Lin and L. S. Gan, Phytochemistry, 2019, 160, 1–10 CrossRef CAS PubMed.
- K. Wende, C. Mügge, K. Thurow, T. Schöpke and U. Lindequist, J. Nat. Prod., 2001, 64(7), 986–989 CrossRef CAS PubMed.
- N. M. Bao, Y. Nian, W. H. Wang, X. L. Liu, Z. T. Ding and M. H. Qiu, Phytochem. Lett., 2015, 12, 200–202 CrossRef CAS.
- S. Kadota, J. X. Li, K. Tanaka and T. Namba, Tetrahedron, 1995, 51(4), 1143–1166 CrossRef CAS.
- Y. Nian, Y. L. Zhang, J. C. Chen, L. Lu, M. H. Qiu and C. Qing, J. Nat. Prod., 2010, 73(2), 93–98 CrossRef CAS PubMed.
- A. Kusano, M. Shibano, G. Kusano and T. Miyase, Chem. Pharm. Bull., 1996, 44(11), 2078–2085 CrossRef CAS PubMed.
- N. Abe, T. Osanai, T. Fujiwara, K. Kameda, T. Matsunaga and K. Okumura, Life Sci., 2006, 78(9), 1021–1028 CrossRef CAS PubMed.
- S. Bellosta, D. Via, M. Canavesi, P. Pfister, R. Fumagalli, R. Paoletti and F. Bernini, Arterioscler. Thromb. Vasc. Biol., 1998, 18(11), 1671–1678 CrossRef CAS PubMed.
- N. Ferri, G. Colombo, C. Ferrandi, E. W. Raines, B. Levkau and A. Corsini, Arterioscler., Thromb., Vasc. Biol., 2007, 27(5), 1043–1049 CrossRef CAS PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83(3), 770–803 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: HREIMS, HRESIMS, IR, 1D and 2D NMR spectra for compounds 1–12. See DOI: 10.1039/d1ra07828c |
| This journal is © The Royal Society of Chemistry 2021 |