Open Access Article
Open Access ArticleVancomycin conjugated iron oxide nanoparticles for magnetic targeting and efficient capture of Gram-positive and Gram-negative bacteria
Mehnaz Rashida,
Md. Ahasanur Rabbia,
Tabassum Arab,
Md. Motahar Hossaina,
Md. Shahidul Islama,
Abdelhamid Elaissaric,
Hasan Ahmad *a and
Md. Mahbubor Rahman
*a and
Md. Mahbubor Rahman *a
*a
aPolymer Colloids & Nanomaterials (PCN) Group, Department of Chemistry, Faculty of Science, University of Rajshahi, Rajshahi 6205, Bangladesh. E-mail: samarhass@yahoo.com; mrchem@ru.ac.bd
bDepartment of Biochemistry and Molecular Biology, Faculty of Science, University of Rajshahi, Rajshahi 6205, Bangladesh
cUniversité Lyon, University Claude Bernard Lyon-1, CNRS, ISA-UMR 5280, Lyon F-69622, France
First published on 10th November 2021
Abstract
Drug conjugated iron oxide magnetite (Fe3O4) nanoparticles are of great interest in the field of biomedicine. In this study, vancomycin (Van) conjugated magnetite (Fe3O4) nanoparticles were envisioned to capture and inhibit the growth of bacteria. Hydrophobic Fe3O4 nanoparticles were synthesized by using co-precipitation of ferrous (Fe2+) and ferric (Fe3+) ions following a surface modification step with oleic acid as stabilizers. Thereafter, a ligand exchange technique was employed to displace oleic acid with hydrophilic dopamine (DOPA) molecules which have a catechol group for anchoring to the iron oxide surface to prepare water dispersible nanoparticles. The surface of the resulting Fe3O4/DOPA nanoparticles contains amino (–NH2) groups that are conjugated with vancomycin via a coupling reaction between the –NH2 group of dopamine and the –COOH group of vancomycin. The prepared vancomycin conjugated Fe3O4/DOPA nanoparticles were named Fe3O4/DOPA/Van and exhibited a magnetic response to an external magnetic field due to the presence of magnetite Fe3O4 in the core. The Fe3O4/DOPA/Van nanoparticles showed bactericidal activity against both Gram positive Bacillus subtilis (B. subtilis) and Streptococcus and Gram-negative bacteria Escherichia coli (E. coli). Maximum inhibition zones of 22 mm, 19 mm and 18 mm were found against B. subtilis, Streptococcus and E. coli respectively. Most importantly, the vancomycin conjugated nanoparticles were effectively bound to the cell wall of the bacteria, promoting bacterial separation and growth inhibition. Therefore, the prepared Fe3O4/DOPA/Van nanoparticles can be promising for effective bacterial separation and killing in the dispersion media.
1. Introduction
Infections caused by drug resistant bacteria (also known as ‘superbugs’) in community places and hospitals have prompted growing concern all over the world.1,2 Limited options remain for treating infections caused by such resistant bacteria. Vancomycin as an antibiotic has been used for treating bacterial infection for many years. This glycopeptide antibiotic binds to the terminal D-alanyl-D-alanine residues of lipid II and nascent peptidoglycan cell wall precursors, thus blocking the bacterial transglucosylases and transpeptidases required for peptidoglycan biosynthesis and cross-linking.3,4 As the bacterial cell wall of Gram-positive bacteria is composed largely of peptidoglycan, vancomycin and the related glycopeptide antibiotics prevent cell division and are bacteriostatic in action.5 Despite of the potential for the retardation of bacterial growth, unfortunately, the use of free vancomycin is less efficient for site specific applications. The drug delivery via a vector to an infection site is more efficient as free drug distributes/diffuses randomly all over the body including the infection site.6,7 The use of free drug also requires higher dose which may cause side effects and toxicity, needs to be reduced by using a carrier platform. In this respect, the antibiotics loaded on nanoparticles or into polymer matrix could be a promising approach for reducing toxicity, and side effects due to greater access and accumulation of the active molecules at the infection site.8Iron oxide nanoparticles (IONPs) have attracted an extensive attention because of their potential applications in biomedical field for magnetic resonance imaging (MRI), magnetic hyperthermia, cancer therapy, drug delivery and biomolecule separation.9–13 However, bare iron oxide nanoparticles have limitations that include colloidal instability, leaching out, toxicity, and degradation in biological medium.14 Therefore, different efforts have been devoted for coating and surface modification of IONPs with different materials such as polymers, silica, and ligand molecules, dextran, and surfactants.15,16 Specifically, Nahar et al. reported poly(N-isopropyl acrylamide-co-acrylic acid) polymer coated IONPs for dual temperature and pH responsive magnetic nanocomposites synthesis.17 In a recent work, Wang and his coworkers synthesized amphiphilic thermally sensitive block helical poly(phenyl isocyanide) polymer to prepare magnetic micelle complex for temperature-controlled drug delivery.18 Polymer coating on IONPs also reduces the generation of reactive oxygen species (ROS) which may cause cellular degradation.19 Besides, silica coating on IONPs has been promising because of its biocompatibility, and ease of further functionalization with other polymers, and molecules.20,21 In addition to that, the surface of IONPs have also been modified with some other organic ligands, e.g., catechol and phosphocholine group containing molecules, that have very high affinity to anchor with metal oxide nanoparticles' surface.22 Dopamine has emerged great interest as capping agent for IONPs due to the stability and strength of the resultant five-membered metallocycle chelate and the ease at which it can be functionalized through amide bonds with other molecules of interest.23–25
The major advantages of drug loaded magnetic nanoparticles are easy accumulation and separation from a dispersion media by using an external magnet. Nonetheless, most of the literatures related to antibacterial IONPs reported mainly antibacterial property of nanoparticles. For example, Pimpha et al. recently reported the preparation of cationic polymer coated IONPs for antimicrobial activity.26 Besides, antibacterial polyguanidine coated magnetite (Fe3O4) nanoparticles have been synthesized for antibacterial activity study against E. coli.27 Lee et al. reported the development of an interesting technique to separate bacteria from blood using magnetic nanoparticles modified with synthetic ligand.28 Likewise, magnetic nanoparticles coated with different polymers have also been reported for bacteria separation.29,30 In addition to that, special attention for the development of very promising IONPs for both capture and enrichment of bacteria has been envisioned.31,32 Despite of the advantages of IONPs for bacteria capture and detection, separation of the remaining living bacteria is a major drawback in detoxification purposes like decontamination of water or targeted killing of bacteria localized inside a particular organ. Therefore, it is desired to envision nanoparticles for simultaneous capture and killing of bacteria from the system to be disinfected. For this purpose, surface of Ag@Fe2O3 yolk–shell nanoparticle conjugated with glucose via dopamine anchor have been developed.33 Instead of glucose molecule conjugation, antibacterial drug can be more effective for bacteria separation and killing as reported earlier.34 More than a decade ago, Kell et al. reported an extensive study on the multi-step preparation of a series of vancomycin modified silica encapsulated IONPs to isolate a variety of bacteria from aqueous solution.35 This is understandable that silica encapsulation of IONPs favored easy functionalization required for anchoring vancomycin, but such practice generally reduces the magnetic property. Interestingly, vancomycin architecture and length of linker between nanoparticles and vancomycin showed significant effect on the magnetic confinement of pathogens. For comparison, commercially available 2.8 μm sized magnetic Dynal beads were also used for modification with vancomycin. Though in neither case experimental evidence in support of magnetic property is provided. Recently, Chen et al., and Zhang et al., reported the potential of vancomycin modified magnetic nanoparticles for enhancing activity against Clostridium difficile, and Staphylococcus aureus and Escherichia coli respectively.36,37 In the former report,36 prostate specific membrane antigen (PSMA) was immobilized on oleic acid coated IONPs for conjugation with vancomycin. The mode of interaction of antigen with oleic acid coated IONPs is not clarified as weaker interaction may initiate leakage of immobilized vancomycin. In the latter one, Zhang and his coworkers developed a magnetic hybrid system comprising polyvinyl alcohol layered silica coated IONPs for entrapping vancomycin and the hybrid was further equipped with cell-penetrating haxapepetide using a multistep preparation method which is very laborious and time consuming.
In this work, antibacterial vancomycin conjugated IONPs synthesis via a facile multiple ligand–receptors interaction is reported for efficient separation and immediate killing of bacteria. In the first step oleic acid is displaced by hydrophilic dopamine (DOPA) molecules and then vancomycin is chemically bonded through amide linkage. The positive side of this facile synthetic route is that the process is easily scalable, environment-friendly, requires less hazardous chemicals and overall, the prepared antibacterial vancomycin conjugated IONPs would retain almost the same initial magnetic property and be biocompatible. Additionally, the hydrophilic vancomycin conjugation would improve water dispersibility and hence, would increase contact area between IONPs and bacteria, which is advantageous for enhanced antibacterial activity.
2. Experimental
2.1 Materials
Ferrous chloride tetrahydrate (FeCl2·4H2O), ferric chloride hexahydrate (FeCl3·6H2O), sodium hydroxide, dopamine hydrochloride (DOPA), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) acid were obtained from Sigma Aldrich, USA and used as received. 25% ammonium hydroxide (NH4OH), acetone, potassium phosphate monobasic and sodium phosphate dibasic were purchased from Acros organics, Belgium. Oleic acid was obtained from Fluka. Disinfectol and chloroform were obtained from Chem-lab, Belgium. Vancomycin hydrochloride was obtained from Aldrich, Belgium. Three different bacterial strains namely Bacillus subtilis, Streptococcus and Escherichia coli were obtained from the Department of Biochemistry & Molecular Biology, Rajshahi University. The strains were preserved at 4 °C on agar slant and sub-cultured at 37 °C for 24 hours on nutrient broth media.2.2 Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of Fe3+
1 molar ratio of Fe3+![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Fe2+. Ferric and ferrous chloride were dissolved in 100 mL of water and heated to 80 °C followed by the addition of 15 mL of ammonium hydroxide, and the reaction was kept under mechanical stirring for 1 hour at ambient condition. Oleic acid (10 mL) was added to the reaction system to introduce a coating of oleic acid on magnetite nanoparticles denoted as Fe3O4-OA, and the reaction mixture was mechanically stirred for another 2 hours. Finally, the nanoparticles were purified by five times washing with water and ethanol, and the particles were dried and stored for further use.
Fe2+. Ferric and ferrous chloride were dissolved in 100 mL of water and heated to 80 °C followed by the addition of 15 mL of ammonium hydroxide, and the reaction was kept under mechanical stirring for 1 hour at ambient condition. Oleic acid (10 mL) was added to the reaction system to introduce a coating of oleic acid on magnetite nanoparticles denoted as Fe3O4-OA, and the reaction mixture was mechanically stirred for another 2 hours. Finally, the nanoparticles were purified by five times washing with water and ethanol, and the particles were dried and stored for further use.2.3 Characterization of the magnetic Fe3O4/DOPA/Van nanoparticles
The surface modification was confirmed with FTIR on a PerkinElmer Frontier FT-IR instrument in conjunction with a MKII Golden Gate set-up equipped with a diamond crystal. The size, shape and overall morphology of the NPs were evaluated with dynamic light scattering (DLS) technique and transmission electron microscopy (TEM). DLS was performed on an Autosizer 4800 Spectrometer from Malvern Instruments and TEM images were obtained with a Phillips CM120 microscope (CMEABG, University of Claude Bernard Lyon I, Villeurbanne, France). Magnetic behavior of the nanoparticles was investigated by vibrating sample magnetometer (VSM) at room temperature on the automatic bench of magnetic measurements at CNRS-IRCE Lyon, France. XRD measurements were carried out on a Bruker (Germany) D8 ADVANCE for the analysis of nanoparticle structures. Powder samples were measured using Cu Kα radiation (λ ≈ 1.5406 Å), AQ6 a tube voltage of 33 kV and a tube current of 45 mA. The intensities were measured at 2θ values from 10° to 90° at a continuous scan rate of 1° min−1 with a position-sensitive detector aperture at 25 °C. The obtained patterns were further processed by semi-quantitative phase analysis software to eliminate the noises to identify peaks.2.4 Antibacterial activity study
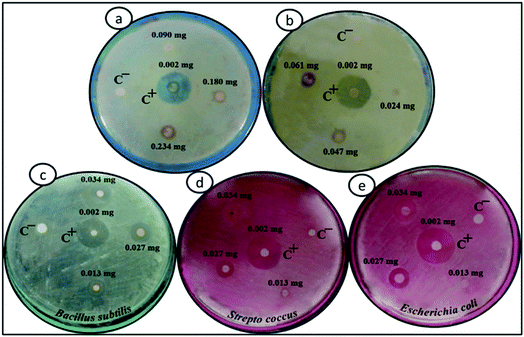
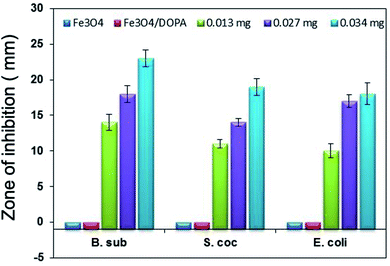
The antibacterial activities of Fe3O4, Fe3O4/DOPA and Fe3O4/DOPA/Van nanoparticles were measured by a disc diffusion assay method. Three different microbial pathogens, Gram positive (B. subtilis, S. aureus) and Gram negative (E. coli) bacteria were tested in this study. Typically, 60 μL of bacterial solution was spread homogeneously on the surface of solid nutrient agar media in sterilized Petri dish. Sterile filter paper disks (10 mm) were then placed on the surface of cultured test bacteria at five different spots. Three different volumes i.e., 5, 10 and 15 μL of Fe3O4/DOPA/Van nanoparticles suspension (2.5 mg mL−1) were dropped on filter paper disk by a micropipette. 10 μL of vancomycin solution (0.002 mg mL−1) was dropped for positive control and 10 μL phosphate buffered saline (PBS) was dropped for negative control experiment. In addition to that, bare Fe3O4 and dopamine modified Fe3O4 nanoparticles were also tested to compare with the Fe3O4/DOPA/Van nanoparticles. The plates were then placed in a refrigerator at 4 °C for 4 hours to permit sample diffusion and then incubated at 37 ± 1 °C for 24 hours to allow bacterial growth. Each experiment was carried out in triplicates and diameter (mm) of zone of inhibition across each filter paper disk was measured. Minimum inhibitory concentrations (MIC) of Fe3O4/DOPA/Van against three pathogens were also measured.2.5 Bacteria separation test
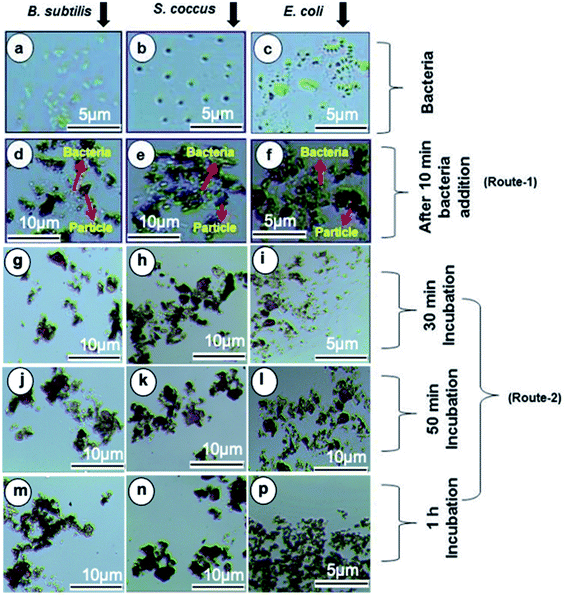
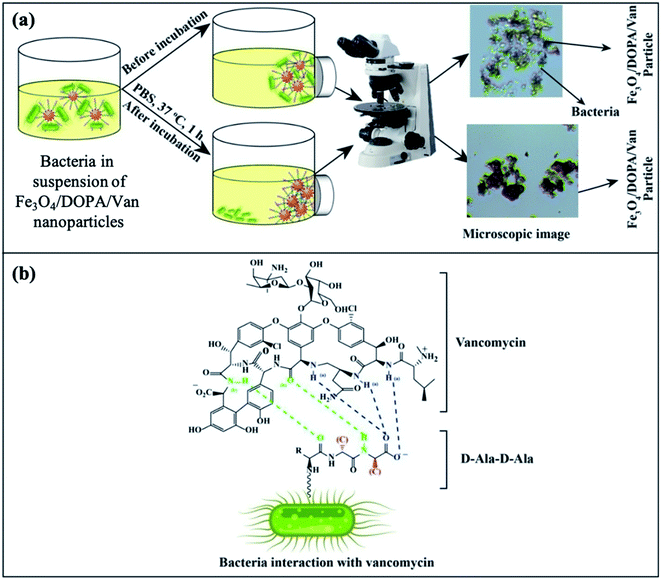
The Gram positive and Gram-negative bacteria (concentration, 1 × 107 CFU per mL) were grown at 37 °C for 24 hours in broth media and then, optical microscopy images of the respective bacteria were taken. The Fe3O4/DOPA/Van nanoparticles were suspended in PBS, (0.01 M, pH 6–7 buffer) to prepare three different 2.65 mg mL−1, 1.325 mg mL−1, and 0.6625 mg mL−1 suspensions. Then 5 mL of each prepared suspensions was taken in test tube. 10 μL of each bacterial stain was added in each test tube and allow the mixture to stand for 10 minutes. Then quickly an external magnet was applied for the separation of bacteria attached to the nanoparticles. After washing twice, the magnetic nanoparticles were resuspended in PBS for optical microscopic analysis by N-800M (China).3. Results and discussion
3.1 Characterization of Fe3O4-OA nanoparticles
The synthesis, surface modification and vancomycin conjugation of IONPs steps are schematically illustrated in Scheme 1. Hydrophobic Fe3O4-OA were synthesized by the coating of oleic acid on Fe3O4 and dispersible in organic solvent, e.g., chloroform, as shown in the digital photograph (upper rightmost) of Scheme 1. Oleic acid ligand exchange was performed to coat Fe3O4 with hydrophilic DOPA to obtain water dispersible hydrophilic Fe3O4-DOPA nanoparticles (lower rightmost). The partitioning of nanoparticles from chloroform to water medium indicated successful ligand exchange from OA on Fe3O4-OA to DOPA on Fe3O4-DOPA nanoparticles. Amine functional group of DOPA was introduced on Fe3O4 to conjugate vancomycin via carbodiimide reaction using EDC and NHS.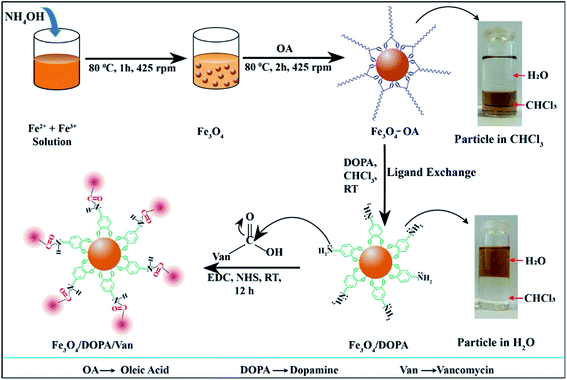 | ||
| Scheme 1 Schematic illustration for the synthesis, oleic acid coating, ligand exchange and vancomycin conjugation of iron oxide nanoparticles to obtain Fe3O4/DOPA/Van nanoparticles. | ||
The prepared Fe3O4-OA nanoparticles were readily dispersed in the chloroform. The suspension was stable for months in storage at 4 °C without noticeable sedimentation. Fig. 1a shows the size histogram obtained from DLS results indicating narrow size distribution. Though, the average hydrodynamic diameter of Fe3O4-OA nanoparticles was found to be 39.3 nm from DLS, but TEM image indicated the particle size in the range of 10–20 nm (Fig. 1b). Two-fold increase in particles size for DLS is due to hydrodynamic measurement and little aggregations of nanoparticles. This may be due in part to the presence of the oleic acid layer surrounding the particles.
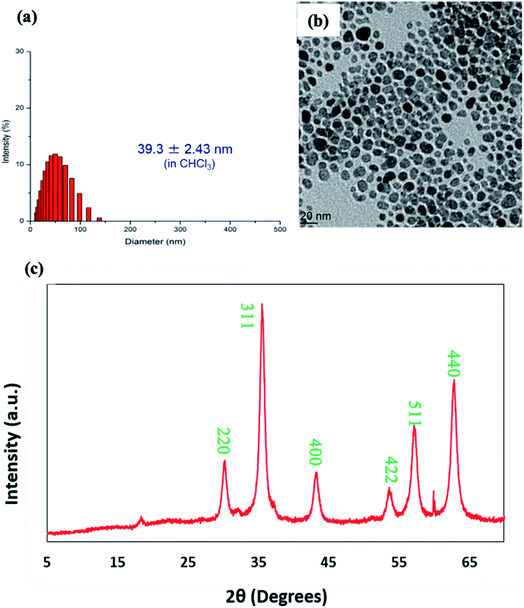 | ||
| Fig. 1 (a) Size distribution measured by DLS, (b) TEM image and (c) XRD pattern of oleic acid coated iron oxide nanoparticles (Fe3O4-OA). | ||
The XRD pattern of Fe3O4-OA nanoparticles shown in Fig. 1c demonstrates the characteristic diffraction signal at two theta (2θ) value of 18° corresponding to amorphous segments of oleic acid coating. Sharp and strong diffraction signals at 2θ values of around 30.1°, 35.5°, 42.8°, 53.8°, 56.7° and 63.8° can be indexed to crystal planes of (220), (311), (400), (422), (511) and (440) respectively and matched well with the data base of crystalline iron oxide magnetite Fe3O4 nanoparticles.39,40
The surface coating of Fe3O4 with oleic acid was further studied and compared with pure oleic acid and uncoated Fe3O4 nanoparticles by FTIR spectra analysis (Fig. 2). The characteristic peaks at around 2920 and 2850 cm−1 were observed for asymmetrical and symmetrical stretching of –CH2 present in both oleic acid (Fig. 2a) and oleic acid coated IONPs (Fig. 2c). Similar peaks were not present in uncoated nanoparticles (Fig. 1b), indicating the coating of oleic acid on the iron oxide nanoparticles. Owing to overlapping with –CH2 stretching, no distinct –OH stretching peak for pure oleic acid was observed.41 However, a peak at 932 cm−1 corresponds to the bending (out of plane) of –OH bond of carboxylic acid was visible. Additionally, the carboxylic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching represented by the sharp peak at 1709 cm−1 in oleic acid (Fig. 2a) is not present in IONPs-OA (Fig. 2c) due to the anchoring on surface of the nanoparticles. This result indicates that the IONPs are coated with a monolayer of oleic acid.42 The disappearance of the above carboxylic stretching in MNP-OA is due to the anchoring of –COOH on the surface of nanoparticles. Besides, the absence of –OH peaks at 1516 and 1411 cm−1 attributed to the carboxylate (–COO−) stretching indicating ligand binding on nanoparticle surface.43,44 Lastly, the peaks at the lower wavenumbers, 620, 680, and 718 cm−1 represent the Fe–O stretching in both uncoated and oleic acid coated magnetic nanoparticles.45
O stretching represented by the sharp peak at 1709 cm−1 in oleic acid (Fig. 2a) is not present in IONPs-OA (Fig. 2c) due to the anchoring on surface of the nanoparticles. This result indicates that the IONPs are coated with a monolayer of oleic acid.42 The disappearance of the above carboxylic stretching in MNP-OA is due to the anchoring of –COOH on the surface of nanoparticles. Besides, the absence of –OH peaks at 1516 and 1411 cm−1 attributed to the carboxylate (–COO−) stretching indicating ligand binding on nanoparticle surface.43,44 Lastly, the peaks at the lower wavenumbers, 620, 680, and 718 cm−1 represent the Fe–O stretching in both uncoated and oleic acid coated magnetic nanoparticles.45
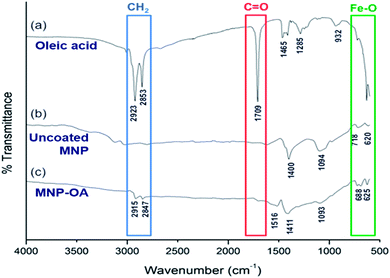 | ||
| Fig. 2 FTIR spectra of (a) pure oleic acid, (b) uncoated magnetic nanoparticles (Fe3O4), and (c) oleic acid coated magnetic nanoparticles (Fe3O4-OA). | ||
TGA curves were acquired for further quantitative demonstration of surface coating with oleic acid on Fe3O4 nanoparticles (Fig. 3). The residual weight of uncoated magnetic nanoparticles is 96.4%. The 3.6% weight loss until 600 °C is attributed to adsorbed and chemisorbed water in uncoated Fe3O4. On the other hand, Fe3O4-OA nanoparticles showed a weight loss of ∼18%. This higher weight loss corresponds to the pyrolysis of organic content (oleic acid), which can therefore be attributed to surface coverage of Fe3O4. At temperatures 350 °C and above, the organic content was thermally decomposed, and the final weight (82%) reflects the inorganic iron oxide content of the sample.
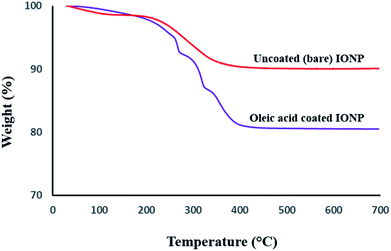 | ||
| Fig. 3 Thermograms of the uncoated (upper curve) and oleic acid coated (lower curve) iron oxide magnetic nanoparticles. | ||
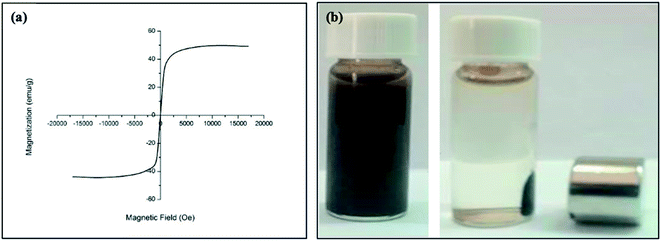
Fig. 4a shows the hysteresis loops for oleic acid coated IONPs which were characterized using a VSM at room temperature. The VSM result demonstrated that the nanoparticles have superparamagnetic character with a saturation of magnetization of 42.5 emu g−1 at 300 K (Fig. 4a). The higher value of saturation of magnetization indicates an excellent magnetic response of IONPs to external magnetic field. The digital photograph of oleic acid coated IONPs suspended in chloroform demonstrates the well dispersibility in absence of magnetic field while the nanoparticles are accumulated easily by a magnet (Fig. 4b).
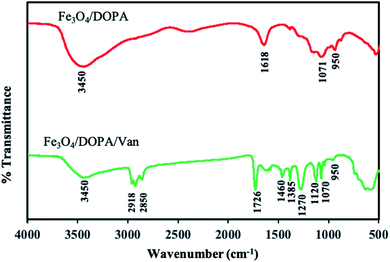
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond) confirmed the conjugation of vancomycin. The absorption bands at 580 and 520 cm−1 were related to Fe–O bonds in the tetrahedral and octahedral sites. The lower bond length of tetrahedral sites resulted higher stretching frequency i.e., the peak at 580 cm−1 corresponds to the intrinsic stretching vibrations of the Fe–O at the tetrahedral sites.48,49 Another broad peak, which is due to merger of multiple peaks, at 1550–1650 cm−1 in Fe3O4/DOPA/Van is attributed to the stretching vibration of amide I and amide II, also revealed vancomycin conjugation on IONPs.50,51
O bond) confirmed the conjugation of vancomycin. The absorption bands at 580 and 520 cm−1 were related to Fe–O bonds in the tetrahedral and octahedral sites. The lower bond length of tetrahedral sites resulted higher stretching frequency i.e., the peak at 580 cm−1 corresponds to the intrinsic stretching vibrations of the Fe–O at the tetrahedral sites.48,49 Another broad peak, which is due to merger of multiple peaks, at 1550–1650 cm−1 in Fe3O4/DOPA/Van is attributed to the stretching vibration of amide I and amide II, also revealed vancomycin conjugation on IONPs.50,51
The separation and fate of bacteria after interaction with Fe3O4/DOPA/Van nanoparticles are schematically illustrated in the Fig. 9a which shows that how bacteria are attached and separated with the aid of Fe3O4/DOPA/Van nanoparticles just immediately after magnetic accumulation. This figure also illustrated that incubation of bacteria with Fe3O4/DOPA/Van nanoparticles for 1 hour leaves fewer number of dead bacteria due to longer contact with nanoparticles. Fig. 9b shows the molecular structure of vancomycin and glycopeptide of bacterial cell wall to elucidate how they interact each other. In the process, the vancomycin molecule containing various amino and amide groups forms hydrogen bonds with the terminal D-alanyl-D-alanine moieties of bacteria.53,54 Therefore, bactericidal action of Fe3O4/DOPA/Van, is accounted primarily from inhibition of cell-wall biosynthesis due to the hydrogen bonding interaction as represented by dotted lines (Fig. 9b) and this ultimately leads to the death of cell.
4. Conclusion
Vancomycin conjugated with dopamine anchored iron oxide magnetite nanoparticles, Fe3O4/DOPA/Van, have been prepared and studied as antibacterial agent against both Gram positive and Gram-negative bacteria. A ligand exchange method facilitated easy transfer of highly crystalline hydrophobic magnetite (Fe3O4-OA) nanoparticles, that were prepared via coprecipitation method and oleic acid coating, into hydrophilic i.e., water dispersible Fe3O4/DOPA nanoparticles using dopamine (DOPA) as a ligand. An important finding of this study is that dopamine, which provides amine (NH2) functionality, played a key role, as evidenced by FTIR analysis, for the conjugation of vancomycin via carbodiimide chemistry. The Fe3O4/DOPA/Van nanoparticles not only showed a maximum 23 mm zone of inhibition but also killed those bacteria adhered to the nanoparticles within one hour. This result revealed that vancomycin conjugated Fe3O4/DOPA/Van nanoparticles showed high sensitivity towards bacteria. Further study to quantify the amount of conjugated vancomycin and their comparative study with the free counterpart, and quantitative efficiency of antibacterial effect are of future interests. The overall results suggests that Fe3O4/DOPA/Van nanoparticles synthesized here possessed enough affinity to the bacterial targets, conducive for killing bacteria in the dispersion media.Authors contribution
The manuscript was written through the contributions of all authors and approved the final version of the manuscript by all authors. M. M. Rahman conceptualized and led the project. M. M. Rahman, M. S. Islam and M. M. Hossain contributed to funding collection. M. M. Rahman, H. Ahmad and A. Elaissari contributed to nanoparticle characterizations and data analysis. M. Rashid, A. Rabbi and T. Ara performed the antibacterial study.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work is financially supported by the Faculty of Science, University of Rajshahi, Bangladesh. Authors also acknowledge the instrument facilities provided by Central Science Laboratory, Rajshahi University, Bangladesh, and Centre Technologique des Microstructures (CTμ), University of Lyon 1, France.References
- https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance.
- C. L. Ventola, P. & T., 2015, 40, 277–283 Search PubMed.
- D. A. Beauregard, D. H. Williams, M. N. Gwynn and D. J. Knowles, Antimicrob. Agents Chemother., 1995, 39, 781–785 CrossRef CAS PubMed.
- J. P. Mackay, U. Gerhard, D. A. Beauregard, D. H. Williams, M. S. Westwell and M. S. Searle, J. Am. Chem. Soc., 1994, 116, 4581–4590 CrossRef CAS.
- A. K. Schaefer, J. E. Melnyk, Z. He, F. D. Rosario and C. L. Grimes, in Immunity and Inflammation in Health and Disease, ed. S. Chatterjee, W. Jungraithmayr and D. Bagchi, Academic Press, 2018, ch. 14, pp. 175–187 Search PubMed.
- M. Ravelingien, S. Mullens, J. Luyten, M. D'Hondt, J. Boonen, B. D. Spiegeleer, T. Coenye, C. Vervaet and J. P. Remon, Eur. J. Pharm. Biopharm., 2010, 76, 366–370 CrossRef CAS PubMed.
- M. Mahmoudian and F. Ganji, Prog. Biomater., 2017, 6, 49–56 CrossRef CAS PubMed.
- S. Radin, J. T. Campbell, P. Ducheyne and J. M. Cuckler, Biomaterials, 1997, 18, 777–782 CrossRef CAS PubMed.
- M. Theerasilp, W. Sungkarat and N. Nasongkla, Bull. Mater. Sci., 2018, 41(42) CAS.
- S. Palanisamy and Y. M. Wang, Dalton Trans., 2019, 48, 9490–9515 RSC.
- N. Gupta, C. Gupta, S. Sharma, B. Rathi, R. K. Sharma and H. B. Bohidar, RSC Adv., 2016, 6, 111099–111108 RSC.
- L. F. E. Huerta-Núñez, G. C. Villanueva-Lopez, A. Morales-Guadarrama, S. Soto, J. Lopez, J. G. Silva, N. Perez-Vielma, E. Sacristan, M. E. Gudino-Zayas and C. A. Gonzalez, J. Nanopart. Res., 2016, 18, 284, DOI:10.1007/s11051-016-3594-8.
- M. M. Rahman and A. Elaissari, Sep. Purif. Technol., 2011, 81, 286–294 CrossRef CAS.
- W. Wu, Q. He and C. Jiang, Nanoscale Res. Lett., 2008, 3, 397, DOI:10.1007/s11671-008-9174-9.
- N. Zhu, H. Ji, P. Yu, J. Niu, M. U. Farooq, M. W. Akram, I. O. Udego, H. Li and X. Niu, Nanomaterials, 2018, 8, 810, DOI:10.3390/nano810081.
- W. Wu, C. Z. Jiang and V. A. L. Roy, Nanoscale, 2016, 8, 19421–19474 RSC.
- M. M. Rahman, Y. Nahar, W. Ullah, A. Elaissari and H. Ahmad, J. Polym. Res., 2015, 22, 33, DOI:10.1007/s10965-015-0673-y.
- Q. Wang, J. Xiao, M. Zhan, X. He, W. Yuan and Y. Li, Polym. Chem., 2021, 12, 2132–2214 RSC.
- Y. Nahar, M. A. Rahman, M. K. Hossain, M. K. Sharafat, M. R. Karim, A. Elaissari, B. Ochiai, H. Ahmad and M. M. Rahman, Mater. Res. Express, 2020, 7, 016102, DOI:10.1088/2053-1591/ab5be1.
- J. Nayeem, M. A. A. Al-Bari, M. Mahiuddin, M. A. Rahman, O. T. Mefford, H. Ahmad and M. M. Rahman, Colloids Surf., A, 2021, 611 DOI:10.1016/j.colsurfa.2020.125857.
- S. Badragheh, M. Zeeb, M. Reza and T. B. Olyai, RSC Adv., 2018, 8, 30550–30561 RSC.
- J. Sherwood, Y. Xu, K. Lovas, Y. Qin and Y. Bao, J. Magn. Magn. Mater., 2017, 427, 220–224 CrossRef CAS.
- K. Wang, J. Fu, S. Wang, M. Gao, J. Zhu, Z. Wang and Q. Xu, J. Colloid Interface Sci., 2018, 516, 263–273 CrossRef CAS PubMed.
- M. Wu, D. Zhang, Y. Zeng, L. Wu, X. Liu and J. Liu, Nanotechnology, 2015, 26, 115102, DOI:10.1088/0957-4484/26/11/115102.
- S. Mumtaz, L.-S. Wang, S. Z. Hussain, M. Abdullah, Z. Huma, Z. Iqbal, B. Creran, V. M. Rotello and I. Hussain, Chem. Commun., 2017, 53, 12306–12308 RSC.
- N. Pimpha, N. Woramongkolchai, P. Sunintaboon and N. Saengkrit, J. Cluster Sci., 2021 DOI:10.1007/s10876-021-01990-0.
- D. T. Nguyen, L. T. Pham, H. T. T. Le, M. X. Vu, H. T. M. Le, H. T. M. Le, N. H. Pham and L. T. Lu, RSC Adv., 2018, 8, 19707–19712 RSC.
- J.-J. Lee, K. J. Jeong, M. Hashimoto, A. H. Kwon, A. Rwei, S. A. Shankarappa, J. H. Tsui and D. S. Kohane, Nano Lett., 2014, 14, 1–5 CrossRef CAS PubMed.
- C. Wang, H. Lin and C. Ye, Front. Environ. Sci. Eng., 2016, 10(6), 8 CrossRef.
- R. Kadam, M. Maas and K. Rezwan, ACS Appl. Bio Mater., 2019, 2, 3520–3531 CrossRef CAS.
- D. Martínez-Matamoros, S. Castro-García, M. Balado, A. Matamoros-Veloza, M. Alonso Camargo-Valero, O. Cespedes, J. Rodríguez, M. L. Lemos and C. Jiménez, RSC Adv., 2019, 9, 13533–13542 RSC.
- H. Gu, P.-L. Ho, K. W. T. Tsang, L. Wang and B. Xu, J. Am. Chem. Soc., 2003, 125, 15702–15703 CrossRef CAS PubMed.
- Z. Wei, Z. Zhou, M. Yang, C. Lin, Z. Zhao, D. Huang, Z. Chen and J. Gao, J. Mater. Chem., 2011, 21, 16344–16348 RSC.
- H. Gu, K. Xu, C. Xu and B. Xu, Chem. Commun., 2006, 941–949 RSC.
- A. J. Kell, G. Stewart, S. Ryan, R. Peytavi, M. Boissinot, A. Huletsky, M. G. Bergeron and B. Simard, ACS Nano, 2008, 2, 1777–1788 CrossRef CAS PubMed.
- Y.-H. Chen, T.-J. Li, B.-Y. Tsai, L.-K. Chen, Y.-H. Lai, M.-J. Li, C.-Y. Tsai, P.-J. Tsai and D.-B. Shieh, Front. Microbiol., 2019, 10, 1141, DOI:10.3389/fmicb.2019.01141.
- W. Zhang, R. Taheri-Ledari, Z. Hajizadeh, E. Zolfaghari, M. R. Ahghari, A. Maleki, M. R. Hamblin and Y. Tian, Nanoscale, 2020, 12, 3855–3870 RSC.
- Y. Liu, T. Chen, C. Wu, L. Qiu, R. Hu, J. Li, S. Cansiz, L. Zhang, C. Cui, G. Zhu, M. You, T. Zhang and W. Tan, J. Am. Chem. Soc., 2014, 136, 12552–12555 CrossRef CAS PubMed.
- S. S. U. Rahman, M. T. Qureshi, K. Sultana, W. Rehman, M. Y. Khan, M. H. Asif and M. Farooq, Results Phys., 2017, 7, 4451–4456 CrossRef.
- U. Kurien, Z. Hu, H. Lee, A. P. Dastoor and P. A. Ariya, RSC Adv., 2017, 7, 45010–45021 RSC.
- Y.-Y. Shi, B. Sun, Z. Zhou, Y.-T. Wu and M.-F. Zhu, Prog. Nat. Sci.: Mater. Int., 2011, 21, 447–454 CrossRef.
- K. Yang, H. Peng, Y. Wen and N. Li, Appl. Surf. Sci., 2010, 256, 3093–3097 CrossRef CAS.
- J. D. Roo, F. V.-D. Broeck, K. D. Keukeleere, J. C. Martins, I. V. Driessche and Z. Hens, J. Am. Chem. Soc., 2014, 136, 9650–9657 CrossRef PubMed.
- S. Mondini, M. Leonzino, C. Drago, A. M. Ferretti, S. Usseglio, D. Maggioni, P. Tornese, B. Chini and A. Ponti, Langmuir, 2015, 31, 7381–7390 CrossRef CAS PubMed.
- H. Veeisi, A. Sedrpoushan, B. Maleki, M. Hekmati, M. Heidari and S. Hemmati, Appl. Organomet. Chem., 2015, 29, 834–839 CrossRef.
- T. Touqeer, M. W. Mumtaz, H. Mukhtar, A. Irfan, S. Akram, A. Shabbir, U. Rashid, I. A. Nehdi and T. S. Y. Choong, Energies, 2020, 13, 177, DOI:10.3390/en13010177.
- H.-M. Yang, H.-J. Lee, K.-S. Jang, C. W. Park, H. W. Yang, W. D. Heo and J.-D. Kim, J. Mater. Chem., 2009, 19, 4566–4574 RSC.
- Y. Xu, J. Sherwood, Y. Qin, R. A. Holler and Y. Bao, Nanoscale, 2015, 7, 12641–12649 RSC.
- J. Sherwood, Y. Xu, K. Lovas, Y. Qin and J. Yuping Bao, Mag. Mag. Mater, 2017, 427, 220–224 CrossRef CAS.
- H.-M. Yang, H. J. Lee, K.-S. Jang, C. W. Park, H. W. Yang, W. D. Heo and J.-D. Kim, J. Mater. Chem., 2009, 19, 4566–4574 RSC.
- H. Chen, T. Chen, J. Hu and C. Wang, Colloids Surf., A, 2005, 268, 24–29 CrossRef CAS.
- D. Liu, L. Ma, L. Liu, L. Wang, Y. Liu, Q. Jia, Q. Guo, G. Zhang and J. Zhou, ACS Appl. Mater. Interfaces, 2016, 8, 24455–24462 CrossRef CAS PubMed.
- B. K. Hubbard and C. T. Walsh, Angew. Chem., Int. Ed., 2003, 42, 730–765 CrossRef CAS PubMed.
- C. T. Walsh, Science, 1999, 284, 442–443 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |