 Open Access Article
Open Access ArticleEffect of DNA modifications on the transition between canonical and non-canonical DNA structures in CpG islands during senescence†
Saki Matsumotoa,
Hisae Tateishi-Karimata a,
Tatsuya Ohyamaa and
Naoki Sugimoto
a,
Tatsuya Ohyamaa and
Naoki Sugimoto *ab
*ab
aFrontier Institute for Biomolecular Engineering Research (FIBER), Konan University, 7-1-20 Minatojima-minamimachi, Kobe 650-0047, Japan. E-mail: sugimoto@konan-u.ac.jp
bGraduate School of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Kobe 650-0047, Japan
First published on 18th November 2021
Abstract
Patterns and levels of DNA modifications play important roles in senescence. Two major epigenetic modifications of DNA, 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), target CpG sites. Importantly, CpG concentrated regions, known as CpG islands, contain GC-rich sequences, which have the potential to fold into non-canonical DNA structures such as i-motifs and G-quadruplexes. In this study, we investigated the effect of 5mC and 5hmC modifications on the transition between a duplex, and i-motif and G-quadruplexes. To examine the transition, we firstly investigated the stability and structure of the i-motif and G-quadruplexes, considering the molecular environment in senescent cells. Analyses of their stability showed that the modifications did not drastically affect the stability. However, noteworthily, the modification can weaken the (de)stabilisation effect on G-quadruplexes caused by cosolute(s) and cations. Circular dichroism analyses indicated that the surrounding environments, including the molecular crowding and the type of cations such as K+ and Na+, regulate the topology of G-quadruplexes, while neither 5mC nor 5hmC had a drastic effect. On the other hand, the modifications changed the transition between duplexes and quadruplexes. Unmodified DNA preferred to fold into quadruplexes, whereas DNA with 5mC and 5hmC preferred to fold into duplexes in the absence of PEG200; on the other hand, DNA with or without modifications tended to fold into i-motifs under crowded conditions. Furthermore, an investigation of quadruplexes forming sequences in CpG islands, which are hyper- or hypomethylated during senescence, followed by gene ontology enrichment analysis for each gene group classified by the presence of quadruplexes, showed a difference in function between genes with and without quadruplexes in the CpG region. These results indicate that it is important to consider the effects of patterns and levels of DNA modifications on the transition between canonical and non-canonical DNA structures to understand gene regulation by epigenetic modification during senescence.
Introduction
DNA epigenetic modification is very important for precisely regulating gene expression via changes in the chromatin structure without changes in the DNA sequence. Such modifications play key roles in biological processes such as differentiation,1 development,2,3 X chromosome inactivation,4,5 and neurodegeneration.6 Failure of DNA epigenetic modifications has been reported to cause various diseases such as cancer7,8 and psychiatric disorders.9 DNA methylation at position 5 of cytosine (5-methylcytosine, 5mC) in CpG dinucleotides is the most studied epigenetic modification. Recently, it was found that changes in the DNA methylation level, including global hypomethylation and region-specific hypermethylation, depend on senescence and aging.10 It has also been reported that methylation levels increase in CpG islands in promoter regions, whereas they decrease in intergenic regions such as repeat regions during senescence.11 Furthermore, it has been found that the level of methylation correlates with the level of individual senescence, and it has hence been utilised as a biomarker for age prediction.12–15 In addition to 5mC, an alternative typical epigenetic modification is hydroxymethylation at position 5 of cytosine (5-hydroxymethylcytosine, 5hmC) in CpG sites.16,17 5hmC is generated via oxidation of 5mC by the ten-eleven translocation family of oxygenases (Fig. 1a). 5hmC was thought to be a mere intermediate of demethylation, but studies in this decade have shown that it plays a key role in epigenetic reprogramming during differentiation,18,19 development,20–22 aging,22–24 carcinogenesis,8,25–29 and the onset of neurodegenerative disorders.30–32CpG islands contains GC-rich sequences, which have the potential to form higher order structures such as an i-motif and G-quadruplexes with several topologies of parallel, hybrid, and antiparallel (Fig. 1b). In general, the modification of 5mC in CpG sites suppresses transcription,33,34 whereas that of 5hmC enhances transcription.35 Such modifications should be recognised by proteins to affect transcription. Hence, understanding the impact of these modifications on the transition between a duplex and quadruplexes such as an i-motif and G-quadruplexes in cellular environments, which are molecularly crowded, is important for elucidating the mechanisms that regulate gene transcriptional activity. The effects of modifications of 5mC on the stability and structure of a duplex, an i-motif, and G-quadruplexes have been investigated. Higher number of 5mC stabilise duplexes.36–38 Single or double 5mC stabilise i-motif, whereas three or more 5mC and single 5hmC destabilise i-motif.39,40 It has also been suggested that the position and not the number of 5mC modulates the stability of i-motif.41 5mC tends to stabilise the G-quadruplex.42,43 For example, quadruple 5mC modifications induce a topological change from a hybrid to parallel conformation in the presence of KCl and stabilise the G-quadruplex (Fig. 1b).43 On the other hand, it has been shown that a single 5hmC modification does not markedly affect the stability and topology of G-quadruplexes.44 Although the effects of 5mC and 5hmC modifications on the stability and structure of duplexes, i-motif, and G-quadruplexes have been partially investigated, the effects of such modifications on the transition between a duplex and quadruplexes are unknown.
Surrounding environments in senescent cells are another critical viewpoint for understanding the effects of modifications on the transition between the duplex and quadruplexes.45 Both molecular crowding conditions and cation conditions are major factors that affect the stability and structure of nucleic acids46–50 and further influence the transition between duplex and quadruplexes.51 Insight into molecular crowding conditions and cation conditions in senescent cells have recently emerged. The extent of molecular crowding in the cytoplasm of cells decreases during cellular senescence.52 The concentrations and compositions of cations in senescent cells are different from those in normal cells because the functions of ion channels are changed abnormally during senescence. In particular, the concentrations of potassium ions seem to be low in senescent cells.53 A few studies have been conducted to determine the effect of molecular crowding on the stability of duplex and i-motif with modifications.38–40,44 It has been observed that molecular crowding does not change the tendency of (de)stabilisation and the structure of a duplex and an i-motif with modifications.38–40 It has also been shown that molecular crowding of 40 vol% acetonitrile does not remarkably change the stability of i-motifs with 5hmC modifications.44 However, little is known about the effect of molecular crowding on the stability and structure of G-quadruplexes with modifications. Moreover, few studies have focused on the transition between a duplex, and i-motif and G-quadruplexes with modifications, considering cellular conditions during senescence, such as molecular crowding and cations.
In this study, we investigated the effect of 5mC and 5hmC modifications on the transition between a duplex and an i-motif and G-quadruplexes in CpG islands during senescence. We used the solution in the presence of NaCl without poly (ethylene glycol) with an average molecular weight of 200 (PEG200) as the model in senescent cells and the solution in the presence of KCl with PEG200 as a model in normal cells, and investigated the stability and conformations of i-motifs and G-quadruplexes with 5mC and 5hmC modifications. Analysis of the stabilities showed that the modifications on both the i-motif and G-quadruplexes did not markedly change their stability. However, the addition of PEG200 and the change of cations from Na+ to K+ significantly stabilised G-quadruplexes regardless of the modifications. Moreover, modifications can weaken the effects of (de)stabilization caused by PEG200 and cations. Circular dichroism (CD) analyses showed that neither 5mC nor 5hmC modifications affected the conformation of i-motifs but both induced topological changes in G-quadruplexes depending on the surrounding environments. However, the effects of the modifications were not observed in the presence of KCl or NaCl and regardless of the presence of PEG200. Interestingly, the modifications regulated the transition between a duplex, and i-motif and G-quadruplexes. In the presence of K+, DNA with 5hmC preferred to fold into duplexes in the absence of PEG200, unlike DNA without modification or with 5mC. The addition of PEG200 suppressed duplex formation; however, DNA with 5hmC still preferred to fold into a duplex. UV melting analysis suggested that this is because the lower thermal stability of the i-motif with 5hmC than that with the i-motif without modifications and i-motif with 5mC, may promote the formation of a duplex with 5hmC. In contrast, DNA with 5mC and 5hmC preferred to fold into duplexes, unlike DNA without modifications in the presence of Na+, which remained unchanged under molecular crowding conditions. Our results indicate that the duplex is more favourable in senescent cells, in which the extent of molecular crowding and the concentration of K+ decrease. To investigate how the presence of quadruplexes affects gene expression during senescence, we classified genes with hyper- or hypomethylated CpG islands in senescent cells according to the presence of quadruplexes in the CpG island. A difference in function was observed in the presence of quadruplexes in the CpG region. Our results indicate the possibility that epigenetic gene expression may be regulated by changes in transition between duplex and quadruplexes of an i-motif and G-quadruplexes depending on the surrounding environments during senescence.
Experimental
Materials
DNA oligonucleotides of high-performance liquid chromatography grade were purchased from Fasmac Co., Ltd. (Japan). Concentrations of single-stranded DNA oligonucleotides were determined using the absorbance at 260 nm and 95 °C and single-stranded extinction coefficients. Absorbance at 260 nm was measured using a Shimadzu 1800 spectrophotometer (Shimadzu, Japan) connected to a temperature controller. PEG200 (Wako Pure Chemical Industries, Japan) was used as a molecular crowding reagent without further purification.Thermodynamic analysis
UV absorbance was measured using a Shimadzu 1800 spectrophotometer (Shimadzu, Japan) equipped with a temperature controller. The UV melting curves of duplex were measured at 260 nm, those of i-motif were measured at 260 nm or 295 nm, and those of G-quadruplexes were measured at 295 nm; both i-motif and G-quadruplexes exhibited a hypochromic transition. All the experiments were conducted in 50 mM MES–LiOH (pH 6.0) and 100 mM KCl or NaCl with 0 or 40 wt% PEG200. The heating rates were 0.5 °C min−1 from 0 °C to 95 °C. To calculate thermodynamic parameters, including the melting temperature (Tm), enthalpy change (ΔH°), entropy change (ΔS°), and free energy at 25 °C for intramolecular i-motif and G-quadruplex formation, the melting curves that were obtained at least three times were fit to the theoretical equation for an intramolecular association.54 Before the measurement, the sample was heated to 95 °C, and then cooled at a rate of 1.0 °C min−1. Intramolecular i-motif and G-quadruplex formation were confirmed by measuring UV melting at oligonucleotide concentrations of 2, 5, 10, and 20 μM.
for intramolecular i-motif and G-quadruplex formation, the melting curves that were obtained at least three times were fit to the theoretical equation for an intramolecular association.54 Before the measurement, the sample was heated to 95 °C, and then cooled at a rate of 1.0 °C min−1. Intramolecular i-motif and G-quadruplex formation were confirmed by measuring UV melting at oligonucleotide concentrations of 2, 5, 10, and 20 μM.
CD measurements
CD spectra were recorded on a JASCO J-1500 spectropolarimeter (JASCO, Japan) equipped with a temperature controller, using a total DNA concentration of 5 μM or 20 μM in 50 mM MES–LiOH (pH 6.0) and 100 mM KCl or NaCl with or without 40 wt% PEG200. The spectra were obtained at 4 °C by capturing at least three scans from 200 to 350 nm for a cuvette with a path length of 0.1 cm. The cuvette-holding chamber was flushed with a constant stream of dry N2 gas to avoid the condensation of water on the cuvette exterior. Before the measurement, the sample was heated to 95 °C, and then cooled at a rate of 1.0 °C min−1.Native gel electrophoresis
Native polyacrylamide gel electrophoresis (PAGE) was conducted on non-denaturing gels containing 10 or 15 wt% polyacrylamide gels in 1× TBE buffer with 30 mM KCl or NaCl. DNA samples (2 μL of 2.5 μM) in 50 mM MES–LiOH (pH 6.0) and 100 mM KCl or 100 mM NaCl containing 0 or 40 wt% PEG200 were loaded and analysed by electrophoresis. The gels were stained with SYBR® Gold (Thermo Fisher Scientific) and imaged using a fluorescent imager (FLA-5100, FUJIFILM). Before the measurement, the samples were heated to 80 °C, and then cooled at a rate of 1.0 °C min−1. The gels were run at 100 V and at 25 °C for 140 min.Identification of i-motif and G-quadruplexes forming sequences
The target sequences of the human genome (GRCh38/hg38)55 as target sequences were obtained from the National Center for Biotechnology Information (NCBI) database. The gene lists for hyper- and hypomethylated genes were generated based on the Δβ value; the β values (intensity of the methylation allele/[intensity of the unmethylated allele + intensity of the methylated allele + 100]) of control cells were subtracted from those of senescent cells.56 Genes with Δβ > 0.2 and Δβ < −0.2 were regarded as hypermethylation and hypomethylation genes, respectively. The start and end positions of these sequences were obtained from the NCBI database, and the target sequences were extracted. To identify CpG islands, we adopted distance-length-based methods using the definition of Gardiner-Garden et al.:57 (1) the sequence length is 200 bases or more; (2) the sequence contains more than 50% of G or C bases; (3) the ratio robserved/expected of the sequence expressed by the following formula is 0.6 or more.where NCpG, Nall, NC, and NG represent the numbers of CpG sites, bases, cytosines, and guanines, respectively. To identify the CpG islands efficiently, the start position of CpG in the target sequences was determined. The sequence lengths were calculated using start position pairs, and pairs having less than 200 bases were removed. The sequences were extracted from the start position pairs, and the fractions of G or C bases and robserved/expected were evaluated. When a sequence contained another sequence or when the same region overlapped in multiple sequences, the sequence with a larger number of G or C bases was adopted. The start and end positions of CpG sites were determined using the regular expression of the CpG pattern. Based on both the positions, CpG sequences were extracted from the GRCh38/hg38 sequence. G-quadruplex forming sequences were determined from the obtained CpG sequences using regular expression. The patterns of (G3–4N1–7G3–4N1–7G3–4N1–7G3–4) were adopted for G-quadruplex forming sequences.
Gene ontology (GO) enrichment analysis
The GO clustering system was utilised to analyse the functions of genes, which were categorised based on whether their CpG islands had quadruplexes. The Database for Annotation, Visualisation and Integrated Discovery (DAVID) v6.8 (https://david.ncifcrf.gov/home.jsp)58,59 was used for GO enrichment analysis. The lists of genes with CpG islands with or without quadruplexes were uploaded, and biological significance was evaluated using the GO functional annotation clustering tool with GOTERM_BP_FAT on the website. Gene enrichment in GO terms from DAVID was performed using Fisher's exact test. The enrichment score was calculated as the geometric mean in the log scale of each p-value for a corresponding GO term cluster. A high score for a gene group indicated that the genes in the group were involved in more enriched GO terms. Because an enrichment score of more than 1.3 corresponds to p-values of less than 0.05, only the enriched GO terms with an enrichment score of more than 1.3 were evaluated.Results and discussion
Effects of 5mC and 5hmC on the stability of DNA i-motifs and G-quadruplexes
To examine the effects of modifications on the transition between the duplexes and quadruplexes, we firstly investigated the stability and structures of i-motifs and G-quadruplexes. We focused on ELOVL2 gene, which has been associated with senescence.14,15 We searched for quadruplex forming sequences in the sense and antisense strands of the CpG island of ELOVL2. The motifs adopted for identification were G3,4N1–7G3,4N1–7G3,4N1–7G3,4 (N refers to any base) for G-quadruplex forming sequences and C3,4N1–7C3,4N1–7C3,4N1–7C3,4 (N refers to any base) for i-motif forming sequences.60 Thus, ten i-motifs and ten G-quadruplexes in both the sense and antisense strands were found (Table S1†). We selected No. 7 in Table S1† as a model sequence to investigate the stability and structure of quadruplexes with modifications, considering the number of CpG sites within the quadruplex forming sequence and its analysability of thermodynamics (Table S1†) because the melting temperatures (Tms) of both the i-motif in one strand and G-quadruplex in the complementary strand are within 40–60 °C, to obtain the exact Tms. Although the position of modification on i-motif forming sequence can contribute the effect of stabilisation of i-motif, we focus on the C10 and C16 in this study because the methylation occurs at cytosine of CpG dinucleotide in mammal. Thus, for i-motif forming sequences, no modification (iM), single methylation at the C10 position (iM10m), double methylation at the C10 and C16 positions (iM10,16m), single hydroxymethylation at the C10 position (iM10hm), and double hydroxymethylation at C10 and C16 positions (iM10,16hm) were introduced (Table 1). For G-quadruplex forming sequences, which are the complementary sequences of iM, no modification (Gq), single methylation at C5 or C11 position (Gq5m and Gq11m), double methylation at C5 and C11 positions (Gq5,11m), single hydroxymethylation at C5 or C11 positions (Gq5hm and Gq11hm), and double hydroxymethylation at C5 and C11 positions (Gq5,11hm) were introduced (Table 1). All the sequences and abbreviations are listed in Table 1. PEG200 was adopted a molecular crowding reagent because it is commonly used and has a correlation with the cellular environments.46,49,61| Abbreviation | Sequence (5′ to 3′) |
|---|---|
| a mC denotes the 5-methylated cytosine, and hmC denotes the 5-hydroxymethylated cytosine of each oligonucleotide sequence. | |
| iM | CCCTCTCCCCGGTCCCGCCCC |
| iM10m | CCCTCTCCCmCGGTCCCGCCCC |
| iM10,16m | CCCTCTCCCmCGGTCCmCGCCCC |
| iM10hm | CCCTCTCCChmCGGTCCCGCCCC |
| iM10,16hm | CCCTCTCCChmCGGTCChmCGCCCC |
| Gq | GGGGCGGGACCGGGGAGAGGG |
| Gq5m | GGGGmCGGGACCGGGGAGAGGG |
| Gq11m | GGGGCGGGACmCGGGGAGAGGG |
| Gq5,11m | GGGGmCGGGACmCGGGGAGAGGG |
| Gq5hm | GGGGhmCGGGACCGGGGAGAGGG |
| Gq11hm | GGGGCGGGAChmCGGGGAGAGGG |
| Gq5,11hm | GGGGhmCGGGAChmCGGGGAGAGGG |
To investigate the effects of modifications on the stability of the i-motif and G-quadruplexes, UV melting analysis of the designed sequences was performed in 50 mM MES–LiOH (pH 6.0) and 100 mM KCl or NaCl with or without 40 wt% PEG200 (Fig. S1 and S2†). We selected the pH 6.0 to systematically examine the stability of both i-motif and G-quadruplex in the same conditions. The thermodynamic parameters (ΔH°, TΔS°, and  ) of the formation of the i-motif and G-quadruplexes as well as the Tms were calculated (Tables S2–S5†). The effects of modifications of methylation and hydroxymethylation on the
) of the formation of the i-motif and G-quadruplexes as well as the Tms were calculated (Tables S2–S5†). The effects of modifications of methylation and hydroxymethylation on the  values for the formation of i-motif and G-quadruplexes were evaluated as
values for the formation of i-motif and G-quadruplexes were evaluated as  , which is
, which is  . Regarding the i-motif, the modifications slightly changed the
. Regarding the i-motif, the modifications slightly changed the 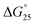 value of i-motifs and the
value of i-motifs and the  values were within −0.2 to +0.3 kcal mol−1 in the presence of K+ without PEG200 (Fig. S3a and Table S2†). The
values were within −0.2 to +0.3 kcal mol−1 in the presence of K+ without PEG200 (Fig. S3a and Table S2†). The  value for iM10m was +0.3 kcal mol−1, whereas the
value for iM10m was +0.3 kcal mol−1, whereas the  values for iM10,16m was −0.1 kcal mol−1. Hydroxymethylation also slightly changed the
values for iM10,16m was −0.1 kcal mol−1. Hydroxymethylation also slightly changed the  values and the
values and the  values were −0.2 and +0.2 kcal mol−1 for iM10hm and iM10,16hm, respectively. In the presence of K+ with 40 wt% PEG200, methylation did not change the stability regardless of the number of modifications, and the
values were −0.2 and +0.2 kcal mol−1 for iM10hm and iM10,16hm, respectively. In the presence of K+ with 40 wt% PEG200, methylation did not change the stability regardless of the number of modifications, and the  values were within −0.1 to 0.0 kcal mol−1 (Fig. S3b and Table S2†). The position of the methyl group did not inhibit the formation of hydrogen bonds in the C–C base pairs of the i-motif. Moreover, in general, C bases in the i-motif are not stacked well. Thus, methyl groups and neighbouring C bases do not interact with each other in CH–π interactions. Therefore, methylation in the i-motif does not significantly affect the stability of the i-motifs. In contrast, hydroxymethylation induced slight destabilisation. The effects of modifications of hydroxymethylation on the
values were within −0.1 to 0.0 kcal mol−1 (Fig. S3b and Table S2†). The position of the methyl group did not inhibit the formation of hydrogen bonds in the C–C base pairs of the i-motif. Moreover, in general, C bases in the i-motif are not stacked well. Thus, methyl groups and neighbouring C bases do not interact with each other in CH–π interactions. Therefore, methylation in the i-motif does not significantly affect the stability of the i-motifs. In contrast, hydroxymethylation induced slight destabilisation. The effects of modifications of hydroxymethylation on the 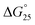 values for the formation of the i-motif were also evaluated as
values for the formation of the i-motif were also evaluated as 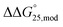 . The
. The  values for iM10hm and iM10,16hm were +0.3 and +0.2 kcal mol−1, respectively (Fig. S3b and Table S2†). This result is in close agreement with the results of previous single or double 5hmC modifications.39,40 The destabilisation caused by hmC might have been due to the significantly lower nucleobase pKa than those of cytosine and mC, which showed that protonation of N3 in hmC is less favourable.36,39 In the presence of Na+ without PEG200, modifications changed the
values for iM10hm and iM10,16hm were +0.3 and +0.2 kcal mol−1, respectively (Fig. S3b and Table S2†). This result is in close agreement with the results of previous single or double 5hmC modifications.39,40 The destabilisation caused by hmC might have been due to the significantly lower nucleobase pKa than those of cytosine and mC, which showed that protonation of N3 in hmC is less favourable.36,39 In the presence of Na+ without PEG200, modifications changed the  values of i-motifs slightly by −0.2 to +0.3 kcal mol−1 (Fig. S3c and Table S3†). The
values of i-motifs slightly by −0.2 to +0.3 kcal mol−1 (Fig. S3c and Table S3†). The  values were +0.3 kcal mol−1 for iM10m and −0.1 kcal mol−1 for iM10,16m. Single hmC of iM10hm was slightly stabilised and the
values were +0.3 kcal mol−1 for iM10m and −0.1 kcal mol−1 for iM10,16m. Single hmC of iM10hm was slightly stabilised and the value was −0.2 kcal mol−1, whereas double hmC of iM10,16hm did not change the stability, in which the
value was −0.2 kcal mol−1, whereas double hmC of iM10,16hm did not change the stability, in which the  value was +0.1 kcal mol−1. This result suggested that the type and number of modifications did not change the stability of the i-motifs systematically. In the presence of Na+ and PEG200, both methylation and hydroxymethylation slightly destabilised i-motif. The
value was +0.1 kcal mol−1. This result suggested that the type and number of modifications did not change the stability of the i-motifs systematically. In the presence of Na+ and PEG200, both methylation and hydroxymethylation slightly destabilised i-motif. The  values for M10m and M10,16m were 0.0 and +0.1 kcal mol−1 and those for both M10hm and M10,16hm were +0.2 kcal mol−1 (Fig. S3d and Table S3†). As the stability of i-motif depends on the pH, we also investigated the stability of i-motif at pH 5.0 and pH 7.0. UV meltings of 20 μM of iM, iM10m, and iM10hm in the buffer of MES–LiOH (pH 5.0) in 100 mM KCl or 100 mM NaCl showed that all the sequences showed drastic stabilisation compared with that at pH 6.0. Tm values were obtained by differentiation and the result showed that the methylation did not change the Tm of i-motif as much as at pH 6.0 while hydroxymethylation decreased the Tm of i-motif in both 100 mM KCl and 100 mM NaCl (Fig. S4a and b†). To analyse the thermodynamic parameters, we also performed UV melting analyses using 10, 5, and 2 μM of iM, iM10m, and iM10hm at pH 5.0 (Fig. S4c–h†). However, the melting curves showed two peaks, which were not observed at pH 6.0. This result indicates that coexistence of intra- and intermolecular i-motif formation and we could not obtain the thermodynamic parameters of i-motifs at pH 5.0. Unfortunately, the UV melting curves of i-motifs at pH 7.0 could not be observed because of low melting temperature (Fig. S5†).
values for M10m and M10,16m were 0.0 and +0.1 kcal mol−1 and those for both M10hm and M10,16hm were +0.2 kcal mol−1 (Fig. S3d and Table S3†). As the stability of i-motif depends on the pH, we also investigated the stability of i-motif at pH 5.0 and pH 7.0. UV meltings of 20 μM of iM, iM10m, and iM10hm in the buffer of MES–LiOH (pH 5.0) in 100 mM KCl or 100 mM NaCl showed that all the sequences showed drastic stabilisation compared with that at pH 6.0. Tm values were obtained by differentiation and the result showed that the methylation did not change the Tm of i-motif as much as at pH 6.0 while hydroxymethylation decreased the Tm of i-motif in both 100 mM KCl and 100 mM NaCl (Fig. S4a and b†). To analyse the thermodynamic parameters, we also performed UV melting analyses using 10, 5, and 2 μM of iM, iM10m, and iM10hm at pH 5.0 (Fig. S4c–h†). However, the melting curves showed two peaks, which were not observed at pH 6.0. This result indicates that coexistence of intra- and intermolecular i-motif formation and we could not obtain the thermodynamic parameters of i-motifs at pH 5.0. Unfortunately, the UV melting curves of i-motifs at pH 7.0 could not be observed because of low melting temperature (Fig. S5†).
In the i-motif forming sequences used in this study, the modification is likely to be located at the 3′ end of a continuous cytosine or in the loop of i-motif. Within the i-motif forming sequence, there are several cytosines and the effect of modification on the stability of i-motif may be different depending on the position of modification. Especially, modification at cytosine in the stem region of i-motif may perturb stability of the base pairing. Although we selected the cytosine of CpG dinucleotide within i-motif forming sequence as we focus on the senescence of human cells, systematic studies on cytosines other than CpG dinucleotide site are desirable in the near future because modification at any cytosine occurs in other organisms.
To examine the effects of solution conditions, such as molecular crowding and cations, the 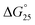 values were evaluated as follows:
values were evaluated as follows:  and
and 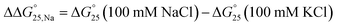 (Fig. S6 and S7†). The
(Fig. S6 and S7†). The  values were small and within ±0.4 kcal mol−1 in the presence of K+ or Na+ (Fig. S6†). The tendency of (de)stabilisation by PEG200 was similar regardless of the type of cation. This result indicated that PEG200 did not drastically affect the stability of the i-motifs. The contributions of cations to the stability of the i-motifs cannot be observed. The
values were small and within ±0.4 kcal mol−1 in the presence of K+ or Na+ (Fig. S6†). The tendency of (de)stabilisation by PEG200 was similar regardless of the type of cation. This result indicated that PEG200 did not drastically affect the stability of the i-motifs. The contributions of cations to the stability of the i-motifs cannot be observed. The  values were quite small and within −0.2 to +0.1 kcal mol−1 (Fig. S7†). This result may be owing to the lack of conformational changes in the i-motifs due to changes in solution conditions.
values were quite small and within −0.2 to +0.1 kcal mol−1 (Fig. S7†). This result may be owing to the lack of conformational changes in the i-motifs due to changes in solution conditions.
As for G-quadruplexes, any pattern of modification, including single and double 5mC and 5hmC, tended to destabilise the G-quadruplexes to varying degrees in the presence of K+ without PEG200 (Fig. S8a and Table S4†). The  values for Gq5m, Gq11m, and Gq5,11m were +1.1, +0.2, and +1.4 kcal mol−1, respectively. Upon hydroxymethylation, the stability of only Gq5hm remained unchanged and the
values for Gq5m, Gq11m, and Gq5,11m were +1.1, +0.2, and +1.4 kcal mol−1, respectively. Upon hydroxymethylation, the stability of only Gq5hm remained unchanged and the  value was −0.1 kcal mol−1, whereas Gq11hm and Gq5,11hm were destabilised and the
value was −0.1 kcal mol−1, whereas Gq11hm and Gq5,11hm were destabilised and the  values were +1.7 and +0.8 kcal mol−1, respectively. Unfortunately, the degree of destabilisation could not be explained by the type, position, or number of modifications, because the detailed structures of G-quadruplexes are still unknown. It is considered that the destabilisation caused by modification might be due to conformational changes in the loop region of G-quadruplexes. The addition of 40 wt% PEG200 resulted in unclear melting curves because the Tm values were higher than the measurement range and no thermodynamic parameters could be calculated. In the presence of Na+ without PEG200, significant changes in stability were not observed for any of the G-quadruplexes, compared with those in the presence of 100 mM KCl (Fig. S8b†). The
values were +1.7 and +0.8 kcal mol−1, respectively. Unfortunately, the degree of destabilisation could not be explained by the type, position, or number of modifications, because the detailed structures of G-quadruplexes are still unknown. It is considered that the destabilisation caused by modification might be due to conformational changes in the loop region of G-quadruplexes. The addition of 40 wt% PEG200 resulted in unclear melting curves because the Tm values were higher than the measurement range and no thermodynamic parameters could be calculated. In the presence of Na+ without PEG200, significant changes in stability were not observed for any of the G-quadruplexes, compared with those in the presence of 100 mM KCl (Fig. S8b†). The  values were within −0.2 to +0.1 kcal mol−1. The addition of 40 wt% PEG200 slightly destabilised G-quadruplexes with 5mC modifications and the
values were within −0.2 to +0.1 kcal mol−1. The addition of 40 wt% PEG200 slightly destabilised G-quadruplexes with 5mC modifications and the 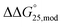 values for Gq5m, Gq11m, and Gq5,11m were +0.3, +0.5, and +0.9 kcal mol−1, respectively. In contrast, the contribution of 5hmC to stability varied depending on the position and number of modifications. Gq5hm and Gq5,11hm were destabilised and the
values for Gq5m, Gq11m, and Gq5,11m were +0.3, +0.5, and +0.9 kcal mol−1, respectively. In contrast, the contribution of 5hmC to stability varied depending on the position and number of modifications. Gq5hm and Gq5,11hm were destabilised and the  values for both Gq5hm and Gq5,11hm were +0.8 kcal mol−1, whereas the stability of Gq11m was increased slightly and the
values for both Gq5hm and Gq5,11hm were +0.8 kcal mol−1, whereas the stability of Gq11m was increased slightly and the 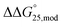 values for Gq11m was −0.2 kcal mol−1 (Fig. S8c and Table S5†). In short, modifications of both 5mC and 5hmC slightly affected the stability of G-quadruplexes in the presence of K+, but not in the presence of Na+.
values for Gq11m was −0.2 kcal mol−1 (Fig. S8c and Table S5†). In short, modifications of both 5mC and 5hmC slightly affected the stability of G-quadruplexes in the presence of K+, but not in the presence of Na+.
To examine the effect of PEG200 and cations on the stability of G-quadruplexes with modifications, both the  and
and  values were calculated (Fig. 2). Evaluation of
values were calculated (Fig. 2). Evaluation of  values in the presence of Na+ showed that the addition of PEG200 significantly stabilised G-quadruplexes (Fig. 2a). When changes in enthalpy (
values in the presence of Na+ showed that the addition of PEG200 significantly stabilised G-quadruplexes (Fig. 2a). When changes in enthalpy (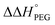 , which is ΔH° [40 wt% PEG200] − ΔH° [0 wt% PEG200]) and entropy (Δ(TΔS°)PEG, which is TΔS° [40 wt% PEG200] − TΔS° [0 wt% PEG200]) upon the addition of PEG200 were evaluated, it was clear that the stabilisation by PEG200 was because of the favourable enthalpic contributions that exceeded the unfavourable entropic contributions (Table S5†). This result is in accordance with previous reports on stabilisation by molecular crowding on a thrombin DNA aptamer, which can fold into an antiparallel G-quadruplex structure, because of the favourable enthalpic contribution.46,49,62 Noteworthily, the extent of stabilisation was different depending on the modifications. Except for Gq11hm, the effect of stabilisation by PEG200 was weakened by the modifications. One possible explanation for this result is that methylation or hydroxymethylation changes the degree of hydration during folding of G-quadruplexes. Moreover, in the evaluation of the
, which is ΔH° [40 wt% PEG200] − ΔH° [0 wt% PEG200]) and entropy (Δ(TΔS°)PEG, which is TΔS° [40 wt% PEG200] − TΔS° [0 wt% PEG200]) upon the addition of PEG200 were evaluated, it was clear that the stabilisation by PEG200 was because of the favourable enthalpic contributions that exceeded the unfavourable entropic contributions (Table S5†). This result is in accordance with previous reports on stabilisation by molecular crowding on a thrombin DNA aptamer, which can fold into an antiparallel G-quadruplex structure, because of the favourable enthalpic contribution.46,49,62 Noteworthily, the extent of stabilisation was different depending on the modifications. Except for Gq11hm, the effect of stabilisation by PEG200 was weakened by the modifications. One possible explanation for this result is that methylation or hydroxymethylation changes the degree of hydration during folding of G-quadruplexes. Moreover, in the evaluation of the  values, significant destabilisation was observed for all G-quadruplexes in the presence of 100 mM NaCl, compared with that in the presence of 100 mM KCl. A comparison of
values, significant destabilisation was observed for all G-quadruplexes in the presence of 100 mM NaCl, compared with that in the presence of 100 mM KCl. A comparison of  (ΔH° (100 mM NaCl) − ΔH° (100 mM KCl)) and Δ(TΔS°)Na (TΔS° (100 mM NaCl) − TΔS° (100 mM KCl)) revealed that this destabilisation was caused by unfavourable enthalpic contributions (Tables S4 and S5†). This preference of G-quadruplexes for KCl has been investigated previously.63,64 The effect of destabilisation by the change of cation from K+ to Na+ was also weakened by the modifications, except for Gq5hm (Fig. 2b). The stability changes of G-quadruplexes owing to coexisting cations depends on the affinity between the cation and the G-quartet. As the stability change owing to coexisting cations was buffered in the presence of PEG200, PEG200 may alter the interactions of the cation with the DNA strand other than the G-quartet. It has been reported that a single strand of DNA changes to a compact structure in the presence of PEG, which reduces the uptake of cations during the formation of the DNA structure.65,66 The addition of a methyl group may significantly alter the single-stranded state of the DNA and affect its binding to the cation.
(ΔH° (100 mM NaCl) − ΔH° (100 mM KCl)) and Δ(TΔS°)Na (TΔS° (100 mM NaCl) − TΔS° (100 mM KCl)) revealed that this destabilisation was caused by unfavourable enthalpic contributions (Tables S4 and S5†). This preference of G-quadruplexes for KCl has been investigated previously.63,64 The effect of destabilisation by the change of cation from K+ to Na+ was also weakened by the modifications, except for Gq5hm (Fig. 2b). The stability changes of G-quadruplexes owing to coexisting cations depends on the affinity between the cation and the G-quartet. As the stability change owing to coexisting cations was buffered in the presence of PEG200, PEG200 may alter the interactions of the cation with the DNA strand other than the G-quartet. It has been reported that a single strand of DNA changes to a compact structure in the presence of PEG, which reduces the uptake of cations during the formation of the DNA structure.65,66 The addition of a methyl group may significantly alter the single-stranded state of the DNA and affect its binding to the cation.
 | ||
Fig. 2 The comparison of effects of molecular crowding and cations. (a) The  in 100 mM NaCl solution and (b) The in 100 mM NaCl solution and (b) The  values in 0 wt% PEG200. values in 0 wt% PEG200. | ||
Our results revealed that i-motifs were not remarkably stabilised or destabilised, whereas G-quadruplexes were slightly (de)stabilised. The addition of 40 wt% PEG200 stabilised the G-quadruplexes, as previously reported. Furthermore, G-quadruplexes preferred KCl solution to NaCl solution, regardless of the presence of modifications of methylation and hydroxymethylation. Noteworthily, the effects of these stabilisations or destabilisations caused by PEG200 and cations were weakened by modifications, with a few exceptions. This suggests that structural changes caused by environmental changes in cells, such as senescent cells, may be suppressed by this modification. Moreover, these effects of mitigating stabilisation may change the transition between the duplex and quadruplexes.
Effects of 5mC and 5hmC on the structures of DNA i-motif and G-quadruplexes
Next, we investigated the structures of i-motif and G-quadruplexes themselves with modifications using CD spectra analyses. All the CD spectra of i-motif with modifications displayed positive peaks centred at approximately 290 nm and negative peaks centred at approximately 265 nm (Fig. S9 and S10†), indicating that all the designed C-rich sequences formed i-motif regardless of the presence of KCl and NaCl, and in the presence of PEG200, showing that both modifications and the surrounding environments did not induce any structural changes in i-motif with modifications.For G-quadruplexes with modifications, CD spectra in the presence of 100 mM KCl showed positive peaks at 265 nm and negative peaks at 240 nm, indicating the formation of G-quadruplex with parallel topology (Fig. S11†). The parallel topology remained unchanged upon the addition of 40 wt% PEG200; however, the intensities of Gq, Gq5,11m, Gq5hm, and Gq11hm significantly decreased, indicating aggregation. The results of native PAGE supported the finding that Gq, Gq5,11m, Gq5hm, and Gq11hm aggregated in the presence of K+ (Fig. S12†). A single band was observed for G-quadruplexes with modifications in the absence of PEG200. On the other hand, this band could not be observed after the addition of 40 wt% PEG200, suggesting the formation of a highly ordered structure from the G-rich sequence. Next, we investigated the effect of cations on the structure of G-quadruplexes with modifications, because the concentration of K+ decreased by changes in the expression levels of channel proteins during senescence.53 In the presence of 100 mM NaCl instead of 100 mM KCl, the CD spectra of all the sequences showed positive peaks at 265 nm and negative peaks at 240 nm, indicating the formation of G-quadruplex with a parallel topology (Fig. S13†). Interestingly, the addition of 40 wt% PEG200 changed the topology of G-quadruplexes with modifications from parallel to hybrid, for which the CD spectra showed positive peaks at 265 nm and 295 nm. The significant stabilisation by PEG200 in the presence of Na+ may be caused by the topological changes, and the difference in the extent of stabilisation by modifications may be due to the different proportions of the constituents of G-quadruplexes with different topologies. To obtain further insight into the topological changes caused by PEG200, we examined the CD spectra for 0–40 wt% PEG200 (Fig. S14†). Varying the concentration of PEG200 from 0 to 40 wt% did not induce systematic changes in the CD spectra, such as the observation of the isosbestic point, indicating that the topological change was not a two state transition. This result was caused by the existence of G-quadruplexes with several topologies in the presence of NaCl, regardless of the presence of PEG200. The mixture of G-quadruplexes with several topologies could not be identified using CD spectra; the elucidation of exact conformations requires structural analysis using NMR spectroscopy or X-ray crystallography in the future. The results of native PAGE indicated that G-quadruplexes with modifications folded into monomeric structures in 100 mM NaCl regardless of the presence of PEG200 (Fig. S15†).
In summary, our results clearly showed that the surrounding environments were crucial to both the stability and topology of G-quadruplexes, but not to those of i-motif. Moreover, modifications can weaken the effects of (de)stabilisation of G-quadruplexes caused by PEG200 and cations, although this mitigating effect unfortunately could not be explained by the conformational changes caused by PEG200 and cations. This mitigating effect may contribute to the transition between a duplex and quadruplexes during senescence.
Effects of 5mC and 5hmC on the transition between duplex and quadruplexes such as i-motif and G-quadruplexes
Since the effects of modifications on the thermal stability and structure of i-motifs and G-quadruplexes had been revealed, we next investigated how these characteristics regulate the transition from a duplex to quadruplexes such as an i-motif and G-quadruplexes. We firstly examined the structures of a mixture of iM and Gq with no modification, a mixture of iM10,16m and Gq5,11m with double 5mC, and a mixture of iM10,16hm and Gq5,11hm with double 5hmC using native PAGE in the presence of K+ (Fig. 3). Native PAGE in 100 mM KCl showed that mixtures of iM and Gq and iM10,16m and Gq5,11m produced two bands derived from an i-motif and a duplex (Fig. 3a and b). The ratio of duplex and i-motif was calculated from the intensity of the bands in Fig. 3 as follows: the intensity of the duplex or i-motif was divided by the sum of the intensity of all the band in the lane. The ratio of duplex was 0.66 for a mixture of iM and Gq and 0.72 for that of iM10,16m and Gq5,11m. And the ratio of i-motif was 0.36 for a mixture of iM and Gq and 0.29 for that of iM10,16m and Gq5,11m. In contrast, a mixture of iM10,16hm and Gq5,11hm showed a single band of the duplex (Fig. 3c) and the ratio of duplex was 0.68.The addition of 40 wt% PEG200 changed the composition of the duplexes and quadruplexes; both the mixture of iM and Gq and that of iM10,16m and Gq5,11m mostly folded into an i-motif rather than a duplex. There are two reasons that PEG200 promotes the formation of i-motif. One is the stabilization of i-motif by PEG200. Another is the aggregation of G-quadruplex in the presence of KCl and PEG200. In previous section, the decrease of the intensity of CD spectra of G-quadruplex forming sequence in the presence of K+ and PEG200 was observed though to varying degrees (Fig. S11†). Moreover, the disappearance of the band for G-quadruplex on the native PAGE in the presence of K+ and PEG200 (Fig. S12†). The aggregation of G-quadruplex was also confirmed by the native PAGE of Gq, Gq5,11m, and Gq5,11hm with 0 to 40 wt% PEG200, in which the intensity of the band for G-quadruplex decreased as the concentration of PEG200 increased (Fig. S16†). These results suggested that the addition of PEG200 could promote aggregation of G-quadruplex forming sequences and affect the transition between a duplex and quadruplexes. A mixture of iM10,16hm and Gq5,11hm still preferred to fold into a duplex, unlike the duplex without modifications and with 5mC, although the amount of i-motif largely increased. The ratio of duplex was 0.26 for the mixture of iM and Gq and 0.24 for that of iM10,16m and Gq5,11m. The ratio of i-motif was 0.68 for that of iM and Gq and 0.70 for that of iM10,16m and Gq5,11m. On the other hand, the ratio of duplex was 0.30 and that of i-motif was 0.49 for the mixture of iM10,16hm and Gq5,11hm. One explanation for the preference of the duplex to quadruplexes is that the stabilising effect of PEG200 on G-quadruplexes was weakened by hydroxymethyl modification, which may have facilitated the transition to duplexes. UV melting curves at 260 and 295 nm supported the preference for duplex formation upon hydroxymethylation, except for the mixtures of iM and Gq and iM10,16m and Gq5,11m. In the absence of PEG200, the duplexes without modifications and those with 5mC tended to form i-motifs in that order, because the representative melting curve of the i-motif at 260 and 295 nm was observed (Fig. 4a and b). On the other hand, DNA with 5hmC did not show any curves of the i-motif, which indicated that DNA with 5hmC prefers to form duplexes in the absence of PEG200. In the presence of 40 wt% PEG200, the UV melting curves derived from i-motifs were clearly observed regardless of the modifications, indicating that PEG200 stabilised i-motifs and reduced the existence ratio of duplexes (Fig. 4c and d). DNA with 5hmC still preferred to fold into duplexes, owing to the lower stability of the i-motif (Table S6†). The Tm of the i-motif with 5hmC in the presence of the complementary strand was 37.5 °C, which was 2.0 °C lower than that of unmodified DNA and 3.6 °C lower than that of DNA with 5mC. Since the stability of the i-motif with 5hmC was not changed by the modifications (Table S2†), the presence of a complementary G-quadruplex forming strand may have affected the stability of the i-motifs with modifications.
Next, we investigated the transition between a duplex and quadruplexes in the presence of Na+. The result of native PAGE in the presence of Na+ showed clear single bands in all the lanes, indicating the formation of a duplex regardless of the presence of 40 wt% PEG200 (Fig. 5). There were no differences between the modifications in the ability to form duplexes. We also examined the formation of duplexes in the presence of Na+ using UV melting curves (Fig. 6). The UV melting curves at 260 nm showed clear melting. DNA duplexes with 5mC were slightly stabilised and those with 5hmC were slightly destabilised compared with unmodified DNA, regardless of the presence of PEG200, although these differences were very small (Table S7†). The UV melting curves at 295 nm for duplexes of unmodified DNA and DNA with 5mC showed two melting curves, indicating that these duplexes tended to fold into both a duplex and quadruplex, unlike DNA with 5hmC, and the quadruplex was more likely to be formed in the order of unmodified DNA and DNA with 5mC (Fig. 6a and b). The addition of 40 wt% PEG200 resulted in the low temperature melting curves of all the unmodified DNAs, indicating the formation of an i-motif. These results showed that in the presence of Na+, unmodified DNA was more likely to fold i-motifs than DNA with 5mC and 5hmC, but all DNAs predominantly fold duplexes regardless of the presence of PEG200.
In summary, both native PAGE and UV melting showed that modification of 5hmC promoted the formation of a duplex in the presence of K+, and molecular crowding of PEG200 increased the existence of i-motifs regardless of modifications. The preference for duplex formation of the 5hmC duplex in the presence of PEG200 may be caused by the effect of mitigating stabilisation, as revealed by the thermodynamic analysis described in the previous section. In contrast, 5mC and 5hmC modifications facilitated the formation of a duplex in the presence of Na+, regardless of the presence of PEG200. In senescent cells, wherein both the extent of molecular crowding and the concentration of potassium are low, unmodified DNA can fold into quadruplexes, whereas modified DNA with 5mC and 5hmC prefers to fold duplexes (Fig. 7, right). In contrast, in young cells, wherein both the extent of molecular crowding and cation concentration are high, unmodified DNA and DNA with 5mC prefer to fold quadruplexes, but only DNA with 5hmC can fold duplexes, although the abundance of duplexes remains small (Fig. 7, left). Thus, the modification in young cells that are unrecognised by proteins such as transcription factors due to the formation of i-motif and G-quadruplexes may be recognised during senescence. Our results obtained here suggest the importance of considering structural changes in GC-rich sequences in CpG islands to understand the mechanisms of epigenetic gene regulation.
 | ||
| Fig. 7 Schematic representation of the transition between duplex and i-motif and G-quadruplexes (a) without modifications, (b) with 5mC, and (c) with 5hmC. | ||
Analysis of i-motif and G-quadruplex forming sequences in CpG sites related to senescence
To understand how the change in transition between duplexes and quadruplexes with modifications contributes to senescence, we analysed the quadruplexes forming sequences in CpG islands, which are hyper- or hypomethylated during replicative senescence. The genes with hyper- and hypomethylated CpG islands were generated based on the Δβ value; see the experimental section for details of the Δβ value. The genes to be analysed were based on a previous study by Sakaki, M. et al.,56 in which changes in DNA methylation during replicative senescence were investigated by methylome and transcriptome analyses. Based on their gene lists, we extracted genes with CpG islands. Thus, 2725 genes with hypermethylated CpG islands and 1627 genes with hypomethylated CpG islands were extracted. These genes were classified based on the presence of quadruplexes in their CpG islands. The motifs adopted for identification were G3,4N1–7G3,4N1–7G3,4N1–7G3,4 (N refers to any base) for G-quadruplex forming sequences and C3,4N1–7C3,4N1–7C3,4N1–7C3,4 (N refers to any base) for i-motif forming sequences (Fig. 8).60 Among the genes with hypermethylated CpG islands, there were 1955 genes with quadruplexes in their CpG islands and 770 genes without quadruplexes in their CpG islands. On the other hand, among genes with hypomethylated CpG islands, there were 978 genes with quadruplexes in their CpG islands and 649 genes without quadruplexes in their CpG islands (Fig. 8). To understand the difference in function between genes with and without quadruplexes in their CpG islands, the gene lists were analysed by GO clustering analysis using DAVID. Each gene list was uploaded to DAVID and analysed using GOTERM_BP_FAT (biological process) and functional annotation clustering (see Experimental section for details). In this clustering, the more common genes share GO terms, the higher chance they will be grouped together. The clusters with enrichment scores higher than 1.3 are summarised in Tables 2 and 3 in order of higher enrichment score. | ||
| Fig. 8 The scheme of sequence analyses in CpG island that are hypermethylated and hypomethylated during senescence. | ||
| Cluster | Representative GO terms | Enrichment score | |
|---|---|---|---|
| With quadruplexes | 1 | Organ morphogenesis | 8.4 |
| 2 | Positive regulation of transcription from RNA polymerase II promoter | 8.3 | |
| 3 | Negative regulation of biosynthetic process | 8.0 | |
| 4 | Embryonic limb morphogenesis | 6.7 | |
| 5 | Enzyme linked receptor protein signaling pathway | 5.9 | |
| No quadruplexes | 1 | mRNA cleavage | 1.9 |
| 2 | mRNA cleavage involved in mRNA processing | 1.5 | |
| 3 | Small molecule catabolic process | 1.4 | |
| 4 | 3′-Phosphoadenosine 5′-phosphosulfate metabolic process | 1.4 |
| Cluster | Representative GO terms | Enrichment score | |
|---|---|---|---|
| With quadruplexes | 1 | Cellular component morphogenesis | 2.6 |
| 2 | Regulation of cellular component organisation | 2.4 | |
| 3 | Response to oxygen levels | 2.4 | |
| 4 | Drug transmembrane transport | 2.1 | |
| 5 | Regulation of fibroblast growth factor receptor signaling pathway | 2.1 | |
| No quadruplexes | 1 | Xenobiotic metabolic process | 2.2 |
| 2 | Amine metabolic process | 2.1 | |
| 3 | Defense response | 1.6 | |
| 4 | Organonitrogen compound biosynthetic process | 1.6 | |
| 5 | Cellular response to lithium ion | 1.4 |
For genes with hypermethylated CpG islands, 463 enriched functional clusters from 1955 genes with quadruplexes in their CpG islands were identified using DAVID. The top five clusters categorised by the presence of quadruplexes are summarised in Table 2. Among these, the ‘organ morphogenesis’ cluster showed the highest enrichment score (8.4), followed by the ‘positive regulation of transcription from RNA polymerase II promoter’ (8.3) and ‘negative regulation of biosynthetic process’ (8.0) clusters (Table 2). Because genes related to morphogenesis should be repressed and activities of gene expression may be positively or negatively regulated during senescence, this result agrees with hypermethylation causing a decrease in gene expression.33,34 In addition, 249 clusters from 770 genes without quadruplexes in their CpG islands were also identified using DAVID. Among these, the ‘mRNA cleavage’ cluster showed the highest enrichment score (1.9), followed by the ‘mRNA cleavage involved in mRNA processing’ (1.5), ‘small molecule catabolic process’ (1.4), and 3′-phosphoadenosine 5′-phosphosulfate metabolic process’ (1.4) clusters (Table 2). The terms related to metabolic processes of nucleic acids were mostly enriched, and these functions are likely to be repressed in senescent cells. For instance, with regard to nucleic acid metabolism, it has been reported that the expression of deoxyribonuclease is low in senescent cells, resulting in the accumulation of genomic DNA fragments in the cytoplasm, which leads to the senescence-associated secretory phenotype.67 Noteworthily, the functions of genes in both categories seemed to be related to the promotion of senescence; however, differences in functions were observed. The different functional terms enriched in the presence or absence of quadruplexes suggest that not only the presence or absence of methylation but also that of quadruplexes in the CpG islands may contribute to the regulation of gene expression in senescent cells.
For genes with hypomethylated CpG islands, 303 and 195 clusters were identified from 978 genes with quadruplexes in their CpG islands and 649 genes without quadruplexes in their CpG islands, respectively. Among clusters for genes with CpG islands containing quadruplexes, the ‘cellular component morphogenesis’ cluster showed the highest enrichment score (2.6), followed by the ‘regulation of cellular component organisation’ (2.4), ‘response to oxygen levels’ (2.4), ‘drug transmembrane transport’ (2.1), and ‘regulation of fibroblast growth factor receptor signalling pathway’ (2.1) clusters (Table 3). In contrast, among clusters for genes without quadruplexes in their CpG islands, the ‘xenobiotic metabolic process’ cluster showed the highest enrichment score (2.2), followed by the ‘amine metabolic processes’ (2.1), ‘defence response’ (1.6), ‘organonitrogen compound biosynthetic process’ (1.6), and ‘cellular response to lithium ion’ (1.4) clusters (Table 3). The terms which were enriched in both categories covered a wide range, and this result did not indicate that the genes were all likely to be upregulated in senescent cells. However, there was a difference between terms enriched in both categories, depending on the presence or absence of quadruplexes. Hence, this result indicates the importance of the formation of quadruplexes in CpG islands during senescence for the regulation of gene expression. Together with the results of the transition analysis, this result suggests that gene expression can be regulated by the transition between duplexes and quadruplexes during senescence. Genes with quadruplex forming sequences in CpG islands where structural transition may occur can be recognised very easily by proteins when the formation of quadruplexes is unravelled by environmental changes.
Conclusions
We investigated the effects of 5mC and 5hmC modifications on the transition from a duplex to an i-motif and G-quadruplexes in the chemical environments inside senescent cells. We first examined the stability and structure of i-motif and G-quadruplexes with modifications. Analysis of stabilities revealed that neither 5mC nor 5hmC modifications markedly modulated the stability of the i-motif and G-quadruplexes, regardless of the extent of molecular crowding and the composition of cations. In contrast, we found that the characteristics of the surrounding environments, such as molecular crowding and cations, modulated the thermal stability of G-quadruplexes with modifications. Moreover, modifications tend to weaken the effects of (de)stabilisation by molecular crowding and cations, with a few exceptions. To examine the mechanisms of stabilisation of G-quadruplexes with modifications by the surrounding environments, we investigated the structure of G-quadruplexes. Both cations and molecular crowding modulated the topology of G-quadruplexes, but there were no drastic differences among modifications. However, the modifications regulated the transition between a duplex and quadruplexes such as an i-motif and G-quadruplexes. In the presence of K+, only DNA with 5hmC preferred to fold into duplexes and molecular crowding decreased the amount of duplex by promoting the formation of i-motif. This can be explained by the low Tm of the i-motif with hydroxymethylation in the presence of complementary strands in a solution containing K+ and PEG200. Moreover, it is also because the stabilisation effects on G-quadruplexes caused by K+ are smaller than those on DNA without modifications. On the other hand, DNA with 5mC and 5hmC modifications tended to fold into duplexes, unlike DNA without modifications, regardless of molecular crowding in the presence of Na+. Our results highlight the importance of considering not only the patterns and levels of epigenetic DNA modifications but also the formation of non-canonical DNA structures to understand the mechanism of epigenetic gene regulation. An analysis of quadruplexes in CpG islands, which were hyper- or hypomethylated during senescence, supports this hypothesis. Genes with hyper- or hypomethylated CpG islands during senescence may have different functions depending on the presence of quadruplexes in the CpG islands. The findings of this study will provide a theoretical basis for proposals on how non-canonical DNA structures with modifications modulate their functions in response to the surrounding environments during senescence.Future perspective
Our results suggest that gene expression can be regulated by the dissociation of G-quadruplex formation on CpG islands, in response to changes in the intracellular environment during senescence. This suggests that the steps of cellular senescence, such as cell cycle arrest that triggered cell enlargement and reduced intracellular crowding, may alter gene expression, leading to individual aging. There are three possible effects of G-quadruplex formation on CpG islands. The first is an alteration in gene expression due to the loss of recognition of methyl and hydroxymethyl cytosine. When G-quadruplexes were unfolded and folded to duplex form due to senescence, methyl and hydroxymethyl groups that were previously unrecognised by steric hindrance of G-quadruplex can be recognised. As a result, when methyl groups are recognised, a decrease in the expression of previously expressed genes can be observed. On the other hand, when the hydroxymethyl group is recognised, the expression of this gene is increased by senescence. The second is an alteration in gene expression due to the inhibition of new methylation, hydroxymethylation, and demethylation. The transition of G-quadruplexes to a duplex during senescence may result in the methylation of previously unmethylated CpG sites. This may suppress the expression of previously expressed genes. However, the effect of the G-quadruplex of CpG islands on transcription should also be considered. The third is a change in gene expression due to the recruitment of G-quadruplex binding proteins. Here, the recruitment of G-quadruplex binding proteins may result in the release of proteins that have been assembled by G-quadruplexes in the CpG region. For example, in epigenetics, it has been suggested that the human methylated protein, DNMT1, binds to the G-quadruplex in the CpG region and methylates CpG sites around the G-quadruplex forming sequence.68 When the transitioning from G-quadruplex to duplex with senescence, DNMT1, which was recruited by G-quadruplex, may dissociate, and a decrease in methylation rate around the G-quadruplex forming sequence may occur and even an increase in gene expression may be observed. It would be meaningful to examine these hypotheses to understand the effect of G-quadruplex formation on CpG islands during senescence. Validation of these hypotheses can promote the development of drug discovery to inhibit senescence by using ligands or peptides that induce or stabilise G-quadruplexes. In addition, as shown in this study, the transition of duplexes to quadruplexes is affected by the surrounding environment. Therefore, it may be possible to develop a strategy for senescence by maintaining G-quadruplex formation by changing the intracellular environment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Ms S. Kamiyama for their assistance in performing the experiments. The research in our laboratories was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and the Japan Society for the Promotion of Science (JSPS), especially a Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (JSPS KAKENHI Grant JP17H06351), a Grant-in-Aid for Scientific Research (A) (19H00928), a Fostering Joint International Research (B) (18KK0164), a Grant-in-Aid for Research Activity Start-up (19K23639), a Transformative Research Areas (B) of “Dimensional response genome” (21H05109), and a Grant-in-Aid for Early-Career Scientists (21K14742), by the Hirao Taro Foundation of Konan Gakuen for Academic Research and by the Chubei Itoh Foundation.Notes and references
- Y. Atlasi and H. G. Stunnenberg, Nat. Rev. Genet., 2017, 18, 643–658 CrossRef CAS PubMed.
- C. Luo, P. Hajkova and J. R. Ecker, Science, 2018, 361, 1336–1340 CrossRef CAS PubMed.
- M. V. C. Greenberg and D. Bourc'his, Nat. Rev. Mol. Cell Biol., 2019, 20, 590–607 CrossRef CAS PubMed.
- H. Fang, C. M. Disteche and J. B. Berletch, Front. Cell Dev. Biol., 2019, 7, 219 CrossRef PubMed.
- C. G. Duncan, S. A. Grimm, D. L. Morgan, P. R. Bushel, B. D. Bennett, N. C. S. Program, J. D. Roberts, F. L. Tyson, B. A. Merrick and P. A. Wade, Sci. Rep., 2018, 8, 10138 CrossRef PubMed.
- J. Y. Hwang, K. A. Aromolaran and R. S. Zukin, Nat. Rev. Neurosci., 2017, 18, 347–361 CrossRef CAS PubMed.
- M. A. Dawson and T. Kouzarides, Cell, 2012, 150, 12–27 CrossRef CAS PubMed.
- K. Skvortsova, C. Stirzaker and P. Taberlay, Essays Biochem., 2019, 63, 797–811 CrossRef CAS PubMed.
- E. Tzika, T. Dreker and A. Imhof, Front. Genet., 2018, 9, 361 CrossRef PubMed.
- F. H. Xiao, H. T. Wang and Q. P. Kong, Front. Genet., 2019, 10, 107 CrossRef CAS PubMed.
- L. Zane, V. Sharma and T. Misteli, Trends Cell Biol., 2014, 24, 686–694 CrossRef CAS PubMed.
- S. Bocklandt, W. Lin, M. E. Sehl, F. J. Sanchez, J. S. Sinsheimer, S. Horvath and E. Vilain, PLoS One, 2011, 6, e14821 CrossRef CAS PubMed.
- S. Horvath, Genome Biol., 2013, 14, R115 CrossRef PubMed.
- P. Garagnani, M. G. Bacalini, C. Pirazzini, D. Gori, C. Giuliani, D. Mari, A. M. Di Blasio, D. Gentilini, G. Vitale, S. Collino, S. Rezzi, G. Castellani, M. Capri, S. Salvioli and C. Franceschi, Aging Cell, 2012, 11, 1132–1134 CrossRef CAS PubMed.
- S. E. Jung, S. M. Lim, S. R. Hong, E. H. Lee, K. J. Shin and H. Y. Lee, Forensic Sci. Int.: Genet., 2019, 38, 1–8 CrossRef CAS PubMed.
- S. Kriaucionis and N. Heintz, Science, 2009, 324, 929–930 CrossRef CAS PubMed.
- M. Tahiliani, K. P. Koh, Y. Shen, W. A. Pastor, H. Bandukwala, Y. Brudno, S. Agarwal, L. M. Iyer, D. R. Liu, L. Aravind and A. Rao, Science, 2009, 324, 930–935 CrossRef CAS PubMed.
- M. T. Bocker, F. Tuorto, G. Raddatz, T. Musch, F. C. Yang, M. Xu, F. Lyko and A. Breiling, Nat. Commun., 2012, 3, 818 CrossRef PubMed.
- S. Uribe-Lewis, T. Carroll, S. Menon, A. Nicholson, P. J. Manasterski, D. J. Winton, S. J. A. Buczacki and A. Murrell, Sci. Rep., 2020, 10, 546 CrossRef CAS PubMed.
- M. A. Hahn, R. Qiu, X. Wu, A. X. Li, H. Zhang, J. Wang, J. Jui, S. G. Jin, Y. Jiang, G. P. Pfeifer and Q. Lu, Cell Rep., 2013, 3, 291–300 CrossRef CAS PubMed.
- R. Lister, E. A. Mukamel, J. R. Nery, M. Urich, C. A. Puddifoot, N. D. Johnson, J. Lucero, Y. Huang, A. J. Dwork, M. D. Schultz, M. Yu, J. Tonti-Filippini, H. Heyn, S. Hu, J. C. Wu, A. Rao, M. Esteller, C. He, F. G. Haghighi, T. J. Sejnowski, M. M. Behrens and J. R. Ecker, Science, 2013, 341, 1237905 CrossRef PubMed.
- V. Lopez, A. F. Fernandez and M. F. Fraga, Ageing Res. Rev., 2017, 37, 28–38 CrossRef CAS PubMed.
- K. E. Szulwach, X. Li, Y. Li, C. X. Song, H. Wu, Q. Dai, H. Irier, A. K. Upadhyay, M. Gearing, A. I. Levey, A. Vasanthakumar, L. A. Godley, Q. Chang, X. Cheng, C. He and P. Jin, Nat. Neurosci., 2011, 14, 1607–1616 CrossRef CAS PubMed.
- C. X. Song, K. E. Szulwach, Y. Fu, Q. Dai, C. Yi, X. Li, Y. Li, C. H. Chen, W. Zhang, X. Jian, J. Wang, L. Zhang, T. J. Looney, B. Zhang, L. A. Godley, L. M. Hicks, B. T. Lahn, P. Jin and C. He, Nat. Biotechnol., 2011, 29, 68–72 CrossRef CAS PubMed.
- C. G. Lian, Y. Xu, C. Ceol, F. Wu, A. Larson, K. Dresser, W. Xu, L. Tan, Y. Hu, Q. Zhan, C. W. Lee, D. Hu, B. Q. Lian, S. Kleffel, Y. Yang, J. Neiswender, A. J. Khorasani, R. Fang, C. Lezcano, L. M. Duncan, R. A. Scolyer, J. F. Thompson, H. Kakavand, Y. Houvras, L. I. Zon, M. C. Mihm Jr, U. B. Kaiser, T. Schatton, B. A. Woda, G. F. Murphy and Y. G. Shi, Cell, 2012, 150, 1135–1146 CrossRef CAS PubMed.
- C. X. Song, S. Yin, L. Ma, A. Wheeler, Y. Chen, Y. Zhang, B. Liu, J. Xiong, W. Zhang, J. Hu, Z. Zhou, B. Dong, Z. Tian, S. S. Jeffrey, M. S. Chua, S. So, W. Li, Y. Wei, J. Diao, D. Xie and S. R. Quake, Cell Res., 2017, 27, 1231–1242 CrossRef CAS PubMed.
- W. Li, X. Zhang, X. Lu, L. You, Y. Song, Z. Luo, J. Zhang, J. Nie, W. Zheng, D. Xu, Y. Wang, Y. Dong, S. Yu, J. Hong, J. Shi, H. Hao, F. Luo, L. Hua, P. Wang, X. Qian, F. Yuan, L. Wei, M. Cui, T. Zhang, Q. Liao, M. Dai, Z. Liu, G. Chen, K. Meckel, S. Adhikari, G. Jia, M. B. Bissonnette, X. Zhang, Y. Zhao, W. Zhang, C. He and J. Liu, Cell Res., 2017, 27, 1243–1257 CrossRef CAS PubMed.
- K. Misawa, S. Yamada, M. Mima, T. Nakagawa, T. Kurokawa, A. Imai, D. Mochizuki, K. Morita, R. Ishikawa, S. Endo and Y. Misawa, J. Cancer, 2019, 10, 5306–5314 CrossRef CAS PubMed.
- G. D. Guler, Y. Ning, C. J. Ku, T. Phillips, E. McCarthy, C. K. Ellison, A. Bergamaschi, F. Collin, P. Lloyd, A. Scott, M. Antoine, W. Wang, K. Chau, A. Ashworth, S. R. Quake and S. Levy, Nat. Commun., 2020, 11, 5270 CrossRef CAS PubMed.
- S. Al-Mahdawi, S. A. Virmouni and M. A. Pook, Front. Neurosci., 2014, 8, 397 Search PubMed.
- S. I. Sherwani and H. A. Khan, Gene, 2015, 570, 17–24 CrossRef CAS PubMed.
- Y. Zhang, Y. Zhang, D. Chen, C. Wang, L. Chen, C. Gao, W. Fan, J. Shi, J. Zhang and B. Li, Front. Mol. Neurosci., 2019, 12, 214 CrossRef CAS PubMed.
- A. P. Bird and A. P. Wolffe, Cell, 1999, 99, 451–454 CrossRef CAS PubMed.
- A. Bird, Genes Dev., 2002, 16, 6–21 CrossRef CAS PubMed.
- D. Q. Shi, I. Ali, J. Tang and W. C. Yang, Front. Genet., 2017, 8, 100 CrossRef PubMed.
- A. Thalhammer, A. S. Hansen, A. H. El-Sagheer, T. Brown and C. J. Schofield, Chem. Commun., 2011, 47, 5325–5327 RSC.
- L. Nardo, M. Lamperti, D. Salerno, V. Cassina, N. Missana, M. Bondani, A. Tempestini and F. Mantegazza, Nucleic Acids Res., 2015, 43, 10722–10733 CrossRef CAS PubMed.
- M. Tsuruta, Y. Sugitani, N. Sugimoto and D. Miyoshi, Int. J. Mol. Sci., 2021, 22, 947 CrossRef CAS PubMed.
- B. Xu, G. Devi and F. Shao, Org. Biomol. Chem., 2015, 13, 5646–5651 RSC.
- Y. P. Bhavsar-Jog, E. Van Dornshuld, T. A. Brooks, G. S. Tschumper and R. M. Wadkins, Biochemistry, 2014, 53, 1586–1594 CrossRef CAS PubMed.
- P. Skolakova, Z. Badri, S. Foldynova-Trantirkova, J. Rynes, J. Sponer, M. Fojtova, J. Fajkus, R. Marek, M. Vorlickova, J. L. Mergny and L. Trantirek, Biochim. Biophys. Acta, Gen. Subj., 2020, 1864, 129651 CrossRef CAS PubMed.
- W. Yoshida, M. Terasaka, S. Laddachote and I. Karube, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862, 1933–1937 CrossRef CAS PubMed.
- J. Lin, J. Q. Hou, H. D. Xiang, Y. Y. Yan, Y. C. Gu, J. H. Tan, D. Li, L. Q. Gu, T. M. Ou and Z. S. Huang, Biochem. Biophys. Res. Commun., 2013, 433, 368–373 CrossRef CAS PubMed.
- R. K. Morgan, M. M. Molnar, H. Batra, B. Summerford, R. M. Wadkins and T. A. Brooks, J. Nucleic Acids, 2018, 2018, 9281286 Search PubMed.
- N. Sugimoto, T. Endoh, S. Takahashi and H. Tateishi-Karimata, Bull. Chem. Soc. Jpn., 2021, 94, 1970–1998 CrossRef CAS.
- D. Miyoshi, H. Karimata and N. Sugimoto, J. Am. Chem. Soc., 2006, 128, 7957–7963 CrossRef CAS PubMed.
- M. C. Miller, R. Buscaglia, J. B. Chaires, A. N. Lane and J. O. Trent, J. Am. Chem. Soc., 2010, 132, 17105–17107 CrossRef CAS PubMed.
- S. E. Kim, I. B. Lee, C. Hyeon and S. C. Hong, J. Phys. Chem. B, 2014, 118, 4753–4760 CrossRef CAS PubMed.
- S. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS PubMed.
- D. Bhattacharyya, G. Mirihana Arachchilage and S. Basu, Front. Chem., 2016, 4, 38 Search PubMed.
- T. V. Chalikian, L. Liu and J. R. B. Macgregor, Biophys. Chem., 2020, 267, 106473 CrossRef CAS PubMed.
- G. E. Neurohr, R. L. Terry, J. Lengefeld, M. Bonney, G. P. Brittingham, F. Moretto, T. P. Miettinen, L. P. Vaites, L. M. Soares, J. A. Paulo, J. W. Harper, S. Buratowski, S. Manalis, F. J. van Werven, L. J. Holt and A. Amon, Cell, 2019, 176, 1083–1097 CrossRef CAS PubMed.
- M. Strickland, B. Yacoubi-Loueslati, B. Bouhaouala-Zahar, S. L. F. Pender and A. Larbi, Front. Physiol., 2019, 10, 158 CrossRef PubMed.
- H. Tateishi-Karimata, S. Nakano and N. Sugimoto, Curr. Protoc. Nucleic Acid Chem., 2013, 53, 7.19.1–7.19.17 Search PubMed.
- K. H. Miga, Y. Newton, M. Jain, N. Altemose, H. F. Willard and W. J. Kent, Genome Res., 2014, 24, 697–707 CrossRef CAS PubMed.
- M. Sakaki, Y. Ebihara, K. Okamura, K. Nakabayashi, A. Igarashi, K. Matsumoto, K. Hata, Y. Kobayashi and K. Maehara, PLoS One, 2017, 12, e0171431 CrossRef PubMed.
- M. Gardiner-Garden and M. Frommer, J. Mol. Biol., 1987, 196, 261–282 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman, Q. Tan, J. R. Collins, W. G. Alvord, J. Roayaei, R. Stephens, M. W. Baseler, H. C. Lane and R. A. Lempicki, Genome Biol, 2007, 8, R183 CrossRef PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908–2916 CrossRef CAS PubMed.
- S. Takahashi, J. Yamamoto, A. Kitamura, M. Kinjo and N. Sugimoto, Anal. Chem., 2019, 91, 2586–2590 CrossRef CAS PubMed.
- T. Fujimoto, S. Nakano, D. Miyoshi and N. Sugimoto, J. Nucleic Acids, 2011, 2011, 857149 Search PubMed.
- E. Largy, J.-L. Mergny and V. Gabelica, in The Alkali Metal Ions: Their Role for Life, ed. A. Sigel, H. Sigel and R. K. O. Sigel, Springer International Publishing, Cham, 2016, pp. 203–258 Search PubMed.
- T. Siebenmorgen and M. Zacharias, Biophys. J., 2017, 112, 2280–2290 CrossRef CAS PubMed.
- K. Hisae, N. Shu-ichi and S. Naoki, Bull. Chem. Soc. Jpn., 2007, 80, 1987–1994 CrossRef.
- S.-i. Nakano, L. Wu, H. Oka, H. T. Karimata, T. Kirihata, Y. Sato, S. Fujii, H. Sakai, M. Kuwahara, H. Sawai and N. Sugimoto, Mol. BioSyst., 2008, 4, 579–588 RSC.
- A. Takahashi, T. M. Loo, R. Okada, F. Kamachi, Y. Watanabe, M. Wakita, S. Watanabe, S. Kawamoto, K. Miyata, G. N. Barber, N. Ohtani and E. Hara, Nat. Commun., 2018, 9, 1249 CrossRef PubMed.
- S.-Q. Mao, A. T. Ghanbarian, J. Spiegel, S. Martínez Cuesta, D. Beraldi, M. Di Antonio, G. Marsico, R. Hänsel-Hertsch, D. Tannahill and S. Balasubramanian, Nat. Struct. Mol. Biol., 2018, 25, 951–957 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07201c |
| This journal is © The Royal Society of Chemistry 2021 |






