Open Access Article
Open Access ArticleImproving the catalytic performance of Pichia pastoris whole-cell biocatalysts by fermentation process†
Denggang Wang ,
Meiqi Chen,
Xin Zeng,
Wenjie Li,
Shuli Liang* and
Ying Lin*
,
Meiqi Chen,
Xin Zeng,
Wenjie Li,
Shuli Liang* and
Ying Lin*
Guangdong Key Laboratory of Fermentation and Enzyme Engineering, School of Biology and Biological Engineering, South China University of Technology, Panyu, Guangzhou 510006, People's Republic of China. E-mail: wangdenggang_1@foxmail.com; feylin@scut.edu.cn; shuli@scut.edu.cn
First published on 11th November 2021
Abstract
Whole-cell biocatalysts have a wide range of applications in many fields. However, the transport of substrates is tricky when applying whole-cell biocatalysts for industrial production. In this research, P. pastoris whole-cell biocatalysts were constructed for rebaudioside A synthesis. Sucrose synthase was expressed intracellularly while UDP-glycosyltransferase was displayed on the cell wall surface simultaneously. As an alternative method, a fermentation process is applied to relieve the substrate transport-limitation of P. pastoris whole-cell biocatalysts. This fermentation process was much simpler, more energy-saving, and greener than additional operating after collecting cells to improve the catalytic ability of whole-cell biocatalysts. Compared with the general fermentation process, the protein production capacity of cells did not decrease. Meanwhile, the activity of whole-cell biocatalysts was increased to 262%, which indicates that the permeability and space resistance were improved to relieve the transport-limitations. Furthermore, the induction time was reduced from 60 h to 36 h. The fermentation process offered significant advantages over traditional permeabilizing reagent treatment and ultrasonication treatment based on the high efficiency and simplicity.
1. Introduction
Steviol glycosides (SGs) are a group of sweetening agents, which have been applied to food and beverages for their advantages of high-intensity sweetness, low-calorie, non-nutritive nature, and high stability.1,2 The therapeutic properties of SGs have been studied on antihyperglycemic, antihypertensive, anti-inflammatory, antitumor, antidiarrheal, diuretic, and immunomodulatory effects.3,4 More than 30 steviol glycosides have been discovered in Stevia rebaudiana in the past few years. The stevioside (ST) and rebaudioside A(RA) are the main contents in commercially steviol glycoside products.5 In the previous study, ST is 250–300 times sweeter than sucrose with an unflavored aftertaste.6 As a sweetener with better taste, the high purity of RA is more popular than St in the market. To extract RA from Stevia rebaudiana, there are some techniques for physical separation such as high-speed counter-current chromatography,7 preparative high-performance liquid chromatography (HPLC),8 and adsorptive separation.9 However, ST is difficult to be separated from the products due to the similar physical properties of the two substances. Therefore, it is necessary to focus on the conversion of ST to RA, and enzymatic catalysis is an efficient method to be considered.Enzymes are the principal cost of industrial-scale production in the biochemical industry.10 On the one hand, the steps of purifying enzyme are omitted of whole-cell biocatalysts, and the cost would be significantly saved.11 On the other hand, whole-cell biocatalysts are more stable than free enzymes, which are promising alternatives. Due to the protection of cell walls and membranes, whole-cell biocatalysts can be applied in harsh conditions and unconventional reaction systems,12,13 and the cost is only one-tenth of a purified enzyme system.10 In addition, whole-cell biocatalysts are commonly used when cofactors are involved in the reaction,14 such as the reactions catalyzed by oxidoreductases that depend on nicotinamide cofactors15,16 and the redox-neutral multi-step reactions coupling of enzymes.17,18
Undeniably, whole-cell biocatalysts have limitations. Low selectivity may lead to by-products,14 which can be overcome by using dead whole-cell biocatalysts19 or the selective knockout of host-related gene.20 One of the main limiting factors is transport-limited caused by cell walls and cytomembranes.21 Generally, there are three main strategies to improve the permeability of whole-cell biocatalysts reliving the limitation of transport: (i) adding permeabilizing reagent, such as surfactant,22,23 organic solvent,24 chelating agent,25 and other similar agents.26,27 (ii) Physical processing, such as ultrasonic treatment,28 osmotic pressure,29 etc.30 (iii) Molecular biology, including changing the structure and composition of cell membranes,31 expressing potential transporters,32 and expressing membrane-active peptides.33 Both advantages and limitations are present in each strategy. For example, adding permeabilizing reagents introduces unwanted substances, physical processes are not conducive to scale-up, and molecular biology strategy is individualized and complicated to operate. Therefore, novel methods to improve transport-limited should be developed.
P. pastoris is widely applied in the production of industrial enzymes.34,35 P. pastoris whole-cell biocatalysts have been reported to apply to various fields, including biodiesel production,36 esters production,37 phosphatidylserine production,38 and synthesis of isomalto-oligosaccharides.39 In the previous study, P. pastoris could be used for RA synthesis as a whole-cell biocatalysts.40,41 Two forms of whole-cell biocatalysts could be constructed depending on whether the proteins are expressed in intracellular or displayed on the surface. When the protein was expressed intracellular, the permeability of the biocatalysts should be improved to increase activity.36 On the contrary, the surface display system was benefited from substrate transportation and protein stability, but the low activity was needed to be improved.22
In this study, a fermentation process was applied to improve the capacity of P. pastoris whole-cell biocatalysts. A recombinant P. pastoris, co-expressing sucrose synthase (SUS) intracellular and surface displaying UDP-glycosyltransferase (UGT), was constructed to synthesize RA. Based on general high cell density fermentation, the initial cell density and methanol feed strategies were explored to study the relationship between the fermentation process and transport-limited. In addition, the improvement of whole-cell biocatalysts by fermentation process optimization was compared with traditional strategies, including permeabilizing reagent and ultrasonication treatment.
2. Materials and methods
2.1. Gene cloning, plasmid construction and strain transformation
P. pastoris GS115 (his4−) (Invitrogen) expressing UDP-glycosyltransferase or sucrose synthase driven by the PAOX1 promoter was used as the wild-type (WT) strain, and all overexpression strains were driven from this strain. Escherichia coli TOP10 was used as the host for plasmid construction. The sucrose synthase gene was cloned from mung bean, which was cloned into a pPICZA (Invitrogen) expression system. Moreover, the UDP-glycosyltransferase gene UTG76G1 was from Stevia rebaudiana, which was fused with the P. pastoris cell wall protein GCW61 and the α-factor signal sequence, then incorporated into a pPIC9K (Invitrogen) Codon optimization of these genes was conducted on http://www.jcat.de, and the optimized sequences were sent for chemical synthesis at Jierui (Shanghai, China). Detailed information on the construction of integration plasmids is shown in ESI Table 1.† All the primers used in this study are listed in ESI Table 2.†The recombination strains containing BleoR and KanR maker were cultivated in yeast extract peptone dextrose (YPD) agar plate, 100 μg mL−1 zeocin, and 200 μg mL−1 geneticin (G418) were added as selection pressure, respectively. For shake-flask culture, single colonies were picked from the agar plate and inoculated into test tubes containing 5 mL buffered glycerol-complex (BMGY) medium for overnight growth at 30 °C in a 250 rpm orbital shaker, then the precultures were inoculated into shake flasks containing 25 mL buffered methanol-complex (BMMY) to an initial OD600 of 1.
BMMY medium, containing 10 g L−1 yeast extract, 20 g L−1 tryptone, 13.4 g L−1 yeast nitrogen base (YNB), and various concentrations of methanol. A 10% (v/v) phosphate buffered saline (PBS, 1 M, pH 6.0) was used to maintain pH. Yeast extract peptone dextrose (YPD) medium containing 10 g L−1 yeast extract, 20 g L−1 tryptone, and 20 g L−1 glucose.
2.2. Feed-batch fermentation
After overnight cultivation in YPD medium at 30 °C, the seed cultures were inoculated (8%, v/v) into a 15 L bioreactor (FUS-15L; Ncbio, Shanghai, China) with 7.5 L basal salt medium (BSM) that contained 27 mL L−1 85% H3PO4, 40 g L−1 glycerol, 18 g L−1 K2SO4, 14.9 g L−1 MgSO4·7H2O, 4.13 g L−1 KOH and 0.93 g L−1 CaSO4, supplemented with 4.35 mL L−1 PTM1 trace salts (65 g L−1 FeSO4·7H2O, 20 g L−1 ZnCl2, 6 g L−1 CuSO4·5H2O, 3 g L−1 MnSO4·H2O, 0.5 g L−1 CoCl2, 0.2 g L−1 MoNa2O4·2H2O, 0.2 g L−1 biotin, 0.09 g L−1 KI, 0.02 g L−1 H3BO3 and 5.0 mL L−1 98% H2SO4).The fermentation process was divided into three stages. Stage I was the cell growth phase in which dissolved oxygen (DO) was maintained at >30% of air saturation by control of the agitation rate (200–800 rpm, stage I). And 1.8 L 50% (w/w) glycerol (containing 12 mL L−1 PTM1 solution) was feed (stage II) to maintain dissolved oxygen >30%. The temperature controlled to 30 °C by recirculating water and the pH maintained at 5.5 by feeding 25% ammonium hydroxide at stages I and II. Stage III was the protein production phase by the addition of methanol (containing 12 mL L−1 PTM1 solution) to induce, the culture temperature-controlled to 25 °C, pH adjusted to 6.0, maintained DO > 20% of air saturation with an agitation rate of 800 rpm. The optimization experiments of culture temperature, pH value and methanol concentration are shown in ESI Table 3.† All culture strategies in the 15 L bioreactor are listed in ESI Table 4.†2.3. Biomass analysis
The biomass was characterized by dry cell weight (DCW, g L−1) and absorbance at 600 nm. A 10 mL sample was collected every 4 h (stages I and II) or 6 h (stage III) then centrifuged at 4200 g at 4 °C for 10 min. 100 mM PBS 7.0 was used to resuspend the cells then repetition the centrifugation procedure. The samples were washed three times and resuspended in 10 mL 100 mM PBS (pH 7.0), and 1 mL cell resuspension was used to measurement of OD600 and 9 mL to measure dry cell weight. The tubes containing centrifuged cells were dried at 105 °C until a constant weight was maintained. The relationship between OD600 and DCW was represented by the equation: 1 OD600 L−1 = 0.24 gDCW L−1.2.4. Measurement of SUS, UGT and catalytic activity
The fermentation broth samples were centrifuged at 8200 g at 4 °C for 5 min, washed three times in 100 mM PBS (pH 7.0), then the OD600 was measured then maintained at OD600 = 60.1 mL sample as described above was pipetted into a pre-cooled cell disruption tube containing 700 mg glass beads (0.5 mm), then adding 3 μL of protease inhibitor cocktail (Sigma), which was subjected to high-speed homogenization for 1 min, then placed in an ice bath for 1 min. This process of high-speed homogenization and the ice bath was repeated 8 times. The cell lysates were collected to measure SUS and UGT enzyme activities, and the catalytic activities of the double-enzyme coupling reaction.The activities of UGT were measured by quantifying the synthetic rate of uridine diphosphate glucose (UDPG) and consumption rate of uridine diphosphate (UDP) generation using HPLC. According to eqn (1), one unit of enzyme activity was defined as generating 1 μmol UDPG (or consuming 1 μmol UDP) per minute. The activities of SUS were measured by quantifying the synthetic rate of UDP and consumption rate of UDPG generation using HPLC. According to eqn (2), one unit of enzyme activity was defined as generating 1 μmol UDP (or consuming 1 μmol UDPG) per minute. The catalytic activity of whole-cell was determined by measuring RA generation by HPLC. According to eqn (1) and (2), one unit of activity was defined as generating 1 μg RA per hour. The reaction starts by adding all the substances of the reaction system. At the end of the reaction, 100 μL 0.9 M phosphoric acid solution was added to stop the reaction for 5 minutes, and then 100 μL 2 M NaOH solution was added to neutralize the reaction solution for liquid phase detection. The SUS reaction system included 50 mM PBS 7.0, 50 mM sucrose, 2 mM UDP, and 3 mM MgCl2, reaction time was 20 min. The UGTs reaction system included 50 mM PBS 7.0, 2 mM UDPG, 3 mM MgCl2, and 10 g L−1 ST, The reaction time was 20 min. The coupling reaction system included 50 mM PBS 7.0, 50 mM sucrose, 1 mM UDP, 1.5 mM MgCl2, and0 g L−1 ST, the reaction time was 6 h.
All reaction systems and conditions are shown in ESI Table 5.†
 | (1) |
 | (2) |
The UDP and UDPG were measured using a Waters 2695 HPLC system equipped with a ZORBAX SB-C18 column (4.6 × 150 mm) and a UV-detector (262 nm), with a column temperature of 35 °C. The mobile phase was 0.2 M phosphate buffer (0.1 M Na2HPO3 and 0.1 M KH2PO3, pH 7.0), at a flow rate of 1 mL min−1. ST and RA were determined by a Waters 2695 HPLC system equipped with a Sapphire C18 column (4.6 × 250 mm) and a UV-detector (210 nm), with a column temperature of 40 °C. The mobile phase was phosphate buffer (1.38 g L−1 Na2HPO3, pH 2.6): acetonitrile at a ratio of 0.68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.32, and flow rate of 1 mL min−1.
0.32, and flow rate of 1 mL min−1.
2.5. The surface display volume analysis
Strains containing a FLAG tag could be used to measure immune response. Flow cytometric analysis was performed as described by Wang et al.42 1 mL cell suspension (OD600 = 40) was centrifuged at 4200 g for 1 min. The cells were washed three times with 10 mM PBS (pH 7.4) then resuspended in 10 mM PBS containing 1% (m/v) bovine serum albumin (BSA) for 30 min to block non-specific binding to the cells surface. A monoclonal antibody (2 μg μL−1) toward the FLAG tag (DYKDDDDK) was used as the primary antibody. Each tube cell's suspension was diluted to OD600 = 5.1 μL aliquot of antibody was added to 200 μL of cells suspension and then incubated on a shaker for 2 h at 37 °C, 80 rpm. The cells were washed three times, resuspended in PBS (pH 7.4) then 200 μL of PBS containing 1% BSA and Alexa Fluor 488 conjugated goat anti-mouse IgG antibody (10 ng μL−1) added. After incubation for 1 h at 37 °C, 80 rpm (in the dark), the cells were washed three times and resuspended in 1.5 mL PBS (pH 7.4). The cell suspension was analyzed by flow cytometry (Beckman Coulter, Fullerton, CA). Ten thousand cells per sample were counted and analyzed using Exp032 software (Beckman Coulter).2.6. Measurement of propidium iodide(PI) staining
The PI staining adopted the method as described before.43 1 mL cell resuspension (OD600 = 40) was centrifuged at 4200 g for 1 min. The cells were collected and washed three times in 10 mM PBS (pH 7.4), and the OD600 adjusted to 5.10 μL aliquot of 5 mmol PI was added to 200 μL of cell suspension then incubated on a shaker for 20 min at 37 °C. Cells were washed three times then resuspended in 1.5 mL PBS (pH 7.4). The cell suspension was analyzed by flow cytometry, as described in Section 2.5.2.7. Reactive oxygen species (ROS) detection
The fluorescent probes DHE (dihydroethidium) was used to determine levels of ROS as described in ref. 44. Briefly, cells of an OD600 of 0.4 were harvested by centrifugation and resuspended in 2 mL PBS. DHE was added to cells at a final concentration of 10 μg mL−1 and incubated at 30 °C for 30 min. Flow cytometry analysis was performed using a FACS Canto with the settings DHE red filter (600–650 nm).2.8. Measurement of methanol concentration and intracellular metabolites
The extraction of intracellular metabolites was performed as described by Pena et al.45 4 mL volume of ethanol was added to a cell suspension then heated to 85 °C, vortex-mixed for 20 s, incubated for 100 s, vortex-mixed again for 10 s then incubated for a further 80 s. After a total of approximately 3 min in the water bath, the sample was vortex-mixed for 10 s then placed onto dry ice for 3 min. Each sample was then centrifuged at 4000 g at −9 °C for 10 min after which the extract was decanted into a 15 mL tube. 1 mL volume of each extract was pipetted into a 2 mL centrifuge tube then dried using a vacuum centrifuge. Dried samples were then dissolved in 500 μL H2O for analysis by HPLC (Waters 2695) equipped with a Carbomix H-NP10:8% column (7.8 × 300 mm, 10 μm, non-porous) and a differential refraction detector (RID, Waters 2414) at a detector temperature of 30 °C. A column temperature of 55 °C was used. The mobile phase was 2.5 mM sulfuric acid at a flow rate of 0.6 mL min−1.3. Results and discussion
3.1. Construction of P. pastoris whole-cell biocatalysts
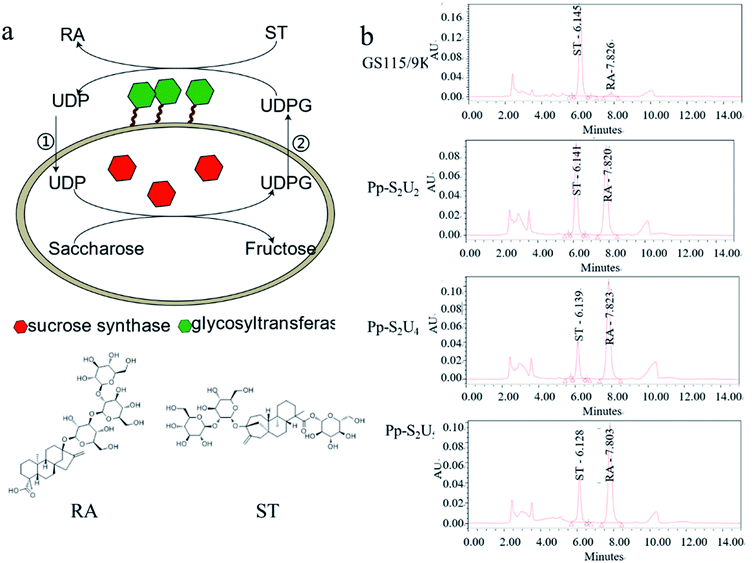
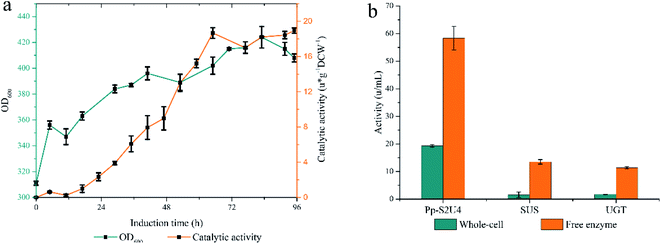
The pattern of glycosylation influences the sweetness and taste perception of steviol glycosides. As the most abundant steviol glycosides, ST can be catalyzed to synthesize RA by UGT with UDP-glucose (UDPG) as an activated sugar donor. Considering the high price of UDPG, sucrose synthase was expressed to form a cycle to regenerate UDPG in the reaction system. In this study, the UGT was displayed on the cell's surface, and SUS was intracellularly expressed together in P. pastoris to synthesize RA efficiently (Fig. 1a). Recombinant yeasts containing different copies of UGT and SUS genes have been constructed to determine the optimal combination of copy numbers, As Fig. 1b. The combination of two copies SUS and four copies UGT (Pp-S2U4) was the optional ratio for a whole-cell biocatalyst, which would be used in the subsequent research.3.2. Pp-S2U4 catalyzes RA synthesis at inefficiency due to transport-limited
Since UGT was displayed on the surface and SUS was located intracellularly of the whole-cell biocatalyst, UDP and UDPG would cross the cell barrier twice for a complete cycle. The intact cell membrane and wall were the rate-limiting factor in the circulation of reaction. The catalytic activity of whole-cell Pp-S2U4 was only 18.7 ± 0.45 μ g−1 DCW−1 after induction 60 h culture by general high-density fermentation process, which would be due to the permeability and space resistance of cells (Fig. 2a).To testify the effect of the permeability and space resistance on the catalytic activity of Pp-S2U4, Pp-S2U4 cells were broken by bead ruptor as the free enzyme to compare the activities of cascade reaction, SUS, and UGT. Comparing the activity of the free enzyme with the whole-cell biocatalyst, the catalysis activity increased to 58.3 ± 4.30 μ g−1 DCW−1 from 19.3 ± 0.45 μ g−1 DCW−1, the SUS activity increased to 13.5 ± 0.81 μ g−1 DCW−1 from 1.5 ± 1.01 μ g−1 DCW−1. The UGT activity increased from 1.7 ± 0.06 μ g−1 DCW−1 to 11.3 ± 0.42 μ g−1 DCW−1 (Fig. 2b). When the inhibition of the cell barrier was removed, free enzyme exhibited a significant increase in catalytic activities. The results indicated that the catalytic capacity of the Pp-S2U4 could be improved by lifting transport-limited.
3.3. Determination of fermentation conditions in the manufacture of Pp-S2U4 strain
As a whole-cell biocatalyst, the fermentation process of Pp-S2U4 may affect its functions and properties. The basal conditions during fermentation, including pH, temperature, and methanol concentration, were investigated in the shaker flasks. The properties of enzymes were closely related to pH. SUS was a weakly acidity enzyme with an optimum pH values of 5.5–7.5,46 and UGT was a neutral enzyme. Therefore, both optimal pH of two enzymes should be considered in the culture of the Pp-S2U4 strain. To determine the optimal pH value of fermentation process, Pp-S2U4 strain was cultured at pH values ranging from 5.0–7.5. The double-enzyme cascade reaction showed the most significant catalytic activity of the Pp-S2U4 strain were culture at pH 5.5 and 6.0, representing 30.79 ± 3.19 and 30.28 ± 0.93 μ g−1 DCW−1 after 30 h induction (Fig. 3a). Considering the optimal pH of UGT, pH 6.0 was finally selected as the proper pH condition for the culture process.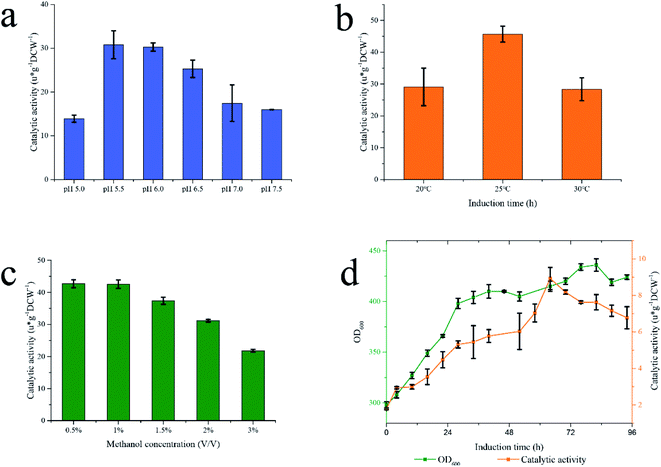 | ||
| Fig. 3 Fermentation in different conditions to determinate the general conditions. (a) pH. (b) Temperature (c) methanol concentration. (d) High-density fermentation by BMMY. | ||
The temperature would affect the yield of recombinant proteins produced by P. pastoris. Low temperatures would enhance cell viability and reduce the degradation of recombinant proteins.47,48 The maximum catalytic activity of Pp-S2U4 was 45.68 ± 2.49 μ g−1 DCW−1 cultured at 25 °C after 24 h induction (Fig. 3b), which indicated that 25 °C should be used in the following experiment.
As the source of carbon, energy, and induction agent, methanol is an essential factor in the production of recombinant proteins for P. pastoris.49 However, high methanol concentrations caused an insufficient oxygen supply that would damage the P. pastoris cells.50 To examine the effect of methanol concentration on the culture of Pp-S2U4 strain, initial methanol concentrations ranged from 0.5%–3% (v/v), pH 6.0 and 25 °C. As shown in Fig. 3c, the catalytic activity of Pp-S2U4 gradually declined with increasing methanol concentrations. The maximum catalytic activity was 42.56 ± 0.15 μ g−1 DCW−1 after 30 h induction with 0.5% (v/v) methanol concentration. The catalytic activities show insufficient difference while the methanol concentration was less than 1% (v/v), which suggested that methanol accumulation was unfavorable to Pp-S2U4 strain fermentation.
Comparing the catalytic activities of Pp-S2U4 cultured in the shake flasks and the high-density fermentation process, the shake flasks were more effective than the fermentation cylinder. The culture conditions of shake flasks fermentation and high-density fermentation process were shown in Table 1, with significant differences in methanol concentration, culture medium, and average metabolic rate. The above data indicated that high methanol concentration did not promote the catalytic activity of Pp-S2U4 (so the option of methanol concentration could be ruled out in the subsequent studies.) which allowed us to rule out the option of methanol concentration. In other words, a DO-stat feedback mode to feed methanol in scale fermentation was suitable for the Pp-S2U4 strain.
| Fermentation method | Culture medium | Methanol concentration | Average metabolic rate of methanol (g g−1 DCW−1 h−1) | pH | Temperature |
|---|---|---|---|---|---|
| High-density fermentation | BSM | DO-stat ∼0 | 0.047 ± 0.0103 | 6 | 20–30 |
| Shake-flask fermentation | BMMY | Add 1% (v/v) per 24 h 1% → 0 | 0.104 ± 0.0081 | 6 | 25 |
BMMY and BSM medium were applied in the high-density fermentation to choose a better medium (Fig. 3d). When the Pp-S2U4 strain was cultured with BMMY, the highest catalytic activity was only 8.92 ± 0.61 μ g−1 DCW−1 after 64 h induction, 46.2% of using the BSM for fermentation. These data suggested that BSM was a better culture medium than BMMY to culture the Pp-S2U4 strain in larger-scale production. BMMY was a nutrient-rich medium that could provide more nutrients for the cell to adapt to the changes and maintain cell integrity. However, the BSM was a basic salt medium that could only provide inorganic salt ions. When culture by BSM medium, cells will be under stress for all requirements need to be synthesized by themselves, which may cause defects of cell membranes or cell walls. When the cell barrier was incomplete in whole-cell biocatalyst, the substrate transport-limited may be lifted, which explained the high activity of Pp-S2U4 cultured with BSM. In summary, the culture conditions were identified as pH 6.0, 25 °C, BSM as the medium, and DO-stat feedback mode. The difference in methanol metabolic rate will be discussed in the 15 L bioreactor.
3.4. Fermentation strategies I: low initial cell densities improved the catalytic performance
Based on the results above, a new fermentation strategy was designed to study the culture conditions of Pp-S2U4 strain to increase its catalytic activity by improving the transport-limited. Four patterns a, b, c, and d, as the initial cell densities of induction were OD600 = 45.5, 86.6, 182, and 325, were performed, respectively (Fig. 4). The DO-stat feedback mode was used to make sure there was no methanol accumulation in the culture medium and the same stirring speed, ventilation rate, pressure, and methanol flow rate to make sure the same DO (Dissolved Oxygen). As shown in ESI Fig. 2,† the differences in cell concentration (d > c > b > a) among the four batches reflected differences in methanol metabolism(a > b > c > d).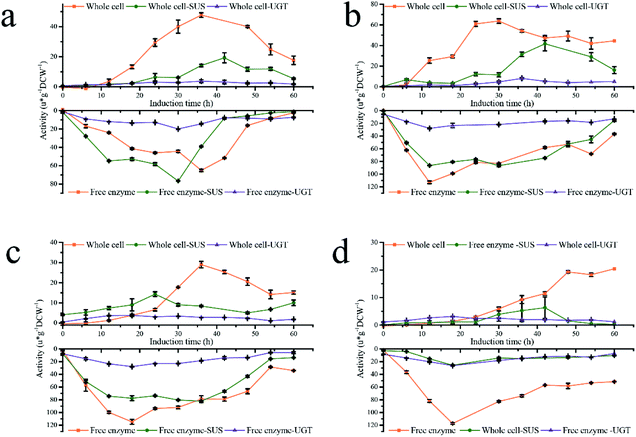 | ||
| Fig. 4 Activity curve of four fermentation strategies. (a) a pattern. (b) b pattern. (c) c pattern. (d) d pattern. | ||
Both activities of coupled enzymatic reaction or single enzyme reaction of the four patterns showed bell-shaped distribution, no matter whole-cell biocatalyst or free enzyme were used. However, the whole-cell and free enzyme acquired the activities peak at different times, and the whole-cell was always later than free enzyme due to transport-limited. The maximum catalytic activity, maximum single enzyme SUS and UGT activity in the whole-cell state were observed in the b pattern, which was 63.67 ± 2.16, 41.62 ± 6.83, 5.29 ± 0.41 μ g−1 DCW−1, respectively. However, the free enzyme had a different result (Table 2). In a free enzyme system, the transport-limited was removed entirely, and the activities of SUS and UGT were rapidly accumulated to the peak in a short time, the longest of which was only 30 h (a pattern). When the initial cell densities OD600 ≥ 86.6 (b, c, and d patterns), there was no significant difference in the activity peaks of double-enzyme coupling reaction and UGT activity, and the time nodes were almost the same in all four patterns. In a pattern, the activities of double-enzyme coupling reaction and UGT are lower than that initialed with OD600 ≥ 86.6, which indicated that low cell densities (high methanol metabolic rate) would decrease the expression level of UGT. Although the SUS enzyme activity peak of a, b, and c patterns had an insufficient difference, a significant reduction was observed in the d pattern, which showed that high cell densities (low methanol metabolic rate) would decrease the expression of SUS. The variation trend of activities was consistent at low initial cell densities (a and b patterns) for both whole-cells and free enzymes. Especially in the a pattern, the catalytic activity peaks of whole-cell and free enzyme were reached almost simultaneously. Moreover, the time gap of reaching the catalytic activity peaks of the whole-cell and the free enzyme was widened with increasing cell density because of time-inconsistent between protein production and transport-limited removal. Therefore, low initial cell densities (a, b patterns) in the fermentation process were conducive to improving the whole-cell biocatalysts Pp-S2U4.
| Pattern | Catalytic reaction | Sucrose synthase | UDP-glycosyltransferase | ||||
|---|---|---|---|---|---|---|---|
| Activity (μ g−1 DCW−1) | Induction time (h) | Activity (μ g−1 DCW−1) | Induction time (h) | Activity (μ g−1 DCW−1) | Induction time (h) | ||
| a | Whole cell | 47.76 ± 1.54 | 36 | 19.29 ± 3.35 | 42 | 3.69 ± 0.97 | 36 |
| Free enzyme | 65.09 ± 1.25 | 36 | 76.69 ± 0.133 | 30 | 14.37 ± 0.65 | 30 | |
| b | Whole cell | 63.67 ± 2.16 | 30 | 41.62 ± 6.83 | 42 | 8.10 ± 0.82 | 36 |
| Free enzyme | 112.78 ± 2.03 | 12 | 86.42 ± 0.44 | 12 | 27.89 ± 0.62 | 12 | |
| c | Whole cell | 29.00 ± 1.61 | 36 | 14.3 ± 1.17 | 24 | 3.81 ± 0.09 | 18 |
| Free enzyme | 114.81 ± 3.97 | 18 | 82.16 ± 0.40 | 18 | 27.81 ± 0.46 | 18 | |
| d | Whole cell | 20.44 ± 0.26 | 60 | 6.30 ± 3.88 | 60 | 2.50 ± 0.08 | 60 |
| Free enzyme | 117.39 ± 1.33 | 18 | 25.74 ± 0.83 | 18 | 26.27 ± 0.49 | 18 | |
Compared the activities ratio of whole-cell to the free enzyme (lyse-cell) (WC/LC) in four patterns (ESI Fig. 4†), the ratio value of SUS, UGT, and double-enzyme represented the degree of elimination of permeability limited, space resistance, and total transport-limited, respectively. The more limitations were removed, the higher value of the ratio. The WC/LC of SUS, UGT, and double-enzyme was gradually increased with the fermentation process, and the a and b patterns were greater than the c and d patterns. All results show that low initial cell densities were beneficial to remove transport-limited. Considering enzymes' production and transport-limited removal, the b pattern was the optimal fermentation process in our research.
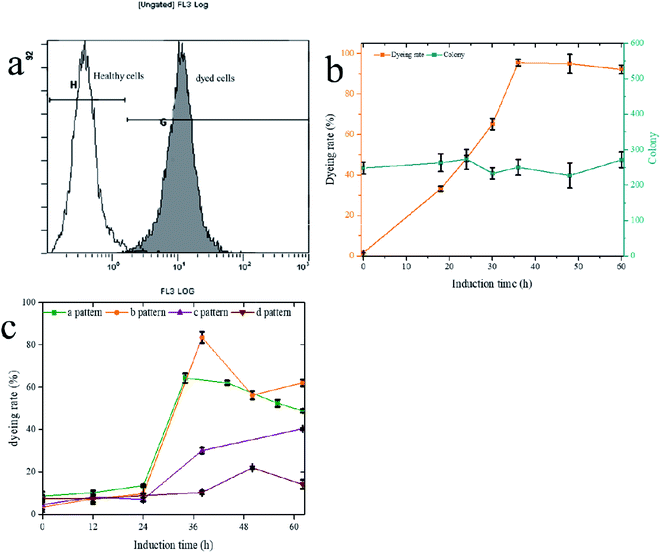
PI staining is often used to identify cell viability, which cannot pass through the membranes of living cells to dye the nucleoplasm. Therefore, the healthy and PI staining cells have two different signals, which is the basis for the flow cytometry test of viability (Fig. 5a). However, the PI staining cells were not necessarily dead, and the method also can be used to identify the cell integrity.43,51 AS verified by plate counts (Fig. 5b), the number of living cells did not decrease when the staining signal was increased. The change of cell surface structure led to cell perforation, allowing PI to enter and stain the chromosomes. The trend of stained perforated cells increased with the induction time extension, and the highest rate (b pattern) reached 83% (Fig. 5c). Low initial cell densities (a and b patterns) had a higher proportion of PI staining cells than high initial cell densities (c and d patterns), indicating that transport-limited was removed more thoroughly for low initial cell densities. The higher the numerical PI staining cells, the less restriction was evident, showing PI-staining could be used to characterize transport-limited and quickly determine the termination time of fermentation.
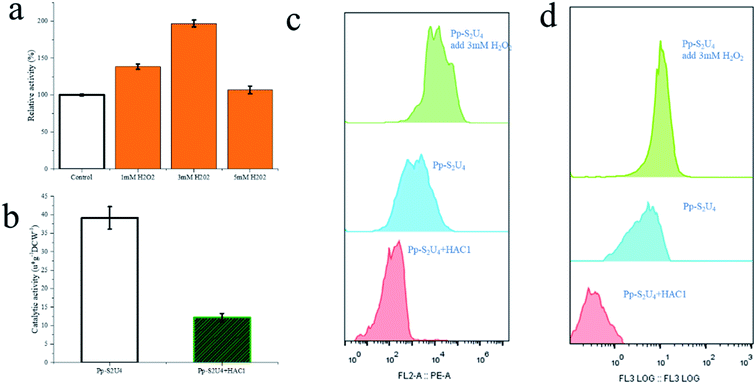
The methanol feed rate was a fixed value, and methanol would not accumulation in fermentation broth (ESI Fig. 5†), which indicated lower cell densities led to a faster rate of methanol metabolism. A faster methanol metabolic rate would accelerate energy metabolism and produce protein speed, which would also accumulate more reactive oxide species (ROS).52–55 The SUS and UGT have had a similar expression level in four patterns show that an appropriate methanol metabolic rate does not affect protein production but may damage the cell membranes and wall by ROS product by itself. The accumulation of ROS, causing damage to the cell wall and cytomembrane,52 indicated cell wall stress. Experiments were designed to verify this hypothesis (Fig. 6). P. pastoris Pp-S2U4 was first cultured in a 15 L bioreactor after 12 h induction, then incubated in 0.1 M PBS (pH 6.0) containing different concentrations of H2O2 (Fig. 6a). Cells of Pp-S2U4 were treated with H2O2 at 1 mM, 3 mM, and 5 mM to improve the catalytic activities. A 1.96 ± 0.05 folds increase in RA production was observed in cells treated with 3 mM H2O2. PI staining results indicated that the cell permeability was also improved (Fig. 6d).
To further demonstrate that the cell wall and cytomembrane damages were caused by ROS result in the improvement of catalytic activity, transcription factor HAC1 was expressed in Pp-S2U4. As a transcription factor, HAC1 could alleviate ER stress, and trigger unfolded protein response (UPR), which would reduce ROS production.56,57 Compared to Pp-S2U4, a 69% catalytic activity decrease and reduced ROS levels were both observed in the HAC1-expressing strain (Fig. 6c and d). This result suggested that the decrease of ROS led to the trans-limited increase, reducing the catalytic activity of the HAC1-expressing strain.
To summarize, ROS accumulation would damage the cell wall and cytomembrane, improving catalytic activity. The fermentation strategy to initial with low intensity of cells accelerated methanol metabolism, which would increase the ROS production, could be an effective solution to improve whole-cell biocatalysts.
3.5. Fermentation strategies II: high rapid metabolism of methanol improved the catalytic performance
To further investigate this fermentation process, a second fermentation strategy based on the strategy I was proposed. Strategy II explored the effect of methanol flow rate (1, 2, 3, and 4 g L−1 h−1) on the catalytic efficiency of Pp-S2U4 cultured in a 15 L bioreactor (Fig. 7a). As displayed in Fig. 7b, a higher methanol feed rate led to faster cell growth. The catalytic activities curve of Pp-S2U4 was presented in Fig. 7c, where the maximum catalytic activity was obtained at 58 h, 42 h, 30 h, and 30 h for methanol flow rates of 1, 2, 3, and 4 g L−1 h−1, respectively. The activities were 42.42 ± 1.10, 58.44 ± 1.17, 63.12 ± 2.3, and 54.26 ± 1.98 μ g−1 DCW−1. With increasing methanol flow rate, the induction time of acquiring the maximum catalytic activity of Pp-S2U4 was gradually decreased from 58 h at 1 g L−1 h−1 to 30 h at 4 g L−1 h−1. This result was similar to the strategy I, where the induction time decreased as the methanol consumption rate increased.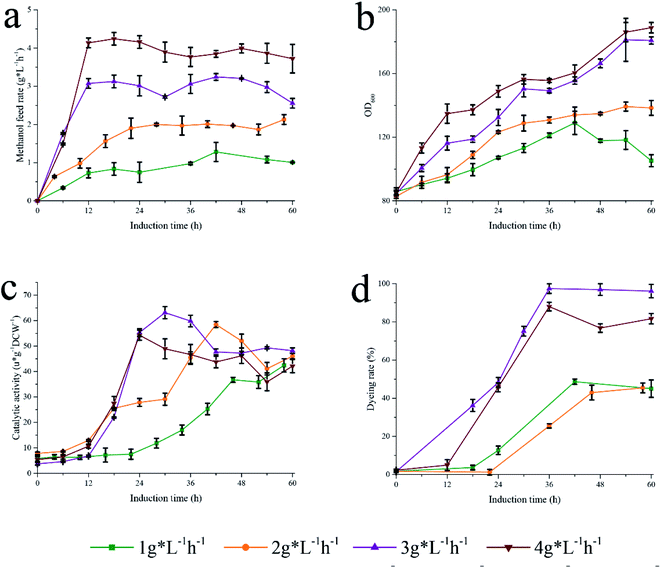 | ||
| Fig. 7 Different methanol feed rate to induce Pp-S2U4. (a) Methanol feed rate. (b) Growth curve. (c) Catalytic activity. (d) PI dyeing rate. | ||
Furthermore, the PI staining of cells was examined after fermentation with strategy II (Fig. 7d). The proportion of PI staining cells to total cells fermented with high methanol consumption was significantly higher than that of cells with low methanol consumption, indicating that rapid methanol consumption increased cell permeability and changed the cell surface properties.
In general, stable cell surface characteristics to regulate substrate permeability and protein production would be achieved by maintaining appropriate methanol flow. However, under different patterns, the fermentation conditions of Pp-S2U4 did not change except for the metabolic rate of methanol. No methanol accumulation was observed, suggesting that the flow of methanol was equal to the consumption. The rapid metabolism of methanol led to an improvement in the surface properties of Pp-S2U4.
3.6. Compared to permeabilizing reagent addition and ultrasonicationtreatment
To demonstrate the advantages of fermentation strategies, the permeabilizing reagents and ultrasonication were used to improve transport-limited of Pp-S2U4 (Fig. 8). Several usual permeabilizing reagents, SDS, Triton-X100, Tween-20, Tween-80, polyethylene glycol 4000(PG-4000), and Sarcosine were added as 0.1% (v/v) and 0.5% (v/v) in Pp-S2U4 for 10 min (Fig. 8a). The catalytic activities were maintained at 85.7 ± 1.85% to 106.1 ± 0.85% of the untreated cell with all the reagents except for SDSs. However, Pp-S2U4 treated with SDS hardly had any catalytic activity, probably because of too much damage to the cellular structure.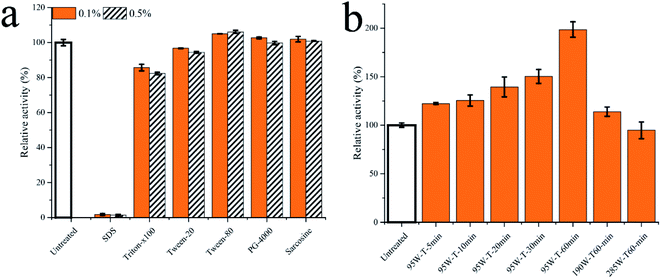 | ||
| Fig. 8 The Pp-S2U4 was treated by permeabilizing reagent and ultrasonication. (a) treated by permeabilizing reagent. (b) treated by ultrasonication. | ||
Ultrasonication is effective in improving surface properties. Pp-S2U4 was treated with different strengths and time lengths of ultrasonication treatment (Fig. 8b). The catalytic activities of all whole-cell biocatalysts improved after ultrasonic treatment. When Pp-S2U4 was treated by 95 W ultrasonication for 1 h, the highest 1.98 ± 0.08 folds increase was observed.
Compared to permeabilizing reagent addition and ultrasonication treatment, improving the catalytic activity of Pp-S2U4 by applying the fermentation strategies I and II had a superior 2.62 ± 0.06 folds increase. With the advantages of short induction time (60 h to 36 h), simple operation, and no extra addition, the fermentation process proposed in this study benefited from reducing the cost and making the production process greener.
4. Conclusion
In this study, the effect of the fermentation process on the P. pastoris whole-cell biocatalyst was investigated and compared with the traditional methods. The catalytic activity of the Pp-S2U4 increased 1.62 folds, and the induction time decreased to 36 h by adjusting initial cell density. A similar result was obtained by changing the methanol flow rate, indicating that the rapid methanol consumption rate was an important factor affecting P. pastoris whole-cell biocatalysts. Compared with permeabilizing reagent treatment and ultrasonication treatment, the fermentation process showed significant advantages that would help to reduce the cost and make the production process greener. In summary, an appropriate fermentation process could improve the efficiency of P. pastoris whole-cell biocatalysts for RA synthesis, which can reduce the fermentation time and minimize the subsequent process.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We are grateful to the National Key Research and Development Program (2018YFA0901700), the National Natural Science Foundation of China (31871739).References
- J. F. Clos, G. E. DuBois and I. Prakash, J. Agric. Food Chem., 2008, 56, 8507–8513 CrossRef CAS PubMed.
- U. Wölwer-Rieck, W. Tomberg and A. Wawrzun, J. Agric. Food Chem., 2010, 58, 12216–12220 CrossRef PubMed.
- V. Chatsudthipong and C. Muanprasat, Pharmacol. Ther., 2009, 121, 41–54 CrossRef CAS PubMed.
- S. K. Goyal, Samsher and R. K. Goyal, Int. J. Food Sci. Nutr., 2010, 61, 1–10 CrossRef CAS PubMed.
- U. Wölwer-Rieck, J. Agric. Food Chem., 2012, 60, 886–895 CrossRef PubMed.
- J. E. Brandle, A. N. Starratt and M. Gijzen, Can. J. Plant Sci., 1998, 78, 527–536 CrossRef CAS.
- X.-Y. Huang, J.-F. Fu and D.-L. Di, Sep. Purif. Technol., 2010, 71, 220–224 CrossRef CAS.
- N. Abu Samah, A. Hisham and S. Rahim, Determination of Stevioside and Rebaudioside A in Stevia Rebaudiana Leaves via preparative High Performance Liquid Chromatography (prep-HPLC), 2013 Search PubMed.
- J. Li, Z. Chen and D. Di, Food Chem., 2012, 132, 268–276 CrossRef CAS PubMed.
- P. Tufvesson, J. Lima-Ramos, M. Nordblad and J. M. Woodley, Org. Process Res. Dev., 2011, 15, 266–274 CrossRef CAS.
- F. Zhang, C. T. Zhu, Q. M. Peng, F. Q. Wang, S. Sheng, Q. Y. Wu and J. Wang, J. Chem. Technol. Biotechnol., 2020, 95, 384–393 CrossRef CAS.
- Y. D. Hu, X. B. Liu, A. T. M. Ren, J. D. Gu and B. Cao, Chemsuschem, 2019, 12, 5142–5148 CrossRef CAS PubMed.
- C. Xiros, R. L. Shahab and M. H. P. Studer, Appl. Microbiol. Biotechnol., 2019, 103, 3355–3365 CrossRef CAS PubMed.
- J. Wachtmeister and D. Rother, Curr. Opin. Biotechnol., 2016, 42, 169–177 CrossRef CAS PubMed.
- B. Yan, Y. W. Ai, Q. Sun, Y. Ma, Y. Cao, J. W. Wang, Z. Y. Zhang and X. D. Wang, Mol. Cell, 2021, 81, 355–369 CrossRef CAS PubMed.
- S. Kato, A. Onoda, A. R. Grimm, U. Schwaneberg and T. Hayashi, J. Inorg. Biochem., 2021, 216 Search PubMed.
- T. Huang, F. Zhang, B. Wang, W. S. Ye, Q. M. Peng, F. A. Wu and J. Wang, Waste Biomass Valorization, 2020, 11, 5321–5332 CrossRef CAS.
- J. Son, J. H. Jang, I. H. Choi, C. G. Lim, E. J. Jeon, H. B. Bang and K. J. Jeong, Microb. Cell Factories, 2021, 20 Search PubMed.
- A. S. Wells, G. L. Finch, P. C. Michels and J. W. Wong, Org. Process Res. Dev., 2012, 16, 1986–1993 CrossRef CAS.
- G. L. Rosano and E. A. Ceccarelli, Front. Microbiol., 2014, 5, 172 Search PubMed.
- Z. Dai and J. Nielsen, Curr. Opin. Biotechnol., 2015, 36, 8–15 CrossRef CAS PubMed.
- J. R. Bober and N. U. Nair, Nat. Commun., 2019, 10, 4548 CrossRef PubMed.
- P. Cheng, R. Shan, H. R. Yuan, L. F. Deng and Y. Chen, Bioresour. Technol., 2018, 267, 774–777 CrossRef CAS PubMed.
- A. De León, B. García, A. P. Barba de la Rosa, F. Villaseñor, A. Estrada and R. López-Revilla, Process Biochem., 2003, 39, 301–305 CrossRef.
- M. Malik, A. Ganguli and M. Ghosh, Appl. Microbiol. Biotechnol., 2012, 95, 223–231 CrossRef CAS PubMed.
- H. B. Shen, X. Y. Yong, Y. L. Chen, Z. H. Liao, R. W. Si, J. Zhou, S. Y. Wang, Y. C. Yong, P. K. OuYang and T. Zheng, Bioresour. Technol., 2014, 167, 490–494 CrossRef CAS PubMed.
- J.-Y. Yun, J.-E. Lee, K.-M. Yang, S. Cho, A. Kim, Y.-E. Kwon and J.-B. Park, Bioprocess Biosyst. Eng., 2012, 35, 211–216 CrossRef CAS PubMed.
- G. Lourinho and P. Brito, Rev. Environ. Sci. Biotechnol., 2015, 14, 287–316 CrossRef CAS.
- M. Cánovas, T. Torroglosa, H.-P. Kleber and J. L. Iborra, J. Basic Microbiol., 2003, 43, 259–268 CrossRef PubMed.
- Y. Numanoğlu and S. Sungur, Process Biochem., 2004, 39, 705–711 CrossRef.
- T. Shi, X. G. Fan, Y. S. Wu, Q. Ma, Q. Y. Xu, X. X. Xie and N. Chen, Biotechnol. Biotechnol. Equip., 2020, 34, 38–47 CrossRef CAS.
- D. B. Kell, N. Swainston, P. Pir and S. G. Oliver, Trends Biotechnol., 2015, 33, 237–246 CrossRef CAS PubMed.
- R. R. Chen and X. Guo, US Pat., US20110009291, 2011 Search PubMed.
- J. Xue, Y. Wu, K. Shi, X. Xiao, Y. Gao, L. Li and Y. Qiao, Bioresour. Technol., 2019, 280, 88–94 CrossRef CAS PubMed.
- J. L. Zhou, X. B. Liu, F. Yuan, B. Deng and X. B. Yu, ACS Sustain. Chem. Eng., 2020, 8, 4781–4791 CrossRef CAS.
- J. Yan, X. Zheng and S. Li, Bioresour. Technol., 2014, 151, 43–48 CrossRef CAS PubMed.
- Z. Jin, S. Liang, X. Zhang, S. Han, C. Ren, Y. Lin and S. Zheng, Biotechnol. Bioprocess Eng., 2013, 18, 365–374 CrossRef CAS.
- Y. Liu, L. Huang, Y. Fu, D. Zheng, J. Ma, Y. Li, Z. Xu and F. Lu, Food Chem., 2019, 274, 535–542 CrossRef CAS PubMed.
- N. Zhao, Y. Xu, K. Wang and S. Zheng, J. Agric. Food Chem., 2017, 65, 9468–9474 CrossRef CAS PubMed.
- Z. Y. Wang, J. F. Hong, S. Y. Ma, T. Huang, Y. Y. Ma, W. Liu, W. B. Liu, Z. M. Liu and H. Song, Int. J. Biol. Macromol., 2020, 163, 1669–1676 CrossRef CAS PubMed.
- M. Q. Chen, X. Zeng, Q. J. Zhu, D. G. Wang, S. Y. Han, S. L. Liang and Y. Lin, Biochem. Eng. J., 2021, 175 Search PubMed.
- P. Wang, L. Zhang, R. Fisher, M. Chen, S. Liang, S. Han, S. Zheng, H. Sui and Y. Lin, J. Ind. Microbiol. Biotechnol., 2017, 44, 1355–1365 CrossRef CAS PubMed.
- N. Zhang, Y. X. Fan, C. Li, Q. M. Wang, N. Leksawasdi, F. L. Li and S. A. Wang, Appl. Microbiol. Biotechnol., 2018, 102, 4183–4191 CrossRef CAS PubMed.
- M. Delic, A. B. Graf, G. Koellensperger, C. Haberhauer-Troyer, S. Hann, D. Mattanovich and B. Gasser, Microbial Cell., 2014, 1, 376–386 CrossRef CAS PubMed.
- D. A. Pena, B. Gasser, J. Zanghellini, M. G. Steiger and D. Mattanovich, Metab. Eng., 2018, 50, 2–15 CrossRef CAS PubMed.
- K. Schmolzer, A. Gutmann, M. Diricks, T. Desmet and B. Nidetzky, Biotechnol. Adv., 2016, 34, 88–111 CrossRef PubMed.
- F. Hong, N. Q. Meinander and L. J. Jönsson, Biotechnol. Bioeng., 2002, 79, 438–449 CrossRef CAS PubMed.
- P. Laurent, L. Buchon, J. F. Guespin-Michel and N. Orange, Appl. Environ. Microbiol., 2000, 66, 1538 CrossRef CAS PubMed.
- Z. Wang, Y. Wang, D. Zhang, J. Li, Z. Hua, G. Du and J. Chen, Bioresour. Technol., 2010, 101, 1318–1323 CrossRef CAS PubMed.
- O. Trentmann, N. K. Khatri and F. Hoffmann, Biotechnol. Prog., 2004, 20, 1766–1775 CrossRef CAS PubMed.
- H. Mendoza, L. Carmona, P. Assuncao, K. Freijanes, A. de la Jara, E. Portillo and A. Torres, Bioresour. Technol., 2015, 197, 369–374 CrossRef CAS PubMed.
- V. Adam-Vizi, Antioxid. Redox Signaling, 2005, 7, 1140–1149 CrossRef CAS PubMed.
- A. J. Kowaltowski, N. C. de Souza-Pinto, R. F. Castilho and A. E. Vercesi, Free Radic. Biol. Med., 2009, 47, 333–343 CrossRef CAS PubMed.
- J. Wang, K. A. Pareja, C. A. Kaiser and C. S. Sevier, eLife, 2014, 3, 1–21 CrossRef PubMed.
- N. E. Polyakov, T. V. Leshina, T. A. Konovalova and L. D. Kispert, Free Radic. Biol. Med., 2001, 31, 398–404 CrossRef CAS PubMed.
- D. M. Tran, Y. Ishiwata-Kimata, T. C. Mai, M. Kubo and Y. Kimata, Sci. Rep., 2019, 9, 12780 CrossRef PubMed.
- P. K. Babele, P. K. Thakre, R. Kumawat and R. S. Tomar, Chemosphere, 2018, 213, 65–75 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra06253k |
| This journal is © The Royal Society of Chemistry 2021 |