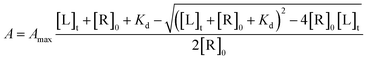Open Access Article
Open Access ArticleNovel alkylaminoethyl derivatives of androstane 3-oximes as anticancer candidates: synthesis and evaluation of cytotoxic effects†
Jovana J. Ajduković *a,
Dimitar S. Jakimovb,
Lucie Rárovác,
Miroslav Strnadd,
Yaraslau U. Dzichenka
*a,
Dimitar S. Jakimovb,
Lucie Rárovác,
Miroslav Strnadd,
Yaraslau U. Dzichenka e,
Sergey Usanove,
Dušan Đ. Škorić
e,
Sergey Usanove,
Dušan Đ. Škorić a,
Suzana S. Jovanović-Šanta
a,
Suzana S. Jovanović-Šanta a and
Marija N. Sakača
a and
Marija N. Sakača
aDepartment of Chemistry, Biochemistry and Environmental Protection, Faculty of Sciences, University of Novi Sad, Trg Dositeja Obradovića 3, 21000 Novi Sad, Serbia. E-mail: jovana.ajdukovic@dh.uns.ac.rs
bOncology Institute of Vojvodina, Faculty of Medicine, University of Novi Sad, Put Dr Goldmana 4, 21204 Sremska Kamenica, Serbia
cDepartment of Experimental Biology, Faculty of Science, Palacký University, Šlechtitelů 27, CZ-78371 Olomouc, Czech Republic
dLaboratory of Growth Regulators, Institute of Experimental Botany of the Czech Academy of Sciences, Faculty of Science, Palacký University, Šlechtitelů 27, CZ-78371 Olomouc, Czech Republic
eInstitute of Bioorganic Chemistry NAS of Belarus, Kuprevicha Street, 5/2, Minsk, 220141, Belarus
First published on 22nd November 2021
Abstract
Steroid anticancer drugs are the focus of numerous scientific research efforts. Due to their high cytotoxic effects against tumor cells, some natural or synthetic steroid compounds seem to be promising for the treatment of different classes of cancer. In the present study, fourteen novel O-alkylated oxyimino androst-4-ene derivatives were synthesized from isomerically pure 3E-oximes, using different alkylaminoethyl chlorides. Their in vitro cytotoxic activity was evaluated against eight human cancer cell lines, as well as against normal fetal lung (MRC-5) and human foreskin (BJ) fibroblasts, to test the efficiency and selectivity of the compounds. Most derivatives displayed strong activity against malignant melanoma (G-361), lung adenocarcinoma (A549) and colon adenocarcinoma (HT-29) cell lines. Angiogenesis was assessed in vitro using migration scratch and tube formation assays on HUVEC cells, where partial inhibition of endothelial cell migration was observed for the 17α-(pyridin-2-yl)methyl 2-(morpholin-4-yl)ethyl derivative. Among the compounds that most impaired the growth of lung cancer A549 cells, the (17E)-(pyridin-2-yl)methylidene derivative bearing a 2-(pyrrolidin-1-yl)ethyl substituent induced significant apoptosis in these cells. In combination with low cytotoxicity toward normal MRC-5 cells, this molecule stands out as a good candidate for further anticancer studies. In addition, in vitro investigations against cytochrome P450 enzymes revealed that certain compounds can bind selectively in the active sites of human steroid hydroxylases CYP7, CYP17A1, CYP19A1 or CYP21A2, which could be important for the development of novel activity modulators of these enzymes and identification of possible side effects.
Introduction
New global cancer data (from December 2020) suggests that cancer diagnoses have increased to 19.3 million, while cancer mortality has risen to 10 million. The International Agency for Research on Cancer (IARC) estimated that one in five people worldwide will suffer from cancer during their lifetime, where lung and prostate cancers are the most commonly diagnosed in men, while breast cancer is the most common form among women.1The main reasons for the failure of available chemotherapy for cancer treatment are the lack of selectivity of conventional drugs, metastatic spreading of initial tumors, multidrug resistance and the heterogeneity of the disease. These disadvantages have inspired medicinal chemists to design and develop safer, target-specific, and effective steroid anticancer agents.2–4 Structurally diverse cytotoxic and cytostatic steroids are very relevant as lead compounds and molecular probes for anticancer drug discovery programs and the elucidation of the molecular mechanisms of anticancer compounds.4
Steroids are currently considered to be relevant scaffolds for the development of new anticancer drugs, thanks to their selectivity, suitable physicochemical properties, and reduced side effects when applied as drugs. Synthetic analogues of natural steroids are widely used in the treatment of cancers of the reproductive tissues.5,6 Besides, several steroids have been reported to exert pronounced anticancer effects in hormone-independent tumors.7 2-Methoxyestradiol, an endogenous metabolite of estradiol without hormonal activity, exhibits potent antiproliferative activity against various tumor cell lines in vitro and inhibits tumor growth in vivo.8 It was also demonstrated to induce programmed cell death in endothelial cells and suppresses cancer-related angiogenesis.9,10
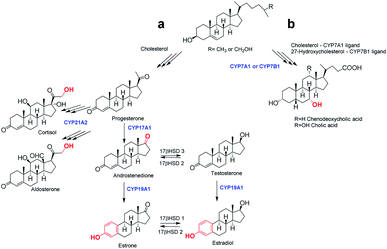
Since many chemotherapeutics used in oncological praxis express nonselective cytotoxicity, today one of the most commonly used targets for the study of the side effects of new drug candidates are cytochrome P450 enzymes (CYP). Human steroid hydroxylases, cholesterol 7α-hydroxylase (CYP7A1), 25-hydroxycholesterol 7α-hydroxylase (CYP7B1), steroid 17α-hydroxylase/17,20-lyase (CYP17A1), aromatase (CYP19A1) and 21-hydroxylase (CYP21A2) (Fig. 1) are key CYP enzymes involved in the biosynthesis of cholesterol, bile acids, neurosteroids, progestins, androgens, estrogens and corticosteroids. Consequently, changes in their endogenous levels and activity are connected with different diseases, including prostate and breast cancer.11–13 Their key importance for normal human physiology is therefore related to their use as targets in the development of highly effective and selective drugs (especially CYP17A1, CYP19A1). Based on this, there are two important aspects of testing the effects of novel compounds against CYPs: their antihormonal effects, and the establishment of new types of biological activity for synthetic steroid derivatives, particularly for prospective anticancer agents, which can help to diminish possible problems during the development of novel drugs.
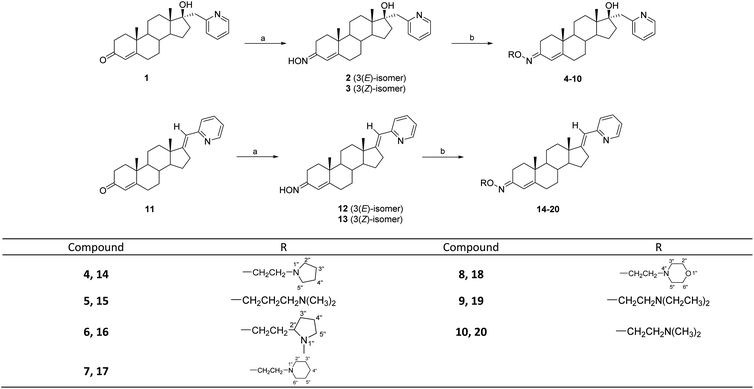
Oxyimino ethers have attracted much interest as important biologically active compounds and precursors for the preparation of a wide variety of drugs and natural products.14–16 Some steroid oximes and oxime-ethers have been shown to have antioxidant,17 antimicrobial,17–19 antineoplastic20–22 or neuromuscular blocking23 activities. O-Alkylated oximes derivatized with an alkylaminoethyl side chain have also been reported as effective candidates for cytotoxic drugs. In view of these findings and in continuation of our previous work7,24–27 on the synthesis and biological activity of androstane derivatives, we have synthesized novel oxime–ether derivatives in 17α-(pyridin-2-yl)methyl series (4–10) and in (17E)-(pyridin-2-yl)methylidene series (14–20), by the reaction of androst-4-ene 3E-oxime 2 and 12, using various alkylaminoethyl chlorides. The biological effects of these new compounds were then studied in vitro on several types of human steroid-converting CYPs and on human cancer cell lines.
Results and discussion
Chemistry
Interest in the development of steroid derivatives remains the focus of many synthetic chemists,28–30 due to their significant antitumor potential. Heterocyclic systems with a nitrogen atom in the steroid nucleus have been consistently confirmed as structural features that have led to improved biological activity for steroid compounds.31–38 Based on relevant literature data, as well as our previous research, the main objective of the present study was to introduce a new structural element with a nitrogen atom in the A ring of 17-substituted androst-4-ene compounds, in order to obtain new derivatives with selective anticancer properties.The 17α-(pyridin-2-yl)methyl 3-oximes 2 and 3, or (17E)-(pyridin-2-yl)methylidene 3-oximes 12 and 13, were synthesized from the corresponding androst-4-en-3-ones 1 and 11,39 the oximation of which was carried out with hydroxylamine-hydrochloride and sodium-acetate in refluxing ethanol, according to the sequence shown in Scheme 1.26 The E/Z configuration in these isomeric oximes has been assigned on the basis of the NMR data reported in the literature, for compounds with a similar A-ring, which are in good correlation with our data.40 In the 1H NMR spectra, protons from the oxyimino groups in E-isomers were found at 10.48 and 10.47 ppm (for compounds 2 and 12), while for the Z-isomers these were registered at lower chemical shifts, at 10.22 and 10.21 ppm (for compounds 3 and 13). This can be explained by the reduced electron density in E isomer, as evident by the observed “de-shielding” effect.
To synthesize new O-alkylated derivatives, only 3E-isomeric oximes 2 and 12 were subjected to further transformation following previously published procedures.23–25 Compounds 2 and 12 were condensed with different alkylaminoethyl chlorides to obtain desired oxime ethers 4–10 and 14–20, respectively (Scheme 1). A total of fourteen alkylaminoethoxyimino androstane derivatives were prepared at different reaction times and yields (Table 1).
| Compound | t (h) | Yield (%) | Compound | t (h) | Yield (%) |
|---|---|---|---|---|---|
| 4 | 48 | 52.4 | 14 | 72 | 35.6 |
| 5 | 48 | 71.8 | 15 | 48 | 43.0 |
| 6 | 72 | 26.0 | 16 | 72 | 9.1 |
| 7 | 48 | 47.2 | 17 | 96 | 53.2 |
| 8 | 96 | 54.9 | 18 | 96 | 56.2 |
| 9 | 48 | 47.9 | 19 | 115 | 14.6 |
| 10 | 49 | 76.3 | 20 | 48 | 30.5 |
All new compounds were fully characterized by IR, 1H NMR and 13C NMR spectra (available in ESI†), as well as by mass spectrometry analysis. In the 1H NMR spectra of all O-alkylated compounds, the appearance of triplets for –C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2N
2N![[double bond splayed right]](https://www.rsc.org/images/entities/char_e00a.gif) group at about 2.80 ppm, and for –OC
group at about 2.80 ppm, and for –OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2– group at about 4.20 ppm was observed. The presence of the pyrrolidine ring in compounds 4 and 14 was confirmed by the 13C NMR spectrum, where C-3′′ and C-4′′ carbons are equivalent and give one signal located at 23.52 or 23.47 ppm, respectively. An intense singlet originating from two N-methyl groups was observed in the 1H NMR spectrum of compounds 5 and 15 at 2.24 ppm. Singlets at 2.36 and 2.35 ppm, corresponding to protons from the N-methyl groups of the pyrrolidine ring, were observed in the 1H NMR spectra of compounds 6 and 16. The presence of the piperidine ring in compounds 7 and 17 is indicated by signals in the 13C NMR spectrum located at 25.80 and 54.85 ppm (for 7), and at 25.59 and 54.76 ppm (for 17), which correspond to the equivalent carbons C-3′′ and C-5′′, as well as C-2′′ and C-6′′, respectively. Further, two multiplets at 2.52 and 3.73 ppm were observed in the 1H NMR spectra of compounds 8 and 18, indicating the presence of equivalent H-3′′ and H-5′′ protons, and H-2′′ and H-6′′ protons, respectively, from the morpholine ring. In the 1H NMR spectra of compounds 9 and 19, triplets at 1.05 and 1.07 ppm, and quartets at 2.63 and 2.65 ppm, respectively, corresponding to protons from two N-ethyl groups were detected. The presence of two nitrogen-bonded methyl groups in compounds 10 and 20 is indicated by intense signals in the 1H NMR spectrum at 2.33 and 2.24 ppm, as well as in the 13C NMR spectrum at 45.85 or at 45.43 ppm, respectively. Finally, in the 1H NMR spectra of all new derivatives 4–10 and 14–20, the absence of a signal at approximately 10 ppm (from the oxyimino group) was observed, which further confirms the presence of substitution on C3-oxime by the corresponding alkyl group.
2– group at about 4.20 ppm was observed. The presence of the pyrrolidine ring in compounds 4 and 14 was confirmed by the 13C NMR spectrum, where C-3′′ and C-4′′ carbons are equivalent and give one signal located at 23.52 or 23.47 ppm, respectively. An intense singlet originating from two N-methyl groups was observed in the 1H NMR spectrum of compounds 5 and 15 at 2.24 ppm. Singlets at 2.36 and 2.35 ppm, corresponding to protons from the N-methyl groups of the pyrrolidine ring, were observed in the 1H NMR spectra of compounds 6 and 16. The presence of the piperidine ring in compounds 7 and 17 is indicated by signals in the 13C NMR spectrum located at 25.80 and 54.85 ppm (for 7), and at 25.59 and 54.76 ppm (for 17), which correspond to the equivalent carbons C-3′′ and C-5′′, as well as C-2′′ and C-6′′, respectively. Further, two multiplets at 2.52 and 3.73 ppm were observed in the 1H NMR spectra of compounds 8 and 18, indicating the presence of equivalent H-3′′ and H-5′′ protons, and H-2′′ and H-6′′ protons, respectively, from the morpholine ring. In the 1H NMR spectra of compounds 9 and 19, triplets at 1.05 and 1.07 ppm, and quartets at 2.63 and 2.65 ppm, respectively, corresponding to protons from two N-ethyl groups were detected. The presence of two nitrogen-bonded methyl groups in compounds 10 and 20 is indicated by intense signals in the 1H NMR spectrum at 2.33 and 2.24 ppm, as well as in the 13C NMR spectrum at 45.85 or at 45.43 ppm, respectively. Finally, in the 1H NMR spectra of all new derivatives 4–10 and 14–20, the absence of a signal at approximately 10 ppm (from the oxyimino group) was observed, which further confirms the presence of substitution on C3-oxime by the corresponding alkyl group.
Cytotoxic activity
The first steps in investigating the potential of newly synthesized O-alkylated androstane compounds for the development of effective anticancer compounds were cytotoxicity assays. The cytotoxic activity of selected compounds was evaluated against several common human tumor cell lines, as well as on normal fetal lung cells (MRC-5) and human foreskin fibroblasts (BJ), in order to determine the antitumor activity and selectivity of the compounds. Cytotoxic activity was assessed in vitro after 72 h treatment using the MTT41 or alamarBlue (resazurin)42 assay. The results were compared with the effect of the widely used nonselective chemotherapeutic drug cisplatin (Cis-Pt) and the steroid aromatase inhibitor formestane (For), which was used as a control for general steroid toxicity.Previously, we reported the cytotoxic activities of parent compounds in the 17α-(pyridin-2-yl)methyl (1) and (17E)-(pyridin-2-yl)methylidene series (11).43 Interestingly, the majority of the newly synthesized compounds in both series appear to be most active against lung adenocarcinoma cells (A549) (Table 2), which is in contrast to results published earlier.26 Of these, both isomeric oximes 12 and 13 of the (17E)-(pyridin-2-yl)methylidene series, and pyrrolidine derivative 14 showed the strongest cytotoxicity (IC50 1.5, 1.8 and 2.0 μM, respectively), and all of these substances were more cytotoxic than cisplatin. Compounds 7 and 18, containing piperidine or morpholine moiety, also displayed significant cytotoxic activity (IC50 5.4 and 5.6 μM), while derivatives 4, 6, 9 and 10 of the 17α-(pyridin-2-yl)methyl series as well as (17E)-(pyridin-2-yl)methylidene derivative 15 with 3-(N,N-dimethylamino)propyl group exhibited moderate cytotoxicity for A549 (IC50 11.8, 14.1, 11.8, 13.0 and 18.9 μM, respectively).
| Assay | IC50 [μM], after 72 h incubation | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MTT | alamarBlue | |||||||||
| Cell line/compound | MCF-7 | MDA-MB-231 | PC-3 | HeLa | HT-29 | A549 | MRC-5 | CEM | G-361 | BJ |
| a N/A – IC50 value was not available due to nonlinear dose dependence or hormetic effect. | ||||||||||
| 2 | >100 | >100 | >100 | 13.8 | >100 | 30.3 | >100 | >50 | 48.0 | >50 |
| 3 | >100 | 38.9 | >100 | >100 | >100 | 52.4 | N/A | >50 | >50 | >50 |
| 4 | 87.5 | >100 | >100 | 22.2 | >100 | 11.8 | >100 | 19.5 | 3.6 | 14.2 |
| 5 | >100 | >100 | >100 | 18.4 | >100 | >100 | >100 | 16.6 | 1.9 | 7.6 |
| 6 | 10.5 | >100 | 49.5 | 68.0 | 18.4 | 14.1 | >100 | 13.0 | 2.6 | 6.1 |
| 7 | >100 | 47.4 | 14.0 | >100 | >100 | 5.4 | >100 | 16.5 | 2.5 | 7.2 |
| 8 | >100 | 30.7 | >100 | >100 | >100 | >100 | N/A | 21.1 | 7.1 | >50 |
| 9 | >100 | 23.9 | 20.2 | N/A | 11.6 | 11.8 | >100 | 14.9 | 2.5 | 7.3 |
| 10 | >100 | >100 | 14.5 | >100 | >100 | 13.0 | >100 | 34.6 | 3.4 | 16.7 |
| 12 | 41.0 | 47.3 | >100 | >100 | 4.4 | 1.5 | >100 | >50 | 45.3 | >50 |
| 13 | 44.9 | 5.2 | 57.7 | >100 | 10.6 | 1.8 | >100 | >50 | 46.6 | >50 |
| 14 | >100 | 4.7 | 77.1 | 22.6 | 3.3 | 2.0 | >100 | 30.4 | 8.9 | 25.3 |
| 15 | 15.1 | 44.6 | >100 | >100 | >100 | 18.9 | >100 | 13.9 | 2.8 | 7.5 |
| 16 | 7.0 | >100 | >100 | >100 | >100 | >100 | >100 | 21.9 | 13.6 | 27.6 |
| 17 | >100 | 19.1 | >100 | 32.2 | 23.9 | 21.3 | >100 | 18.0 | 4.7 | 20.0 |
| 18 | >100 | 26.8 | >100 | 44.1 | 7.8 | 5.6 | >100 | 21.9 | 11.4 | >50 |
| 19 | >100 | >100 | >100 | >100 | >100 | 26.6 | 86.4 | 23.8 | 3.8 | 14.7 |
| 20 | >100 | >100 | 69.5 | >100 | 13.0 | 82.8 | >100 | 22.7 | 10.6 | 34.2 |
| Cis-Pt | 1.6 | 2.6 | 4.5 | 2.1 | 4.1 | 3.2 | 0.2 | 0.8 | 4.5 | 9.6 |
| For | >100 | 19.6 | 26.4 | 3.4 | >100 | 38.6 | >100 | – | – | – |
Colon cancer cells HT-29 were more sensitive to (17E)-(pyridin-2-yl)methylidene series, where 3E-oxime 12, pyrrolidine 14 and morpholine derivative 18 exhibited low micromolar IC50 at 4.4, 3.3 and 7.8 μM, respectively, while compounds 6, 9, 13 and 20 showed moderate cytotoxicity to HT-29 (IC50 18.4, 11.6, 10.6 and 13.0 μM, respectively). Compounds 13 (with 3Z-oxyimino function) and 14 (with pyrrolidine ring) showed strong cytotoxic activity (IC50 5.2 and 4.7 μM, respectively) against estrogen receptor negative (ER−) breast cancer cell line (MDA-MB-231). It is not the first time that we have encountered derivatives that can not only stop the proliferation of triple-negative breast cancer cells but also do it selectively.7 In addition, compound 16 with N-methylpyrrolidin ring expressed significant cytotoxicity (IC50 7.0 μM) against the ER+ breast cancer cell line MCF-7, while formestane, aromatase inhibitor used in clinical praxis in the treatment of estrogen-dependent breast carcinoma, showed no toxicity toward these tumor cells. The cytotoxicity results also revealed that 17α-(pyridin-2-yl)methyl derivative 7 with a piperidine ring and derivative 10 with a (N,N-dimethylamino)ethyl group showed mild cytotoxicity against androgen receptor negative (AR-) prostate cancer PC-3 cells (IC50 14.0 and 14.5 μM, respectively), similar to 3E-oxime 2 and 3-(N,N-dimethylamino)propyl derivative 5, which were moderately but also selectively toxic when tested on HeLa cervix carcinoma cells (IC50 13.8 and 18.4 μM, respectively). Finally, it should be noted that all newly synthesized compounds were confirmed to be non-toxic to the normal MRC-5 cells, while cisplatin was very toxic to these cells.
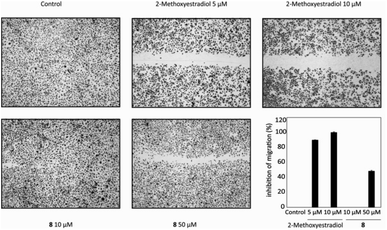
Furthermore, two additional human cancer cell lines (CEM, acute leukemia cell line; G-361, malignant melanoma cell line) and normal human skin fibroblasts (BJ) were also used to evaluate the cytotoxicity of eighteen derivatives after 72 h treatment (Table 2). Seven compounds (4, 5, 6, 7, 9, 15 and 17) were active against CEM cells in the lower micromolar range; seven exhibited moderate cytotoxicity (8, 10, 14, 16, 18, 19 and 20); while four compounds (2, 3, 12 and 13) showed zero activity. The similar series of compounds (4–10, 14–15, 17 and 19) was also very toxic to melanoma G-361 cells after 72 h (<10 μM), and their cytotoxicity was comparable to that of Cis-Pt. Of all O-alkylated compounds, only morpholine derivatives 8 and 18 showed no cytotoxicity against normal human BJ fibroblasts. Moreover, 17α-(pyridin-2-yl)methylandrostene 8 expressed stronger cytotoxic activity against melanoma G-361 cells (IC50 7.1 μM), and therefore it was selected for further experiments to test anti-angiogenic and anti-inflammatory activity in vitro.
To examine the influence of new androstane-derived compounds on angiogenesis in vitro, migration scratch and tube formation assays on HUVEC cells treated with 10 and 50 μM of morpholine derivative 8 for 20 hours were performed. Compound 8, which was selected for this experiment thanks to its strong antiproliferative effect against melanoma G-361 cells, inhibited the migration of endothelial cells after application of 50 μM of this compound by 48% compared to 10 μM 2-methoxyestradiol as a positive control (Fig. 2). However, both tested concentrations did not affect the creation of tube-like structures by HUVECs (data not shown). Derivative 8 had also no influence on the expression of the inflammatory adhesion molecule E-selectin on the cell surface after 4 h of HUVECs' treatment (data not shown).
Apoptosis induction
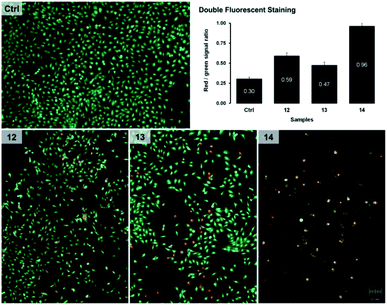
Some of the tested compounds have been shown to have an antiproliferative effect on human tumor cell lines that respond poorly to hormone therapy. Here we tried to detect the presence of apoptosis and compare in, as much as possible, the same conditions for samples, rather than to precisely determine the percentage of apoptosis. Since compounds 12, 13 and 14 showed potent micromolar cytotoxicity against lung adenocarcinoma A549 cells, we investigated whether the selected test compounds induced programmed cell death in this cell line as a model system. Apoptosis in cells was monitored by a fluorescent double-staining method using acridine orange and ethidium bromide dyes after cells treatment for 72 h with equitoxic concentrations of steroid compounds, equal to their IC50s. Results are shown in Fig. 3 as the ratio of fluorescent signals (red to green) for a control sample of untreated cells (Ctrl) and samples treated with selected compounds 12, 13 and 14, and were obtained using the program ImageJ.As shown in Fig. 3, fluorescent signals indicating apoptotic changes were detected in all samples of A549 cells treated with test compounds, where derivative 14 was the most effective in inducing apoptosis. The intensity of apoptotic changes in cells treated with compounds 12–14 can also be observed on microphotographs of specimens stained with fluorescent dyes.
The oxime derivatives 12 and 13, as well as O-alkylated pyrrolidine derivative 14, showed strong cytotoxic activity and the potential to induce apoptosis in A549 lung adenocarcinoma cells, while at the same time being non-toxic to normal lung fibroblasts MRC-5. These findings classify them in the group of rare selective antitumor compounds whose steroid core is likely to have low general cytotoxicity in humans. Although other compounds have shown lower cytotoxicity, they have also been selective, and such properties are desirable in candidates for the development of new antitumor drugs.
In vitro screening toward human steroid hydroxylases
Screening of novel steroid oximes against a palette of human steroid hydroxylases including CYP7A1, CYP7B1, CYP17A1, CYP19A1 and CYP21A2 (Fig. 1) was performed to identify novel ligands of these enzymes. The results obtained in such binding tests may direct further research towards the design of novel compounds or further biomedical studies, depending on whether the compounds bind to the desired CYP enzyme. The key importance of these enzymes for normal human physiology is therefore related to their use as targets in the development of highly effective and selective drugs, or in the identification of possible side effects during administration. Binding affinities of CYPs for new androstane-derived compounds are presented in Table 3.| Compound | CYP7A1 | CYP7B1 | CYP17A1 | CYP19A1 | CYP21A2 |
|---|---|---|---|---|---|
| a Binding was detected, but spectral response is too low.b Kd and ligand type cannot be estimated because of two maxima, corresponding to binding of substrate-like molecule (λ = 393 nm) and inhibitor-like molecule (λ = 433 nm). | |||||
| 5 | Type II binding (inhibitor-like molecule) | — | — | — | — |
| ΔA = 0.004 | |||||
| C = 0–98 μM | |||||
| Kd ∼ 0.2 μMa (R2 = 0.6) | |||||
| 10 | — | Type I + type II bindingb | — | — | — |
| 17 | — | — | — | — | Type I binding (substrate-like molecule) |
| ΔA = 0.004 | |||||
| C = 0–49 μM | |||||
| Kd = 3.6 ± 1.0 μM (R2 = 0.93) | |||||
| 18 | — | Type I + type II bindingb | — | — | — |
| 19 | — | — | Type I binding (substrate-like molecule) | — | — |
| ΔA = 0.012 | |||||
| C = 0–49 μM | |||||
| Kd = 8.6 ± 1.5 μM (R2 = 0.97) | |||||
| 20 | — | — | — | Type I binding (substrate-like molecule) | — |
| ΔA = 0.004 | |||||
| C = 0–15 μM | |||||
| Kd ∼ 0.2 μMa (R2 = 0.77) | |||||
| 2–4, 6–9, 12–16 | No binding | ||||
It was found that novel compounds are able to bind either as substrate-like ligands (leading to the displacement of the water molecule from the Fe coordination sphere) or as inhibitor-like molecules (leading to the replacement of the water molecule from the Fe coordination sphere). In all cases, the amplitude of the spectral response is quite small (compared to known ligands of these enzymes). This means that only a minor fraction of corresponding P450 enzymes (in terms of different protein conformations) can bind substrate molecules.
In the case of CYP7A1 and CYP7B1 enzymes, that are involved in the synthesis of bile acids, compounds 5, 10 and 18 were detected as inhibitors based on spectral data, meaning that the nitrogen atom of these compounds shares unpaired electrons with the Fe2+ of the cofactor. Moreover, in the case of CYP7B1, a dual-type binding mode was detected, which means that there are two possible orientations of the ligands in the active site of the enzyme.
It was also found that some of the tested compounds can bind microsomal steroid hydroxylases CYP17A1, CYP19A1 and CYP21A2, necessary for the biosynthesis of steroid hormones. The most prominent results were obtained for CYP17A1 and 2-(N,N-diethylamino)ethyl derivative 19. The data show that this molecule interacts with CYP17A1 with an affinity of Kd = 8.6 ± 1.5 μM, which is comparable with the affinity of natural ligands of the protein (progesterone and 17α-hydroxyprogesterone).13
In silico analysis of compound 19 binding in the CYP17A1 active site showed that the pose of the steroid molecule is similar to that of abiraterone and galeterone. The pyridine fragment of the modified steroid occupies a hydrophobic pocket, formed by residues V366, A367, I371 and V483, like a benzimidazole moiety of galeterone.13 Derivative 19 is localized in such a way that the steroid core of the molecule interacts with amino acids from α-helix I (A301, G302), while the large substituting group at C3 interacts with amino acid residues A105, I205, R239 and D298 from α-helices B′, F, G and I (Fig. 4). Unfavorable interactions of the highly hydrophobic part of the substitution group with polar residues could be the reason for the relatively low affinity (compared to known ligands of the enzyme) of the modified steroid. It was also found that the oxygen atom of the oxyimino group forms bonds with conserved N202 from α-helix F. It is well-known that such interactions are crucial for the stabilization of CYP17A1 ligands.13
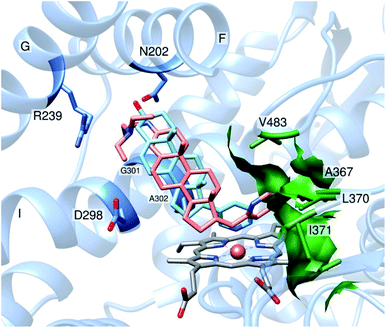 | ||
| Fig. 4 Fragment of active site of CYP17A1 in complex with compound 19 and abiraterone (PDB ID: 3RUK). In the stick and sphere representations, non-carbon atoms are indicated in blue (N) and red (O), Fe atom is indicated as an orange sphere. Molecules are colored by dark grey (heme), salmon (compound 19) and light blue (abiraterone). Surface and key residues, forming hydrophobic pocket are colored by forest green. Key residues, forming bonds with derivative 19, are colored cornflower blue. | ||
Based on these findings, compound 19 could be considered as a promising candidate for the development of highly efficient inhibitors of the enzyme, and in the development of drug candidates for the treatment of androgen-dependent diseases, especially prostate cancer. On the other hand, compounds expressing strong cytotoxicity based on apoptosis induction and showed no binding to steroid-converting CYP, thus bypassing possible side-effects, impose as molecules whose structural features enable their potential in drug development.
Conclusions
An efficient synthesis of four isomerically pure oximes and fourteen new oxime ether steroid derivatives in 17α-(pyridin-2-yl)methyl and (17E)-(pyridin-2-yl)methylideneandrost-4-ene series was performed. In vitro cytotoxic activity of all compounds was first evaluated against eight human cancer cell lines and two normal human fibroblasts. Most of the compounds tested showed significant cytotoxicity against tumor cell lines, especially A549, HT-29 and G-361 cells. In vitro screening also revealed that compounds 12, 13 and 14 proved to be the most promising candidates for further examination, due to their markedly strong cytotoxic effects on A549, HT-29 and MDA-MB-231 cells and their demonstrated pro-apoptotic potential against A549 cells. Compound 8, which was very cytotoxic against melanoma G-361 cells, but non-toxic toward normal human fibroblasts (BJ), was evaluated in vitro for anti-angiogenic and anti-inflammatory activity, where partial inhibition of endothelial cell migration was observed after treatment with 50 μM. Overall, the lung adenocarcinoma cell line (A549) appears to be the most sensitive to the majority of the compounds tested, which is a very encouraging result, due to the fact that lung cancer has recently become the leading cause of death for both women and men. In addition, compounds in the (17E)-(pyridin-2-yl)methylidene series showed stronger cytotoxic effects against the tested tumor cells lines, compared to the 17α-(pyridin-2-yl)methyl derivatives. In vitro screening toward cytochrome P450 enzymes, as possible molecular targets, suggests that compounds 5, 10 and 17–20 can selectively bind to the active site of human steroid hydroxylases CYP7, CYP17A1, CYP19A1 or CYP21A2. This information could be important for the development of highly effective anticancer drug candidates with low side effects based on these newly synthesized compounds.The very high complexity of biological model systems such as cell lines, especially from metastatic cancers, limits us to boldly make a hypothesis. So, the exact mechanism of action of the compounds remains unknown. Indications are that the structure of the compounds plays a significant role since small changes in the structure lead to a different biological effect. Although further studies are necessary, collected data highlight the importance of oxyimino and O-alkylated oxyimino functionality for the development of new candidates for tumor treatment. Combining the alkylaminoethyl side chain and pyridine heterocycle with steroid nucleus seems to be the right step for the development of potent and selective antineoplastic agents.
Experimental
General information
Infrared spectra (wavenumbers in cm−1) were recorded on a PerkinElmer Spectrum Two spectrometer. NMR spectra were recorded on a Bruker AVANCE III HD 400 spectrometer operating at 400 MHz (1H) and 100 MHz (13C). Chemical shifts are given in ppm (δ-scale). For chemical shift calibration, internal TMS or residual solvent signals were used. High-resolution mass spectrometry (HRMS) measurements were recorded on a Thermo LTQ Orbitrap XL instrument using (ESI+) ionisation. Chromatographic separations were performed on silica gel columns (Kieselgel 60, 0.04–0.063 mm and 0.063–0.20 mm, Merck). All the reagents used were of analytical grade.General synthetic procedure for 17α-(pyridin-2-yl)methyl and (17E)-(pyridin-2-yl)methylidene 3-hydroxyimino derivatives 2, 3 and 12, 13
Compound 1 (0.190 g, 0.50 mmol) or 11 (0.262 g, 0.72 mmol) was dissolved in 95% ethanol (30 mL), and sodium acetate (5 equivalents) and hydroxylamine hydrochloride (5 equivalents) were added. The reaction mixture was stirred under reflux for 1.5 h, then poured into water and adjusted to pH 8 with saturated NaHCO3. The white precipitate that formed was filtered off and washed with water. Obtained mixtures of the isomeric E and Z oximes 2 and 3 (molar ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 = 2.8
3 = 2.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by NMR), or 12 and 13 (molar ratio 12
1 by NMR), or 12 and 13 (molar ratio 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13 = 2.6
13 = 2.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by NMR) were then separated by flash chromatography (petroleum ether/ethyl acetate 8
1 by NMR) were then separated by flash chromatography (petroleum ether/ethyl acetate 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 for 2 and 3, or petroleum ether/ethyl acetate 5
5 for 2 and 3, or petroleum ether/ethyl acetate 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 for 12 and 13), yielding pure 3E isomer 2 and pure 3Z isomer 3, as well as pure 3E isomer 12 and pure 3Z isomer 13, as white solid.
2 for 12 and 13), yielding pure 3E isomer 2 and pure 3Z isomer 3, as well as pure 3E isomer 12 and pure 3Z isomer 13, as white solid.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NOH); 13C NMR (100 MHz, DMSO-d6) δ 14.83 (C-18), 18.00 (C-19), 18.99, 21.20, 23.97, 31.73, 32.20 (2× CH2), 34.00, 34.90, 36.71, 37.98, 43.82, 46.61, 49.89, 53.75, 82.96 (C-17), 118.35 (C-4), 121.77 (C-5′, Py), 125.90 (C-3′, Py), 136.77 (C-4′, Py), 148.40 (C-6′, Py), 53.14 (C-3), 154.61 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C25H35N2O2 [M + H]+ calcd 395.26985, found 395.26941.
NOH); 13C NMR (100 MHz, DMSO-d6) δ 14.83 (C-18), 18.00 (C-19), 18.99, 21.20, 23.97, 31.73, 32.20 (2× CH2), 34.00, 34.90, 36.71, 37.98, 43.82, 46.61, 49.89, 53.75, 82.96 (C-17), 118.35 (C-4), 121.77 (C-5′, Py), 125.90 (C-3′, Py), 136.77 (C-4′, Py), 148.40 (C-6′, Py), 53.14 (C-3), 154.61 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C25H35N2O2 [M + H]+ calcd 395.26985, found 395.26941.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NOH); 13C NMR (100 MHz, DMSO-d6) δ 14.83 (C-18), 18.29 (C-19), 21.04, 23.95, 24.79, 31.68, 32.60, 32.82, 33.99, 36.58, 36.62, 38.94, 43.82, 46.65, 49.83, 54.01, 82.93 (C-17), 111.78 (C-4), 121.78 (C-5′, Py), 125.90 (C-3′, Py), 136.77 (C-4′, Py), 148.40 (C-6′, Py), 151.47 (C-3), 156.98 (C-5), 160.77 (C-2′, Py); HRMS (m/z): for C25H35N2O2 [M + H]+ calcd 395.26985, found 395.26968.
NOH); 13C NMR (100 MHz, DMSO-d6) δ 14.83 (C-18), 18.29 (C-19), 21.04, 23.95, 24.79, 31.68, 32.60, 32.82, 33.99, 36.58, 36.62, 38.94, 43.82, 46.65, 49.83, 54.01, 82.93 (C-17), 111.78 (C-4), 121.78 (C-5′, Py), 125.90 (C-3′, Py), 136.77 (C-4′, Py), 148.40 (C-6′, Py), 151.47 (C-3), 156.98 (C-5), 160.77 (C-2′, Py); HRMS (m/z): for C25H35N2O2 [M + H]+ calcd 395.26985, found 395.26968.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NO
NO![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ); 13C NMR (100 MHz, DMSO-d6) δ 18.00 (C-18), 18.97, 19.16 (C-19), 21.49, 25.00, 30.35, 32.13, 32.28, 34.86, 35.55, 35.90, 38.00, 45.73, 53.43, 53.93, 117.63 (C-20), 118.44 (C-4), 120.84 (C-5′, Py), 123.48 (C-3′, Py), 136.61 (C-4′, Py), 149.44 (C-6′, Py), 152.98 (C-3), 154.58 (C-5), 157.38 (C-17), 160.37 (C-2′, Py); HRMS (m/z): for C25H33N2O [M + H]+ calcd 377.25929, found 377.25960.
); 13C NMR (100 MHz, DMSO-d6) δ 18.00 (C-18), 18.97, 19.16 (C-19), 21.49, 25.00, 30.35, 32.13, 32.28, 34.86, 35.55, 35.90, 38.00, 45.73, 53.43, 53.93, 117.63 (C-20), 118.44 (C-4), 120.84 (C-5′, Py), 123.48 (C-3′, Py), 136.61 (C-4′, Py), 149.44 (C-6′, Py), 152.98 (C-3), 154.58 (C-5), 157.38 (C-17), 160.37 (C-2′, Py); HRMS (m/z): for C25H33N2O [M + H]+ calcd 377.25929, found 377.25960.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NO
NO![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) ); 13C NMR (100 MHz, DMSO-d6) δ 18.27 (C-18), 19.17 (C-19), 21.33, 24.77, 24.97, 30.35, 32.64, 32.75, 35.47, 35.84, 36.55, 38.95, 45.77, 53.35, 54.22, 111.27 (C-4), 117.66 (C-20), 120.84 (C-5′, Py), 123.47 (C-3′, Py), 136.60 (C-4′, Py), 149.44 (C-6′, Py), 151.45 (C-3), 156.80 (C-5), 157.38 (C-17), 160.30 (C-2′, Py); HRMS (m/z): for C25H33N2O [M + H]+ calcd 377.25929, found 377.25942.
); 13C NMR (100 MHz, DMSO-d6) δ 18.27 (C-18), 19.17 (C-19), 21.33, 24.77, 24.97, 30.35, 32.64, 32.75, 35.47, 35.84, 36.55, 38.95, 45.77, 53.35, 54.22, 111.27 (C-4), 117.66 (C-20), 120.84 (C-5′, Py), 123.47 (C-3′, Py), 136.60 (C-4′, Py), 149.44 (C-6′, Py), 151.45 (C-3), 156.80 (C-5), 157.38 (C-17), 160.30 (C-2′, Py); HRMS (m/z): for C25H33N2O [M + H]+ calcd 377.25929, found 377.25942.General synthetic procedure for the synthesis of O-alkylated compounds 4–10 and 14–20
To a solution of 3E-oxyimino derivative 2 or 12 (0.28 mmol) in dry butanone (25 mL), anhydrous K2CO3 was added (15 equivalents) and the reaction mixture was refluxed for the next 2 h. Respective hydrochlorides of alkylaminoethyl chlorides (10 equivalents) and KOH (7 equivalents) were added and the reaction mixture was further refluxed with stirring. After reaction completion (Table 1), the mixture was cooled, filtered, and the solvent was removed under reduced pressure. Iced water was added to an oily residue and it was stirred for 1 h, to remove inorganic compounds. The entire rinsing process was repeated twice more. The obtained resinous product was filtered, dissolved in dichloromethane and dried. The solvent was removed under vacuum and the crude product was purified by column chromatography, to afford the corresponding alkylaminoethyloxyimino derivatives 4–10 and 14–20 as light yellow oil, in different yields (Table 1).![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 2.80 (2H, NC
2Py), 2.80 (2H, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 3.05 (d, 1H, J = 14.7 Hz, C
2), 3.05 (d, 1H, J = 14.7 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.20 (t, 2H, J = 6.0 Hz, OC
2Py), 4.20 (t, 2H, J = 6.0 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.78 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.63 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.16 (C-18), 17.83 (C-19), 19.62, 21.08, 23.52 (C-3′′ and C-4′′), 23.91, 32.11, 32.21, 32.54, 34.87, 35.96, 36.59, 38.03, 43.14, 46.39, 50.32, 53.88 (N
2), 5.78 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.63 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.16 (C-18), 17.83 (C-19), 19.62, 21.08, 23.52 (C-3′′ and C-4′′), 23.91, 32.11, 32.21, 32.54, 34.87, 35.96, 36.59, 38.03, 43.14, 46.39, 50.32, 53.88 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 54.78 (C-2′′ and C-5′′), 54.95, 73.02 (O
H2), 54.78 (C-2′′ and C-5′′), 54.95, 73.02 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.39 (C-17), 117.31 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.04 (C-3), 156.34 (C-5), 160.82 (C-2′, Py); HRMS (m/z): for C31H46N3O2 [M + H]+ calcd 492.35900, found 492.35737.
H2), 83.39 (C-17), 117.31 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.04 (C-3), 156.34 (C-5), 160.82 (C-2′, Py); HRMS (m/z): for C31H46N3O2 [M + H]+ calcd 492.35900, found 492.35737.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 2.35 (2H, NC
3)2), 2.35 (2H, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.97 (d, 1H, J = 14.0 Hz, C
2), 2.97 (d, 1H, J = 14.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.06 (d, 1H, J = 14.0 Hz, C
2Py), 3.06 (d, 1H, J = 14.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.08 (t, 2H, J = 6.4 Hz, OC
2Py), 4.08 (t, 2H, J = 6.4 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.61 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.5 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.83 (C-19), 19.46, 21.09, 23.92, 27.47, 32.11, 32.21, 32.53, 34.87, 35.95, 36.59, 38.03, 43.14, 45.47 (N(
2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.61 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.5 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.83 (C-19), 19.46, 21.09, 23.92, 27.47, 32.11, 32.21, 32.53, 34.87, 35.95, 36.59, 38.03, 43.14, 45.47 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3)2), 46.39, 50.32, 53.88 (N
H3)2), 46.39, 50.32, 53.88 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 56.54, 71.96 (O
H2), 56.54, 71.96 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.40 (C-17), 117.38 (C-4), 121.37 (C-5′, Py), 124.78 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.26 (C-3), 156.24 (C-5), 160.82 (C-2′, Py); HRMS (m/z): for C30H46N3O2 [M + H]+ calcd 480.35900, found 480.35855.
H2), 83.40 (C-17), 117.38 (C-4), 121.37 (C-5′, Py), 124.78 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.26 (C-3), 156.24 (C-5), 160.82 (C-2′, Py); HRMS (m/z): for C30H46N3O2 [M + H]+ calcd 480.35900, found 480.35855.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 2.96 (d, 1H, J = 15.0 Hz, C
3), 2.96 (d, 1H, J = 15.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.05 (d, 1H, J = 15.0 Hz, C
2Py), 3.05 (d, 1H, J = 15.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.30 (t, 2H, J = 8.0 Hz, OC
2Py), 4.30 (t, 2H, J = 8.0 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 4.0 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.83 (C-19), 19.63, 21.09, 22.85, 23.19, 29.40, 32.15, 32.17, 32.21, 32.52, 32.66, 34.89, 35.94, 36.59, 38.03, 43.14, 46.38 (N
2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 4.0 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.83 (C-19), 19.63, 21.09, 22.85, 23.19, 29.40, 32.15, 32.17, 32.21, 32.52, 32.66, 34.89, 35.94, 36.59, 38.03, 43.14, 46.38 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 50.32, 52.76, 53.87, 58.87 (C-2′′), 81.31 (O
H3), 50.32, 52.76, 53.87, 58.87 (C-2′′), 81.31 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.40 (C-17), 117.56 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.78 (C-4′, Py), 148.02 (C-6′, Py), 155.02 (C-3), 156.10 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C32H48N3O2 [M + H]+ calcd 506.37465, found 506.37289.
H2), 83.40 (C-17), 117.56 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.78 (C-4′, Py), 148.02 (C-6′, Py), 155.02 (C-3), 156.10 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C32H48N3O2 [M + H]+ calcd 506.37465, found 506.37289.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). IR (film, ν, cm−1): 3325, 2935, 2855, 1596, 1470, 1438, 1302, 1250, 1125, 1050, 852, 836, 754; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.07 (s, 3H, H-19), 2.50 (m, 4H, H-2′′ and H-6′′), 2.70 (t, 2H, J = 6.0 Hz, NC
1). IR (film, ν, cm−1): 3325, 2935, 2855, 1596, 1470, 1438, 1302, 1250, 1125, 1050, 852, 836, 754; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.07 (s, 3H, H-19), 2.50 (m, 4H, H-2′′ and H-6′′), 2.70 (t, 2H, J = 6.0 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.79 (d, 1H, J = 14.4 Hz, C
2), 2.79 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.05 (d, 1H, J = 14.5 Hz, C
2Py), 3.05 (d, 1H, J = 14.5 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.21 (t, 2H, J = 6.0 Hz, OC
2Py), 4.21 (t, 2H, J = 6.0 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.82 (C-19), 19.60, 21.07 (C-4′′), 23.91, 24.10, 25.80 (C-3′′ and C-5′′), 32.10, 32.20, 32.53, 34.85, 35.94, 36.58, 38.02, 43.13, 46.38, 50.31, 53.86 (N
2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.82 (C-19), 19.60, 21.07 (C-4′′), 23.91, 24.10, 25.80 (C-3′′ and C-5′′), 32.10, 32.20, 32.53, 34.85, 35.94, 36.58, 38.02, 43.13, 46.38, 50.31, 53.86 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 54.85 (C-2′′ and C-6′′), 57.71, 71.61 (O
H2), 54.85 (C-2′′ and C-6′′), 57.71, 71.61 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.39 (C-17), 117.27 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.02 (C-6′, Py), 155.47 (C-3), 156.41 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C32H48N3O2 [M + H]+ calcd 506.37465, found 506.37295.
H2), 83.39 (C-17), 117.27 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.02 (C-6′, Py), 155.47 (C-3), 156.41 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C32H48N3O2 [M + H]+ calcd 506.37465, found 506.37295.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). IR (film, ν, cm−1): 3402, 2938, 1633, 1596, 1377, 1118, 1021, 953, 871, 854, 753, 708, 657; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.08 (s, 3H, H-19), 2.55 (m, 4H, H-3′′ and H-5′′), 2.72 (t, 2H, J = 5.6 Hz, NC
1). IR (film, ν, cm−1): 3402, 2938, 1633, 1596, 1377, 1118, 1021, 953, 871, 854, 753, 708, 657; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.08 (s, 3H, H-19), 2.55 (m, 4H, H-3′′ and H-5′′), 2.72 (t, 2H, J = 5.6 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.78 (d, 1H, J = 14.4 Hz, C
2), 2.78 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.05 (d, 1H, J = 14.4 Hz, C
2Py), 3.05 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.73 (m, 4H, H-2′′ and H-6′′), 4.22 (t, 2H, J = 5.6 Hz, OC
2Py), 3.73 (m, 4H, H-2′′ and H-6′′), 4.22 (t, 2H, J = 5.6 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 7.16 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.82 (C-19), 19.64, 21.07, 23.91, 32.09, 32.20, 32.55, 34.83, 35.94, 36.57, 38.05, 43.13, 46.38, 50.31, 53.87 (N
2), 5.77 (s, 1H, H-4), 7.16 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.82 (C-19), 19.64, 21.07, 23.91, 32.09, 32.20, 32.55, 34.83, 35.94, 36.57, 38.05, 43.13, 46.38, 50.31, 53.87 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 53.99 (C-3′′ and C-5′′), 57.49, 66.87 (C-2′′ and C-6′′), 71.49 (O
H2), 53.99 (C-3′′ and C-5′′), 57.49, 66.87 (C-2′′ and C-6′′), 71.49 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.38 (C-17), 117.16 (C-4), 121.37 (C-5′, Py), 124.75 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.73 (C-3), 156.57 (C-5), 160.79 (C-2′, Py); HRMS (m/z): for C31H46N3O3 [M + H]+ calcd 508.35392, found 508.35436.
H2), 83.38 (C-17), 117.16 (C-4), 121.37 (C-5′, Py), 124.75 (C-3′, Py), 136.79 (C-4′, Py), 148.03 (C-6′, Py), 155.73 (C-3), 156.57 (C-5), 160.79 (C-2′, Py); HRMS (m/z): for C31H46N3O3 [M + H]+ calcd 508.35392, found 508.35436.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). IR (film, ν, cm−1): 3325, 2937, 1596, 1472, 1437, 1377, 1250, 1201, 1050, 1000, 963, 836, 754; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.05 (t, 6H, J = 7.2 Hz, N(C
1). IR (film, ν, cm−1): 3325, 2937, 1596, 1472, 1437, 1377, 1250, 1201, 1050, 1000, 963, 836, 754; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.05 (t, 6H, J = 7.2 Hz, N(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CH2)2), 1.07 (s, 3H, H-19), 2.63 (q, 4H, J = 7.2 Hz, N(CH3C
3CH2)2), 1.07 (s, 3H, H-19), 2.63 (q, 4H, J = 7.2 Hz, N(CH3C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2)), 2.78 (m, 1H, C
2)), 2.78 (m, 1H, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 2.80 (t, 2H, J = 4.5 Hz, NC
2Py), 2.80 (t, 2H, J = 4.5 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 3.05 (d, 1H, J = 14.4 Hz, C
2), 3.05 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.16 (t, 2H, J = 4.5 Hz, OC
2Py), 4.16 (t, 2H, J = 4.5 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, J = 4.5 Hz, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.61 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 11.77 (N(
2), 5.77 (s, 1H, J = 4.5 Hz, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.61 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 11.77 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3CH2)2), 14.15 (C-18), 17.81 (C-19), 19.57, 21.07, 23.91, 32.09, 32.20, 32.53, 34.86, 35.95, 36.58, 38.02, 43.14, 46.38, 47.82 (N(CH3
H3CH2)2), 14.15 (C-18), 17.81 (C-19), 19.57, 21.07, 23.91, 32.09, 32.20, 32.53, 34.86, 35.95, 36.58, 38.02, 43.14, 46.38, 47.82 (N(CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2)2), 50.31, 51.26, 53.86 (N
H2)2), 50.31, 51.26, 53.86 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 72.09 (O
H2), 72.09 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.38 (C-17), 117.29 (C-4), 121.36 (C-5′, Py), 124.75 (C-3′, Py), 136.78 (C-4′, Py), 148.02 (C-6′, Py), 155.39 (C-3), 156.34 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C31H48N3O2 [M + H]+ calcd 494.37465, found 494.37502.
H2), 83.38 (C-17), 117.29 (C-4), 121.36 (C-5′, Py), 124.75 (C-3′, Py), 136.78 (C-4′, Py), 148.02 (C-6′, Py), 155.39 (C-3), 156.34 (C-5), 160.80 (C-2′, Py); HRMS (m/z): for C31H48N3O2 [M + H]+ calcd 494.37465, found 494.37502.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). IR (film, ν, cm−1): 3351, 2940, 1596, 1569, 1438, 1375, 1249, 1034, 999, 963, 916, 851, 836, 754, 657; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.07 (s, 3H, H-19), 2.33 (s, 6H, N(C
2). IR (film, ν, cm−1): 3351, 2940, 1596, 1569, 1438, 1375, 1249, 1034, 999, 963, 916, 851, 836, 754, 657; 1H NMR (400 MHz, CDCl3) δ 0.98 (s, 3H, H-18), 1.07 (s, 3H, H-19), 2.33 (s, 6H, N(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 2.66 (t, 2H, J = 5.6 Hz, NC
3)2), 2.66 (t, 2H, J = 5.6 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.79 (d, 1H, J = 14.4 Hz, C
2), 2.79 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 3.05 (d, 1H, J = 14.4 Hz, C
2Py), 3.05 (d, 1H, J = 14.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2Py), 4.18 (t, 2H, J = 5.6 Hz, OC
2Py), 4.18 (t, 2H, J = 5.6 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.81 (C-19), 19.60, 21.07, 23.91, 32.09, 32.20, 32.54, 34.82, 35.95, 36.58, 38.08, 43.14, 45.85 (N(
2), 5.77 (s, 1H, H-4), 7.15 (m, 2H, H-3′ and H-5′, Py), 7.62 (m, 1H, H-4′, Py), 8.45 (d, 1H, J = 3.6 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 14.15 (C-18), 17.81 (C-19), 19.60, 21.07, 23.91, 32.09, 32.20, 32.54, 34.82, 35.95, 36.58, 38.08, 43.14, 45.85 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3)2), 46.38, 50.31, 53.86 (N
H3)2), 46.38, 50.31, 53.86 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 58.10, 71.77 (O
H2), 58.10, 71.77 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 83.39 (C-17), 117.24 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.02 (C-6′, Py), 155.58 (C-3), 156.50 (C-5), 160.81 (C-2′, Py); HRMS (m/z): for C29H44N3O2 [M + H]+ calcd 466.34335, found 466.34560.
H2), 83.39 (C-17), 117.24 (C-4), 121.37 (C-5′, Py), 124.76 (C-3′, Py), 136.79 (C-4′, Py), 148.02 (C-6′, Py), 155.58 (C-3), 156.50 (C-5), 160.81 (C-2′, Py); HRMS (m/z): for C29H44N3O2 [M + H]+ calcd 466.34335, found 466.34560.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 4.22 (t, 2H, J = 6.0 Hz, C
2), 4.22 (t, 2H, J = 6.0 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2O), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.02 (m, 1H, H-5′), 7.27 (m, 1H, H-3′), 7.60 (t, 1H, J = 7.5 Hz, H-4′), 8.56 (d, 1H, J = 4.5 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.79 (C-19), 19.61, 21.42, 23.47 (C-3′′ and C-4′′), 53.97 (N
2O), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.02 (m, 1H, H-5′), 7.27 (m, 1H, H-3′), 7.60 (t, 1H, J = 7.5 Hz, H-4′), 8.56 (d, 1H, J = 4.5 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.79 (C-19), 19.61, 21.42, 23.47 (C-3′′ and C-4′′), 53.97 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 24.96, 29.79, 32.08, 32.48, 34.80, 35.65, 35.81, 38.02, 45.76, 53.28, 53.97, 54.65 (C-2′′ and C-5′′), 72.55 (O
H2), 24.96, 29.79, 32.08, 32.48, 34.80, 35.65, 35.81, 38.02, 45.76, 53.28, 53.97, 54.65 (C-2′′ and C-5′′), 72.55 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.37 (C-20), 118.05 (C-4), 120.32, 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.17 (C-6′, Py), 155.32 (C-3), 156.39 (C-5), 157.57 (C-17), 160.18 (C-2′, Py); HRMS (m/z): for C31H44N3O [M + H]+ calcd 474.34844, found 474.34996.
H2), 117.37 (C-20), 118.05 (C-4), 120.32, 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.17 (C-6′, Py), 155.32 (C-3), 156.39 (C-5), 157.57 (C-17), 160.18 (C-2′, Py); HRMS (m/z): for C31H44N3O [M + H]+ calcd 474.34844, found 474.34996.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 2.37 (t, 2H, J = 7.4 Hz, NC
3)2), 2.37 (t, 2H, J = 7.4 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 4.08 (t, 2H, J = 6.4 Hz, C
2), 4.08 (t, 2H, J = 6.4 Hz, C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2O), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.02 (m, 1H, H-5′), 7.26 (m, 1H, H-3′), 7.59 (t, 1H, J = 7.8 Hz, H-4′), 8.56 (d, 1H, J = 4.7 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.80 (C-19), 19.44, 21.43, 24.97, 27.38, 29.80, 32.09, 32.47, 34.81, 35.66, 35.83, 38.01, 45.38 (N(
2O), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.02 (m, 1H, H-5′), 7.26 (m, 1H, H-3′), 7.59 (t, 1H, J = 7.8 Hz, H-4′), 8.56 (d, 1H, J = 4.7 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.80 (C-19), 19.44, 21.43, 24.97, 27.38, 29.80, 32.09, 32.47, 34.81, 35.66, 35.83, 38.01, 45.38 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3)2), 45.76, 53.28, 53.97 (N
H3)2), 45.76, 53.28, 53.97 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 56.48, 71.93 (O
H2), 56.48, 71.93 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.51 (C-20), 118.05 (C-4), 120.22 (C-5′, Py), 122.78 (C-3′), 135.82 (C-4′, Py), 149.17 (C-6′, Py), 154.99 (C-3), 156.13 (C-5), 157.58 (C-17), 160.20 (C-2′, Py); HRMS (m/z): for C30H44N3O [M + H]+ calcd 462.34844, found 462.35004.
H2), 117.51 (C-20), 118.05 (C-4), 120.22 (C-5′, Py), 122.78 (C-3′), 135.82 (C-4′, Py), 149.17 (C-6′, Py), 154.99 (C-3), 156.13 (C-5), 157.58 (C-17), 160.20 (C-2′, Py); HRMS (m/z): for C30H44N3O [M + H]+ calcd 462.34844, found 462.35004.![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3), 4.10 (m, 2H, OC
3), 4.10 (m, 2H, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.79 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′), 7.28 (m, 1H, H-3′), 7.60 (t, 1H, J = 8.0 Hz, H-4′), 8.56 (d, 1H, J = 4.0 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.83 (C-18), 18.80 (C-19), 19.53, 19.62, 21.44, 21.97, 24.98, 29.71, 29.80, 31.05, 32.12, 32.47, 34.88, 35.67, 35.84, 38.02, 40.50, 45.77 (N
2), 5.79 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′), 7.28 (m, 1H, H-3′), 7.60 (t, 1H, J = 8.0 Hz, H-4′), 8.56 (d, 1H, J = 4.0 Hz, H-6′); 13C NMR (100 MHz, CDCl3) δ 17.83 (C-18), 18.80 (C-19), 19.53, 19.62, 21.44, 21.97, 24.98, 29.71, 29.80, 31.05, 32.12, 32.47, 34.88, 35.67, 35.84, 38.02, 40.50, 45.77 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3), 53.30, 53.99, 57.17 (C-5′′), 59.07 (C-2′′), 81.52 (O
H3), 53.30, 53.99, 57.17 (C-5′′), 59.07 (C-2′′), 81.52 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.49 (C-20), 118.06 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′), 135.83 (C-4′), 149.19 (C-6′), 154.71 (C-3), 155.06 (C-5), 157.59 (C-17), 160.23 (C-2′, Py); HRMS (m/z): for C32H46N3O [M + H]+ calcd 488.36409, found 488.36457.
H2), 117.49 (C-20), 118.06 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′), 135.83 (C-4′), 149.19 (C-6′), 154.71 (C-3), 155.06 (C-5), 157.59 (C-17), 160.23 (C-2′, Py); HRMS (m/z): for C32H46N3O [M + H]+ calcd 488.36409, found 488.36457.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.09 (s, 3H, H-19), 2.54 (m, 4H, H-2′′ and H-6′′), 2.75 (t, 2H, J = 2.8 Hz, NC
1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.09 (s, 3H, H-19), 2.54 (m, 4H, H-2′′ and H-6′′), 2.75 (t, 2H, J = 2.8 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.77–2.90 (m, 2H, H-16a and H-16b), 4.24 (t, 2H, J = 5.6 Hz, OC
2), 2.77–2.90 (m, 2H, H-16a and H-16b), 4.24 (t, 2H, J = 5.6 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′, Py), 7.26 (m, 1H, H-3′, Py), 7.60 (t, 1H, J = 6.0 Hz, H-4′, Py), 8.56 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.79 (C-19), 19.60, 21.43, 24.97 (C-4′′), 25.59, 29.79, 32.09, 32.49, 34.80, 35.66, 35.82, 38.03, 45.77, 53.28, 53.97 (N
2), 5.78 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′, Py), 7.26 (m, 1H, H-3′, Py), 7.60 (t, 1H, J = 6.0 Hz, H-4′, Py), 8.56 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.81 (C-18), 18.79 (C-19), 19.60, 21.43, 24.97 (C-4′′), 25.59, 29.79, 32.09, 32.49, 34.80, 35.66, 35.82, 38.03, 45.77, 53.28, 53.97 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 54.76 (C-2′′ and C-6′′), 57.56, 71.39 (O
H2), 54.76 (C-2′′ and C-6′′), 57.56, 71.39 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.37 (C-20), 118.07 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.18 (C-6′, Py), 155.33 (C-3), 156.41 (C-5), 157.58 (C-17), 160.18 (C-2′, Py); HRMS (m/z): for C32H46N3O [M + H]+ calcd 488.36409, found 488.36478.
H2), 117.37 (C-20), 118.07 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.18 (C-6′, Py), 155.33 (C-3), 156.41 (C-5), 157.58 (C-17), 160.18 (C-2′, Py); HRMS (m/z): for C32H46N3O [M + H]+ calcd 488.36409, found 488.36478.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.09 (s, 3H, H-19), 2.55 (m, 4H, H-3′′ and H-5′′), 2.71 (t, 2H, J = 6.0 Hz, NC
1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.09 (s, 3H, H-19), 2.55 (m, 4H, H-3′′ and H-5′′), 2.71 (t, 2H, J = 6.0 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 2.75-2.93 (m, 2H, H-16a and H-16b), 3.73 (m, 4H, H-2′′ and H-6′′), 4.22 (t, 2H, J = 6.0 Hz, OC
2), 2.75-2.93 (m, 2H, H-16a and H-16b), 3.73 (m, 4H, H-2′′ and H-6′′), 4.22 (t, 2H, J = 6.0 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.77 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′, Py), 7.25 (m, 1H, H-3′, Py), 7.59 (t, 1H, J = 7.6 Hz, H-4′, Py), 8.55 (d, 1H, J = 4.0 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.82 (C-18), 18.79 (C-19), 19.61, 21.43, 24.97, 29.79, 32.08, 32.49, 34.79, 35.65, 35.82, 38.04, 45.76, 53.28 (N
2), 5.77 (s, 1H, H-4), 6.21 (s, 1H, H-20), 7.03 (m, 1H, H-5′, Py), 7.25 (m, 1H, H-3′, Py), 7.59 (t, 1H, J = 7.6 Hz, H-4′, Py), 8.55 (d, 1H, J = 4.0 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.82 (C-18), 18.79 (C-19), 19.61, 21.43, 24.97, 29.79, 32.08, 32.49, 34.79, 35.65, 35.82, 38.04, 45.76, 53.28 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 53.97, 54.01 (C-3′′ and C-5′′), 57.50, 66.91 (C-2′′ and C-6′′), 71.54 (O
H2), 53.97, 54.01 (C-3′′ and C-5′′), 57.50, 66.91 (C-2′′ and C-6′′), 71.54 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.32 (C-20), 118.06 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.18 (C-6′, Py), 155.44 (C-3), 156.46 (C-5), 157.57 (C-17), 160.17 (C-2′, Py); HRMS (m/z): for C31H44N3O2 [M + H]+ calcd 490.34335, found 490.34437.
H2), 117.32 (C-20), 118.06 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.18 (C-6′, Py), 155.44 (C-3), 156.46 (C-5), 157.57 (C-17), 160.17 (C-2′, Py); HRMS (m/z): for C31H44N3O2 [M + H]+ calcd 490.34335, found 490.34437.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.07 (t, 6H, J = 6.4 Hz, N(C
1). 1H NMR (400 MHz, CDCl3) δ 0.92 (s, 3H, H-18), 1.07 (t, 6H, J = 6.4 Hz, N(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3CH2)2), 1.09 (s, 3H, H-19), 2.65 (q, 4H, J = 6.4 Hz, N(CH3C
3CH2)2), 1.09 (s, 3H, H-19), 2.65 (q, 4H, J = 6.4 Hz, N(CH3C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2)2), 2.83 (t, 2H, J = 6.0 Hz, NC
2)2), 2.83 (t, 2H, J = 6.0 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 4.18 (t, 2H, J = 6.0 Hz, OC
2), 4.18 (t, 2H, J = 6.0 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.78 (s, 1H, H-4), 6.22 (s, 1H, H-20), 7.02 (m, 1H, H-5′, Py), 7.26 (m, 1H, H-3′, Py), 7.60 (m, 1H, H-4′, Py), 8.56 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 11.63 (N(
2), 5.78 (s, 1H, H-4), 6.22 (s, 1H, H-20), 7.02 (m, 1H, H-5′, Py), 7.26 (m, 1H, H-3′, Py), 7.60 (m, 1H, H-4′, Py), 8.56 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 11.63 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3CH2)2), 17.81 (C-18), 18.80 (C-19), 19.57, 21.43, 24.97, 29.80, 32.09, 32.49, 34.81, 35.66, 35.82, 38.03, 45.77, 47.79 (N(CH3
H3CH2)2), 17.81 (C-18), 18.80 (C-19), 19.57, 21.43, 24.97, 29.80, 32.09, 32.49, 34.81, 35.66, 35.82, 38.03, 45.77, 47.79 (N(CH3![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2)2), 51.18, 53.29, 53.96 (N
H2)2), 51.18, 53.29, 53.96 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 72.00 (O
H2), 72.00 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.40 (C-20), 118.07 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.19 (C-6′, Py), 155.24 (C-3), 156.17 (C-5), 157.59 (C-17), 160.19 (C-2′, Py); HRMS (m/z): for C31H46N3O [M + H]+ calcd 476.36409, found 476.36522.
H2), 117.40 (C-20), 118.07 (C-4), 120.23 (C-5′, Py), 122.79 (C-3′, Py), 135.83 (C-4′, Py), 149.19 (C-6′, Py), 155.24 (C-3), 156.17 (C-5), 157.59 (C-17), 160.19 (C-2′, Py); HRMS (m/z): for C31H46N3O [M + H]+ calcd 476.36409, found 476.36522.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). 1H NMR (400 MHz, CDCl3) δ 0.94 (s, 3H, H-18), 1.10 (s, 3H, H-19), 2.24 (s, 6H, N(C
1). 1H NMR (400 MHz, CDCl3) δ 0.94 (s, 3H, H-18), 1.10 (s, 3H, H-19), 2.24 (s, 6H, N(C![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 3)2), 2.78 (t, 2H, J = 5.6 Hz, NC
3)2), 2.78 (t, 2H, J = 5.6 Hz, NC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 4.23 (t, 2H, J = 5.6 Hz, OC
2), 4.23 (t, 2H, J = 5.6 Hz, OC![[H with combining low line]](https://www.rsc.org/images/entities/char_0048_0332.gif) 2), 5.79 (s, 1H, H-4), 6.23 (s, 1H, H-20), 7.04 (m, 1H, H-5′, Py), 7.28 (m, 1H, H-3′ Py), 7.61 (t, 1H, J = 7.6 Hz, H-4′, Py), 8.58 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.80 (C-18), 18.79 (C-19), 19.61, 21.42, 24.97, 29.79, 32.08, 32.50, 34.75, 35.65, 35.81, 38.04, 45.43 (N(
2), 5.79 (s, 1H, H-4), 6.23 (s, 1H, H-20), 7.04 (m, 1H, H-5′, Py), 7.28 (m, 1H, H-3′ Py), 7.61 (t, 1H, J = 7.6 Hz, H-4′, Py), 8.58 (d, 1H, J = 4.8 Hz, H-6′, Py); 13C NMR (100 MHz, CDCl3) δ 17.80 (C-18), 18.79 (C-19), 19.61, 21.42, 24.97, 29.79, 32.08, 32.50, 34.75, 35.65, 35.81, 38.04, 45.43 (N(![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H3)2), 45.76, 53.27, 53.95 (N
H3)2), 45.76, 53.27, 53.95 (N![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 57.71, 71.13 (O
H2), 57.71, 71.13 (O![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) H2), 117.25 (C-20), 118.05 (C-4), 120.23 (C-5′, Py), 122.80 (C-3′, Py), 135.84 (C-4′, Py), 149.17 (C-6′, Py), 155.61 (C-3), 156.66 (C-5), 157.56 (C-17), 160.17 (C-2′, Py); HRMS (m/z): for C29H42N3O [M + H]+ calcd 448.33279, found 448.33398.
H2), 117.25 (C-20), 118.05 (C-4), 120.23 (C-5′, Py), 122.80 (C-3′, Py), 135.84 (C-4′, Py), 149.17 (C-6′, Py), 155.61 (C-3), 156.66 (C-5), 157.56 (C-17), 160.17 (C-2′, Py); HRMS (m/z): for C29H42N3O [M + H]+ calcd 448.33279, found 448.33398.Cell culture
Six human tumor cell lines: estrogen receptor positive (ER+) breast adenocarcinoma MCF-7, estrogen receptor negative (ER−) breast adenocarcinoma MDA-MB-231, prostate cancer PC-3, cervical carcinoma HeLa, colon adenocarcinoma HT-29 and lung adenocarcinoma A549, as well as one normal fetal lung fibroblasts cell line MRC-5 were used in the present study (American Type Culture Collection – ATCC). Cells were grown in Dulbecco's Modified Eagle's Medium (DMEM, Sigma) with 4.5% of glucose, supplemented with 10% of fetal calf serum (FCS, Sigma) and antibiotics antimycotics solution (Sigma). Cells were cultured in flasks (Costar, 25 cm2) at 37 °C in 100% humidity with 5% CO2. T-Lymphoblastic leukemia (CEM) and malignant melanoma (G-361) cell lines (European Collection of Authenticated Cell Cultures ECACC, London, UK) were also used for cytotoxicity screening. Human foreskin fibroblasts (BJ) and human umbilical vein endothelial cells (HUVEC) were purchased from the American Type Culture Collection (Manassas, VA, USA). G-361 and BJ cells were cultured in DMEM medium, CEM cells in RPMI 1640 (Sigma, MO, USA) and HUVECs in endothelial cell growth medium (ECPM, Provitro, Berlin, Germany). Media used were supplemented with 10% (G-361, BJ, HUVEC) or 20% (CEM) fetal bovine serum, 2 mM L-glutamine, and 1% penicillin-streptomycin. The cell lines were maintained under standard cell culture conditions at 37 °C and 5% CO2 in a humid environment. Cells were sub-cultured two or three times a week using the standard trypsinization procedure for adherent cells.Cytotoxicity assay
The cytotoxicity of test and reference compounds (cisplatin and formestane) on the above-mentioned cancer and normal cells was determined after 72 h of incubation, using two cytotoxicity tests. The first, MTT test with tetrazolium salt,41 as described in detail by Ajduković et al.26 was performed on MCF-7, MDA-MB-231, PC-3, HeLa, HT-29, A549 and MRC-5 cell lines, with five different concentrations of the tested compounds ranging from 0.01 to 100 μM. The second cytotoxicity assay using resazurin42 (manufacturer's protocol – Sigma Aldrich, St. Louis, MO, USA), was performed as described earlier for protocol with Calcein AM dye44 on CEM, G-361 and BJ cell lines, with six concentrations of tested compounds ranging from 0.2 to 50 μM (dilution 3×). The data shown are the IC50 values calculated from the means of results obtained from at least two or three independent experiments performed in triplicate or quadruplicate.Angiogenesis in vitro
Migration scratch assay was performed with confluent HUVECs. They were scratched and immediately treated with a full medium containing different doses of the substance for 20 h. For tube formation assay, ibidi 15-well μ-slides (Ibidi, Munich, Germany) were coated with Matrigel (Corning, Tewksbury, MA, USA). 1 × 104 HUVECs were treated with compound for 20 h. At the end of both assays, images were captured using a microscope (IX51, Olympus, Japan). Images were analyzed using in-house software. The data were obtained from at least three independent experiments performed in triplicate.Cell-surface ELISA CD62E (E-Selectin, ELAM) and cytotoxicity
Enzyme-linked activity assay (ELISA) was used to detect the levels of cell adhesion molecule ELAM on HUVECs after 30 min of incubation with tested compounds and 4 h of stimulation with TNFα as described earlier.44 Curcumin was used as a positive control. Experiments were repeated three times in triplicate. Calcein AM (Molecular Probes, Invitrogen, Karlsruhe, Germany) cytotoxicity assay after 4 h of treatment in the HUVECs was used to measure the cytotoxicity of compounds for ELAM expression assay as described previously.45 Triplicates of at least three independent experiments were used.Apoptosis assay
The fluorescent double-staining method46,47 with acridine orange (AO) and ethidium bromide (EB) dyes were used to detect cells in which membrane integrity is disturbed as a consequence of the apoptotic process. Cultures of A549 lung adenocarcinoma cells were seeded in 6-well flat bottom tissue culture plates (Sarstedt), with 1 × 105 cells in 5 mL of supplemented DMEM cell culture medium per well. All seeded cultures were incubated initially for 24 h at 37 °C, absolute humidity and 5% CO2 conditions. Experimental cultures were exposed to IC50 concentrations of the investigated steroid derivatives over 72 h. The untreated culture was used as a control sample. After incubation, 100 μL of fluorescent mixture solution (15 mg of AO (Sigma-Aldrich) and 50 mg of EB (Serva) dissolved in the dark in 1 mL of absolute (96%) ethanol, with the addition of 49 mL of distilled H2O, frozen in aliquots) were added to each well and the cells were incubated again with dyes for 15 min. Medium with dyes was discarded and cells were then washed once in wells with 1 mL of phosphate-buffered saline (PBS, Sigma) and immediately examined and photographed (6-well plates were turned bottom-up) using a fluorescence microscope (Olympus BX51) at 10 × 15 and 40 × 15 magnification.Microphotographs with recorded fluorescent signals were analysed in the program ImageJ (NIH Image, http://imagej.nih.gov). Channels for the blue, red and green colour in each image were separated and their density was measured. The samples were compared by the numerical value of the red and green channel density ratio.
In vitro screening toward human cytochromes P450 (CYPs)
where, A – amplitude of the spectral change at [L]t ligand concentration; Amax – amplitude of the spectral change at saturation ligand concentration; [L]t – ligand concentration; [R]0 – protein concentration.
Author contributions
J. Ajduković synthesized the compounds, organized the collaboration and drafted the manuscript. D. Jakimov planned, performed and evaluated in vitro cytotoxicity experiments on MCF-7, MDA-MB-231, PC-3, HeLa, HT-29, A549 and MRC-5 cell lines, including double-fluorescence apoptosis assays. L. Rárová planned and carried out the in vitro experiments on CEM, G-361, BJ and HUVEC cells. Y. Dzichenka and S. Jovanović-Šanta performed in vitro and in silico experiments toward human CYPs. M. Sakač supervised synthesis of new compounds. M. Strnad and S. Usanov supervised biological experiments. D. Škorić performed the NMR experiments and spectral analysis. All authors read, corrected and approved the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support of the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grant No. 451-03-9/2021-14/200125) and the Czech Science Foundation of the Czech Republic (Grant GA19-01383S). The results of in vitro studies toward human cytochromes P450 were obtained in the frame of the Belarus–Serbia bilateral project (Grant No X20SRBG-004/337-00-00612/2019-09/04). We thank dr. Edward T. Petri for editing this manuscript for English grammar and usage, as well as Eva Hrdličková and Anežka Šindlerová for excellent technical assistance.References
- GLOBOCAN 2020. New Global Cancer Data, International Agency for Research on Cancer (IARC), Geneva, Switzerland, 2020, https://www.uicc.org/news/globocan-2020-new-global-cancer-data, accessed June 2021 Search PubMed.
- R. Bansal and P. C. Acharya, Chem. Rev., 2014, 114, 6986 CrossRef CAS PubMed.
- R. G. Fenton and D. L. Longo, in Harrison's Principles of Internal Medicine, ed. A. S. Fauci, E. Braunwald, K. J. Isselbachner, J. D. Wilson, J. B. Martin, D. L. Kasper, S. L. Hauser and D. L Longo, McGraw Hill, New York, 1998, pp. 505–511 Search PubMed.
- J. A. R. Salvador, J. F. S. Carvalho, M. A. C. Neves, S. M. Silvestre, A. J. Leitão, M. M. C. Silva and M. L. Sá e Melo, Nat. Prod. Rep., 2013, 30, 324 RSC.
- S. X. Lin, J. Chen, M. Mazumdar, D. Poirier, C. Wang, A. Azzi and M. Zhou, Nat. Rev. Endocrinol., 2010, 6, 485 CrossRef CAS PubMed.
- N. Sharifi, J. L. Gulley and W. L. Dahut, Endocr. Relat. Cancer, 2010, 17, R305 CAS.
- D. S. Jakimov, V. V. Kojić, L. D. Aleksić, G. M. Bogdanović, J. J. Ajduković, E. A. Djurendić, K. M. Penov Gaši, M. N. Sakač and S. S. Jovanović-Šanta, Bioorg. Med. Chem., 2015, 23, 7189 CrossRef CAS PubMed.
- T. Fotsis, Y. Zhang, M. S. Pepper, H. Adlercreutz, R. Montesano, P. P. Nawroth and L. Schweigerer, Nature, 1994, 368, 237 CrossRef CAS PubMed.
- T. L. Yue, X. Wang, C. S. Louden, S. Gupta, K. Pillarisetti, J. L. Gu, T. K. Hart, P. G. Lysko and G. Z. Feuerstein, Mol. Pharmacol., 1997, 51, 951 CrossRef CAS PubMed.
- T. M. LaVallee, X. H. Zhan, C. J. Herbstritt, E. C. Kough, S. J. Green and V. S. Pribluda, Cancer Res., 2002, 62, 3691 CAS.
- A. V. Yantsevich, Y. V. Dichenko, F. Mackenzie, D. V. Mukha, A. V. Baranovsky, A. A. Gilep, S. A. Usanov and N. V. Strushkevich, FEBS J., 2014, 281, 1700 CrossRef CAS PubMed.
- W. Tempel, I. Grabovec, F. MacKenzie, Y. V. Dichenko, S. A. Usanov, A. A. Gilep, H. W. Park and N. Strushkevich, J. Lipid Res., 2014, 55, 1925 CrossRef CAS PubMed.
- T. A. Sushko, A. A. Gilep, A. V. Yantsevich and S. A. Usanov, Biochem., 2013, 78, 282 CAS.
- M. Shimizu, K. Tsukamoto and T. Fujisawa, Tetrahedron Lett., 1997, 38, 5193 CrossRef CAS.
- J. A. Marco, M. Carda, J. Murga, F. González and E. Falomir, Tetrahedron Lett., 1997, 38, 1841 CrossRef CAS.
- K. C. Nicolaou, H. J. Mitchell, F. L. van Delft, F. Rübsam and R. M. Rodríguez, Angew. Chem., Int. Ed., 1998, 37, 1871 CrossRef CAS.
- I. H. Lone, K. Z. Khan, B. I. Fozdar and F. Hussain, Steroids, 2013, 78, 945 CrossRef CAS PubMed.
- S. A. Khan, A. M. Asiri and K. Saleem, J. Saudi Chem. Soc., 2012, 16, 7 CrossRef CAS.
- M. Abid, K. Husain and A. Azam, Bioorg. Med. Chem. Lett., 2005, 15, 4375 CrossRef CAS PubMed.
- D. P. Jindal, R. Chattopadhaya, S. Guleira and R. Gupta, Eur. J. Med. Chem., 2003, 38, 1025 CrossRef CAS PubMed.
- R. Bansal, S. Guleria, C. Ries and R. W. Hartmann, Arch. Pharm. Chem. Life Sci., 2010, 343, 377 CrossRef CAS PubMed.
- P. C. Acharya and R. Bansal, Arch. Pharm. Chem. Life Sci., 2014, 347, 193 CrossRef CAS PubMed.
- D. B. Garcia, R. G. Brown and J. N. Delgado, J. Pharm. Sci., 1980, 69, 995 CrossRef CAS PubMed.
- E. A. Djurendić, J. J. Ajduković, M. N. Sakač, J. J. Csanádi, V. V. Kojić, G. M. Bogdanović and K. M. Penov Gaši, Eur. J. Med. Chem., 2012, 54, 784 CrossRef PubMed.
- J. J. Ajduković, E. A. Djurendić, E. T. Petri, O. R. Klisurić, A. S. Ćelić, M. N. Sakač, D. S. Jakimov and K. M. Penov Gaši, Bioorg. Med. Chem., 2013, 21, 7257 CrossRef PubMed.
- J. J. Ajduković, K. M. Penov Gaši, D. S. Jakimov, O. R. Klisuric, S. S. Jovanović-Šanta, M. N. Sakač, L. D. Aleksić and E. A. Djurendić, Bioorg. Med. Chem., 2015, 23, 1557 CrossRef PubMed.
- M. P. Savić, J. J. Ajduković, J. J. Plavša, S. S. Bekić, A. S. Ćelić, O. R. Klisurić, D. S. Jakimov, E. T. Petri and E. A. Djurendić, Med. Chem. Comm., 2018, 9, 969 RSC.
- S. Samanta, A. K. Ghosh, S. Ghosh, A. A. Ilina, Y. A. Volkova, I. V. Zavarzin, A. M. Scherbakov, D. I. Salnikova, Y. U. Dzichenka, A. B. Sachenko, V. Z. Shirinian and A. Hajra, Org. Biomol. Chem., 2020, 18, 5571 RSC.
- U. Kiełczewska, J. W. Morzycki, L. Rárová and A. Wojtkielewicz, Org. Biomol. Chem., 2019, 17, 9050 RSC.
- A. Kiss, R. Jójárt, E. Mernyák, S. Bartha, R. Minorics, I. Zupkó and G. Schneider, Steroids, 2021, 176, 108911 CrossRef CAS PubMed.
- B. B. Shingate, B. G. Hazra, D. B. Salunke, V. S. Pore, F. Shirazi and M. V. Deshpande, Eur. J. Med. Chem., 2011, 46, 3681 CrossRef CAS PubMed.
- S. E. Barrie, G. A. Potter, P. M. Goddard, B. P. Haynes, M. Dowsett and M. Jarman, J. Steroid Biochem. Mol. Biol., 1994, 50, 267 CrossRef CAS PubMed.
- G. A. Potter, S. E. Barrie, M. Jarman and M. G. Rowlands, J. Med. Chem., 1995, 38, 2463 CrossRef CAS PubMed.
- M. Jarman, S. E. Barrie and J. M. Llera, J. Med. Chem., 1998, 41, 5375 CrossRef CAS PubMed.
- Y. Z. Ling, J. S. Li, Y. Liu, K. Kato, G. T. Klus and A. M. H. Brodie, J. Med. Chem., 1997, 40, 3297 CrossRef CAS PubMed.
- V. C. O. Njar, K. Kato, I. P. Nnane, D. M. Grigoryev, B. J. Long and A. M. H. Brodie, J. Med. Chem., 1998, 41, 902 CrossRef CAS PubMed.
- N. Zhu, Y. Ling, X. Lei, V. Handratta and A. M. H. Brodie, Steroids, 2003, 68, 603 CrossRef CAS PubMed.
- V. D. Handratta, T. S. Vasaitis, V. C. O. Njar, L. K. Gediya, R. Kataria, P. Chopra, D. Newman, R. Farquhar, Z. Guo, Y. Qiu and A. M. H. Brodie, J. Med. Chem., 2005, 48, 2972 CrossRef CAS PubMed.
- K. M. Penov Gaši, M. Dj. Djurendić Brenesel, E. A. Djurendić, M. N. Sakač, J. J. Canadi, J. J. Daljev, T. Armbruster, S. Andrić, D. M. Sladić, T. T. Božić, I. T. Novaković and Z. D. Juranić, Steroids, 2007, 72, 31 CrossRef PubMed.
- R. H. Mazur, J. Org. Chem., 1963, 28, 248 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55 CrossRef CAS PubMed.
- S. Al-Nasiry, N. Geusens, M. Hassens, C. Luyten and R. Pijnenborg, Hum. Reprod., 2006, 22, 1304 CrossRef PubMed.
- E. Djurendić, J. Daljev, M. Sakač, J. Canadi, S. Jovanović-Šanta, S. Andrić, O. Klisurić, V. Kojić, G. Bogdanović, M. Djurendić-Brenesel, S. Novaković and K. Penov-Gaši, Steroids, 2008, 73, 129 CrossRef PubMed.
- L. Rárová, J. Steigerová, M. Kvasnica, P. Bartůněk, K. Křížová, H. Chodounská, Z. Kolář, D. Sedlák, J. Oklestkova and M. Strnad, J. Steroid Biochem. Mol. Biol., 2016, 159, 154 CrossRef PubMed.
- H. C. Morrogh-Bernard, I. Foitová, Z. Yeen, P. Wilkin, R. de Martin, L. Rárová, K. Doležal, W. Nurcahyo and M. Olšanský, Sci. Rep., 2017, 7, 16653 CrossRef CAS PubMed.
- C. M. Henry, E. Hollville and S. J. Martin, Methods, 2013, 61, 90 CrossRef CAS PubMed.
- A. R. Nikolić, I. Z. Kuzminac, S. S. Jovanović-Šanta, D. S. Jakimov, L. D. Aleksić and M. N. Sakač, Steroids, 2018, 135, 101 CrossRef PubMed.
- T. A. Sushko, A. A. Gilep and S. A. Usanov, Biochem., 2012, 77, 585 CAS.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455 CAS.
- N. M. DeVore and E. E. Scott, Nature, 2012, 482, 116 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: 1H and 13C NMR spectra of newly synthesized compounds. See DOI: 10.1039/d1ra07613b |
| This journal is © The Royal Society of Chemistry 2021 |