DOI:
10.1039/D1RA05798G
(Paper)
RSC Adv., 2021,
11, 40040-40050
Synthesis, crystal structures, DNA interactions, and antitumor activity of two new dinuclear copper(II) complexes with thiazole ligand†
Received
30th July 2021
, Accepted 1st December 2021
First published on 16th December 2021
Abstract
Two new dinuclear copper(II) complexes, [Cu(ambt)2(cnba)4] (1) and [Cu(ambt)2(clba)4] (2) were synthesized with 2-amino-6-methoxybenzothiazole (ambt) as the main ligand. The structures of the two complexes were characterized by single-crystal XRD. The binding between CT-DNA (calf thymus DNA) and the complexes was evaluated by viscometry, electronic absorption, and fluorescence spectroscopy, and the binding constants were calculated using the Stern–Volmer equation. The complexes were intercalatively bound to CT-DNA, and [Cu(ambt)2(clba)4] having a greater binding constant than [Cu(ambt)2(cnba)4]. The two complexes had better antitumor properties against HepG2 (human hepatocellular carcinoma), A549 (human lung carcinoma), and HeLa (human cervical carcinoma) tumor cell lines than their respective ligands and cisplatin. Furthermore, [Cu(ambt)2(clba)4] had a stronger inhibitory ability on the three types of tumor cells than [Cu(ambt)2(cnba)4], which is congruent with the binding power of the complexes with DNA. Flow cytometry revealed that [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] could trigger apoptosis or necrosis, arrest the HepG2 cell cycles, and cause G0/G1-phase cells to accumulate.
1. Introduction
The discovery of cisplatin demonstrated that metal complexes are effective antitumor agents.1 However, traditional platinum drugs have some side effects including nephrotoxicity and ototoxicity, which limit their clinical application.2 Transition metal complexes have sparked interest due to their potential applications in antitumor drugs.3,4 In organisms, DNA is the main genetic material and the carrier of genetic information.5 It can store and transmit genetic information.6 Active metal complexes interact with DNA, untwist and bend its double helix, and destroy its structures by covalent binding of specific bases.7–12 The transition metal represented by copper has excellent coordination properties.13,14 Meanwhile, copper produces a variable valence ROS (Reactive Oxygen Species) during the transformation from Cu(II) to Cu(I) as a fundamental trace factor for the human body, which can cause tumor cells apoptosis and death.15–18 Hence, copper complexes are considered as substitute to cisplatin as antitumor drugs.19–21
Aromatic carboxylic acids are widely used in the construction of complexes due to their excellent structural rigidity and thermal stability.22,23 Their carboxyl groups can form monodentate coordination,24 chelating coordination,25 and bridging coordination.26 Metal aromatic carboxylic acid complexes have attracted the attention of researchers due to their unique catalytic activity,27 optical properties,28 magnetic properties,29 and biological properties.30
The selection of ligand plays an important role in the biological activity of complexes.31 Thiazole derivatives, due to their electron donation properties with their nitrogen-containing heterocyclic, are commonly used as dinuclear32,33 or mononuclear34 ligands to synthesize multi-functional thiazole metal complexes with transition metal.35,36 Thiazole metal complexes are widely used in medicine,37 optical materials,38 and agriculture.39 New antitumor drugs may be developed if aromatic carboxylic acid and thiazole are both used as ligands to synthesize copper(II) complexes. For extending this research area further, the synthesis of two new copper(II) complexes with ambt and 4-chloro-3-nitrobenzoic acid (cnba)/4-chlorobenzoic acid (clba) as ligands was reported in this paper. Their structures were characterized through elemental analysis, IR (infrared), as well as X-ray single crystal diffraction. Electronic absorption spectroscopy, fluorescence spectroscopy, and viscosity tests were employed to explore the interactions of the complexes with CT-DNA. According to the data, both complexes were bound to CT-DNA intercalatively. We evaluated the in vitro cytotoxicity of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] against HepG2, HeLa, and A549 tumor cell lines by using the CCK-8 assay. The data revealed that the complexes had anti-proliferative activity against the three kinds of tumor cells. Finally, HepG2 cell cycle allocation and apoptosis reaction were examined by flow cytometry in response to exposure to copper(II) complexes. The data suggested that both complexes may induce apoptosis or necrosis.
2. Experimental
2.1. Materials and instrumentation
All reagents of analytical grade were applied without further purification. The Chinese Academy of Council Cell Bank of Typical Culture Preservation (Shanghai, China) supplied the tumor cell lines HepG, HeLa, A549, and LO2. Sinopharm Chemical Reagent Co. Ltd (Shanghai, China) provided all chemicals adopted in this study.
The fluorescence test was carried out on the spectrofluorometer (RF-5301PC, Shimadzu Corp., Kyoto, Japan). Infrared (IR) spectra of the ligands as well as the complexes ranging from 4000–400 cm−1 were documented on a FT/IR spectrometer (Spectrum 65, PerkinElmer, Massachusetts, United States) with KBr pellets. Carbon, hydrogen, and nitrogen were quantitated in an elemental analyzer (2400 II, PerkinElmer, Massachusetts, USA), ESI-MS study has been reported on a Time of Flight Mass Spectrometer (G2-XS Tof, Waters Xevo, Massachusetts, USA). Electronic spectra were documented on a spectrophotometer (UV-8000, Yuanxi Corp., Shanghai, China) ranging from 200 to 1100 nm. In vitro cytotoxicity assays were carried out on a microplate reader with multiple functions (Multiskan Spectrum, Thermo Fisher Scientific, Massachusetts, USA). Cell cycle and apoptosis assays were carried out in a flow cytometry system (Beckman, CyAn-ADP, California, United States).
2.2. Synthesis
The preparation of complexes was shown in Scheme 1.
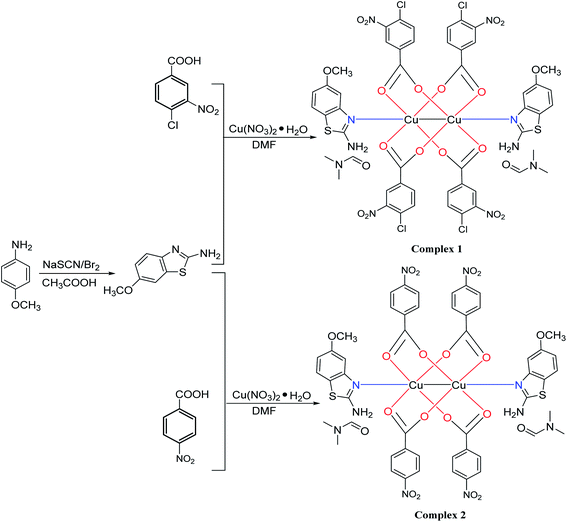 |
| | Scheme 1 Synthetic paths for ambt and [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4]. | |
2.2.1. Synthesis of [Cu(ambt)2(cnba)4] (complex 1). Ambt (0.270 g, 1.5 mmol) and cnba (0.907 g, 4.5 mmol) was mixed and dispersed in 4.0 mL N,N-dimethylformamide (DMF). A solution of copper(II) nitrate (0.18 mmol) in DMF (10.0 mL) was added to the mixture dropwise. Afterwards, the solution became dark green with a pH of 6.0–7.0. It was stirred for 30 minutes at 25 °C before being sealed with a plastic film and stored in a cool place. Well-formed green crystals that were appropriate for XRD were collected after 20 days with a final yield of 54.7%. The calculated values for C50H42Cl4Cu2N10O20S2 (Mw = 1435.94) were 41.82% C; 2.95% H, and 9.75% N, whereas the measured values for C50H42Cl4Cu2N10O20S2 were 41.70% C, 2.89% H, and 9.81% N. The IR (cm−1) data was ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) 3411, ν(
CH) 3411, ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH) 3149 and 2932, ν(C
NH) 3149 and 2932, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1661, 1535 and 780, ν(N
C) 1661, 1535 and 780, ν(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1474, ν(C–N) 1380 and 1218, ν(C–N–C) 1051, 745, and 665, and ν(C–O–C) 921, 830, and 554.
O) 1474, ν(C–N) 1380 and 1218, ν(C–N–C) 1051, 745, and 665, and ν(C–O–C) 921, 830, and 554.
2.2.2. Synthesis of [Cu(ambt)2(clba)4] (complex 2). Ambt (0.180 g, 1.0 mmol), as well as clba (0.167 g, 1.0 mmol) were mixed and dispersed in 10.0 mL DMF. The dissolution of copper(II) nitrate (0.241 g, 1.0 mmol) was made in 5.0 mL of DMF. Copper(II) nitrate solution was slowly put to the mixed solution. After addition, the solution became dark green with a pH of 6.0–6.5. It was stirred for 30 minutes while at 25 °C before being sealed with a plastic film and placed in a cool place. The dark green triangular prism solidified after two months, resulting in well-formed green crystals suitable for XRD. The final yield was 35.13%. The calculated values for C50H46Cu2N10O20S2 (Mw = 1298.17) were 46.26% C; 3.57% H, and 10.79% N, while the measured values were 46.46% C, 3.61% H, and 10.69% N. The IR (cm−1) data was ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) 3413, ν(
CH) 3413, ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH) 3311 and 2935, ν(C
NH) 3311 and 2935, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1646, 1597 and 833, ν(N
C) 1646, 1597 and 833, ν(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1521, ν(C–N) 1416 and 1340, 733, ν(C–N–C) 1221, 1058, and 749, and ν(C–O–C) 1100, 1029, and 544.
O) 1521, ν(C–N) 1416 and 1340, 733, ν(C–N–C) 1221, 1058, and 749, and ν(C–O–C) 1100, 1029, and 544.
2.3. Methods
2.3.2. DNA binding studies. Electronic absorption conveniently explores binding between metal complexes and DNA. The baseline was scanned with Tris–HCl/NaCl buffer solution. We added 2.5 mL complex solution (24 μM for 1 or 12 μM for 2) and 2.0 mM CT-DNA (30 μL for 1 and 10 μL for 2) solution were also added into the sample cuvette and the blank cuvette with a pipette gun, respectively. The solution was left to stand for 5 minutes, before being scanned in the wavelength range of 200–450 nm.The measurement of fluorescence was made on a spectrofluorometer configured with a 1 cm quartz cuvette. A solution of 10.0 mL ethidium bromide (EB, 8 μmol L−1) and 10.0 mL CT-DNA (10 μmol L−1) was allowed to react for 12 h. Afterwards, 2.5 mL EB–CT-DNA mixed solution was added into the sample cuvette. The excitation wavelength and scanning speed were set as 525 nm and 240 nm min−1, respectively. The fluorescence spectrum of the EB–CT-DNA mixed solution at 540–700 nm was determined. To increase the concentration of the complex, a pipette gun was used to slowly add 2 μL of complex solution (50 μM for [Cu(ambt)2(cnba)4] or 5 μM for [Cu(ambt)2(clba)4]) into the EB–CT-DNA mixed system. The solution was mixed thoroughly after each drop and left to stand for 5 minutes to determine the fluorescence emission spectrum.
The measurement of CT-DNA relative viscosity was made with an viscometer (Ubbelohde, Shenyi glass Products Co. Ltd, Shanghai, China) at constant 29.0 ± 0.1 °C with and without the complex. The immersion of viscometer was made in the constant temperature water bath for 5 minutes before each measurement was taken. After carrying out each experiment in triplicate, the mean value was taken. The viscometer was filled with 20.0 mL of CT-DNA solution with 200 μM concentration, then the complex solution (1 mM) was gradually added. The [complex]/[DNA] ratio was 0, 0.05, 0.1, 0.15, 0.20, 0.25, 0.30, and the time required for CT-DNA solution to flow through the capillary was recorded. EB was used as a positive control.
2.3.3. In vitro cytotoxicity assays. The complexes cytotoxicity against HepG, HeLa, A549, and LO2 cell lines were explored with Cell Counting Kit-8. In brief, HepG, HeLa, A549, and LO2 cells were digested, counted, and prepared into cell suspensions at 5 × 104 cells per mL concentration. Each well of the 96-well cell culture plates was filled with 100 μL cell suspensions (5 × 103 cells per well). The cells were inoculated overnight in a 5% CO2 incubator at 37 °C. The complete medium (90% medium + 10% fetal bovine serum) was applied to prepare working solution containing complexes at different concentrations. After mixing, 100 μL corresponding working solution were added into each well, and each concentration was triplicated. The 24 hour, 48 hour, 72 hour and 96 hour inoculation of cells were made under 5% CO2 and 37 °C conditions. The supernatant was discarded and the cells were rinsed once with medium. The cells were cultured continuously for 3 h after 100 μL CCK-8 working solution (vmedium![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) voriginal CCK-8 solution = 10
voriginal CCK-8 solution = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were introduced into each well of the 96-well plates. To calculate the rate of cell proliferation, the OD value (λ = 450 nm) of each well was determined.
1) were introduced into each well of the 96-well plates. To calculate the rate of cell proliferation, the OD value (λ = 450 nm) of each well was determined.
2.3.4. Cell cycle assays. In virtue of the tumor cell cycle, the mechanism of the DNA-binding copper complexes can be identified. The protocol for HepG2 cells of digesting, counting, preparing, and inoculating was similar to that of cytotoxicity assay. After mixing, 2.0 mL corresponding working solution were added into each well. The 24 hour and 48 hour inoculation of cells were made under 5% CO2 and 37 °C conditions. The cells were washed once using PBS (Phosphate Buffer Saline), digested using trypsin, and centrifuged. The cells were then rinsed twice using precooled PBS. Precooled 70% alcohol was added and cells were fixed and stored at −20 °C. After staining, a Beckman CyAn-ADP flow cytometry platform was used to analyze the samples. The analysis of ten thousand cells was made per sample.
2.3.5. Cell apoptosis assay. The protocol for this experiment was similar to that of cell cycle assay. Cells were collected using centrifugation (1000 rpm min−1) after being digested. We discarded the supernatant and the cells were rinsed twice using 1.0 mL calcium-free PBS. Binding buffer (100 μL), annexin V-FITC (5 μL), along with PI solution (5 μL) were introduced into the plates. The cells were inoculated in the dark at 25 °C for 15 minutes. After adding 400 μL binding buffer, 1 × 104 cells were analyzed at 488 nm with a flow cytometry platform.
3. Results and discussion
3.1. Synthesis and characterization
Two novel dinuclear complexes, [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], with ambt as main ligand were synthesized and found to be soluble in DMF and stable in air. The crystallization of [Cu(ambt)2(cnba)4] was shown in the triclinic crystal system with space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , however, the crystallization of [Cu(ambt)2(clba)4] was shown in the monoclinic crystal system with space group C2/c (Fig. 1). The Cu(II) ions in [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] were hexa-coordinated by one oxygen (O) atom from ambt, one Cu(II) atom from the other nuclear, and four oxygen (O) atoms from cnba and clba for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–O distances ranged from 1.9640 (13)–1.9873 (13) and 1.9558 (19)–1.989 (2) Å for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–Cu(II) distance was 2.7290 (4) and 2.6970 (6) for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The O/N–Cu–O/N/Cu bond angles ranged from 78.44(4)–172.83(4)° to 81.09(6)–173.01(6)° for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively (Table S1†).
, however, the crystallization of [Cu(ambt)2(clba)4] was shown in the monoclinic crystal system with space group C2/c (Fig. 1). The Cu(II) ions in [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] were hexa-coordinated by one oxygen (O) atom from ambt, one Cu(II) atom from the other nuclear, and four oxygen (O) atoms from cnba and clba for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–O distances ranged from 1.9640 (13)–1.9873 (13) and 1.9558 (19)–1.989 (2) Å for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–Cu(II) distance was 2.7290 (4) and 2.6970 (6) for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The O/N–Cu–O/N/Cu bond angles ranged from 78.44(4)–172.83(4)° to 81.09(6)–173.01(6)° for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively (Table S1†).
 |
| | Fig. 1 Crystal structures of [Cu(ambt)2(cnba)4] (a) and [Cu(ambt)2(clba)4] (b). | |
ESI-MS spectral study has been performed to determine the composition of the multinuclear two complexes in MeOH (Fig. S5 and S6†). The spectral analyses reveal that [Cu(ambt)2(cnba)4] shows m/z value of 1435.6635 (Mw = 1435.94), [Cu(ambt)2(clba)4] shows m/z value of 1298.1991 (Mw = 1298.17). These data show that the complexes retain their whole structures in MeOH solution phase.
3.2. DNA binding experiments
3.2.1. Electronic absorption. Electronic absorption is one of the most common, easy, and effective approaches to investigate the interactions between metal complexes and DNA.40 Phenomenon such as hyperchromicity or hypochromism, redshift or blueshift of the absorption peak of the electronic absorption spectrum, can be employed to assess the binding mode of the complex with DNA due to great stacking interactions between the aromatic chromophore of the benzoyl or thiazole and the DNA base pairs.41 The electronic absorption peak had hypochromism when the complex interacted with DNA through intercalative binding. However, there was no significant change when the metal complex and DNA were bound in a static or groove surface interaction. The binding strength was analyzed by computing the binding constant (Kb) according to eqn (1):42| | |
[DNA]/(εa − εf) = [DNA]/[(εb − εf) + 1/Kb(εb − εf)]
| (1) |
where εa, εf, and εb mean the appear, free, and bound extinction indicators, respectively. [DNA] means the CT-DNA concentration. Kb represents the ratio of the slope to the intercept. The determined binding constants were 18.8 M−1 and 2.04 × 104 M−1 for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively (Fig. 2). The determined intrinsic equilibrium binding constants (Kb) (103–105 M−1) conformed to literature values.43 Thus, the two complexes and the DNA showed high affinity.
 |
| | Fig. 2 Electronic absorption spectra of [Cu(ambt)2(cnba)4] (a) and [Cu(ambt)2(clba)4] (b) in the absence and presence of CT-DNA. Arrows indicate changes in absorbance in response to increasing DNA level. Inset: [DNA]/(εa − εf) vs. [DNA] plots. | |
3.2.2. Fluorescence spectroscopy. Fluorescence spectrometry is another common approach to assess the binding mode of metal complexes and DNA.44 In this test, EB was used as a fluorescent probe. The fluorescence performance of the probe was significantly improved when EB was embedded into the base pair of DNA. When the complex solution was introduced to the DNA–EB solution, the fluorescence quenching constant Ksv of the complex was computed, and the binding way of the complex with DNA was then analyzed. The fluorescence quenching role of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] on the EB–CT-DNA system was shown in Fig. 3, and strong fluorescence peaks were notable at 585 nm (Fig. 3a and b). The intensity of fluorescence of the EB–CT-DNA system gradually diminished as complexes were added, and a slight red shift occurred, indicating that complexes may squeeze out EB and enter the DNA base pair through the embedding mode. The classical Stern–Volmer equation was used to quantify the fluorescence quenched by CT-DNA in complexes.45| | |
F0/F = 1 + Kqτ0[C] = 1 + Ksv[C]
| (2) |
where F0 denotes fluorescence intensity as complexes are not added, F means the fluorescence intensity after adding complexes, [C] denotes the concentration of the complex, Ksv means the quenching constant, Kq represents the constant rate of bimolecular quenching, and τ0 represents the lifetime of the fluorescent molecule (almost 10−8 s) in the absence of the complex. The F0/F vs. [C] plot is linear, the slope obtained was the quenching constant Ksv, and Kq can be obtained by calculation. As shown in Fig. 3, the quenching constants (Ksv) were 2.81 × 106 M−1 for [Cu(ambt)2(cnba)4] and 1.95 × 107 M−1 for [Cu(ambt)2(clba)4] and Kq1 = 2.81 × 1014 M−1 s−1, Kq2 = 1.95 × 1015 M−1 s−1. The constant rate of quenching (Kq) was evidently greater than the constant restrictive diffusion (Kdif) for biomolecules (2.0 × 1010 M−1 s−1),46 showing that the quenching was a static process.47 Eqn (3) 48 was employed to describe the association of the fluorescence intensity with [C] for static quenching:| |
log[(F0 − F)/F] = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[C] log[C]
| (3) |
where n means the quantity of binding places and Ka denotes the association constant. In Fig. 4, the value of Ka was 1.51 × 106 M−1 and 2.57 × 108 M−1 for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The n values were 0.97 and 1.16 for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively.
 |
| | Fig. 3 EB–DNA system emission spectra with and without [Cu(ambt)2(cnba)4] (a) and [Cu(ambt)2(clba)4] (b). Inset: F0/F vs. [complex] plots. Arrows show variations in intensity as complex concentration escalates. | |
 |
| | Fig. 4 Double-log plots of CT-DNA fluorescence quenching by [Cu(ambt)2(cnba)4] (a) and [Cu(ambt)2(clba)4] (b). | |
3.2.3. Viscosity measurements. The viscosity approach is widely regarded as one of the effective methods for researching the interactions between complexes and CT-DNA.49 The viscosity of CT-DNA solution will not change remarkably if the complex interactions with CT-DNA through the electrostatic, groove binding.50 Partial or non-classical intercalation bends the CT-DNA helix and decreases DNA viscosity, while classical intercalation increases CT-DNA viscosity.51 The relative viscosity was obtained by converting the measured time average value according to the following formula (4):52where t0 designates the time for the buffer solution to flow through the capillary, and t designates the time for the DNA solution containing complex with different concentrations to flow through the capillary. We used (η/η0)1/3 (η0 designates the relative viscosity of DNA solution without complex) to [complex]/[DNA]. The viscosity of DNA increases as the [complex]/[DNA] ratio rise (Fig. 5). Therefore, copper(II) complex binds DNA by intercalation as EB. Absorption and emission spectrum analysis corroborated this conclusion.
 |
| | Fig. 5 Increasing number of the complex and EB had an effect on related CT-DNA viscosity. | |
3.3. Cytotoxicity assay in vitro
The cell lines HepG, HeLa, A549, and LO2 were inoculated with diverse levels of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] for 24 h, 48 h, 72 h, and 96 h. The effects of complexes on HepG, HeLa, A549, and LO2 cells viability and proliferation were shown in Fig. 6 and 7. The IC50 of complexes against HepG, HeLa, A549, and LO2 cells at different time points were shown in Table 2. These data illustrated that both the complexes had strong anti-proliferative activity against HepG, HeLa, A549 cells. At the same time point, both complexes had greater antitumor efficacy against three kinds tumor cell lines than the ligands and the positive control cisplatin but lower efficacy against LO2. However, [Cu(ambt)2(clba)4] had a stronger activity than [Cu(ambt)2(cnba)4]. Cell viability was concentration- and time-dependent, indicating that both complexes gradually entered the cells and caused apoptosis.
 |
| | Fig. 6 Distribution of HepG2 cell cycle after 24 h and 48 h exposure to [Cu(ambt)2(cnba)4]. | |
 |
| | Fig. 7 Distribution of HepG2 cell cycle after 24 h and 48 h exposure to [Cu(ambt)2(clba)4]. | |
Table 2 Value of IC50 of the complexes against HepG, HeLa, A549, and LO2 at various time points
| Complexes |
IC50 (μM) |
| t (h) |
LO2 |
A549 |
HeLa |
HepG2 |
| 1 |
24 |
87.41 ± 1.94 |
61.56 ± 1.08 |
37.27 ± 1.05 |
48.35 ± 0.86 |
| 48 |
50.28 ± 1.70 |
25.73 ± 0.79 |
19.77 ± 1.03 |
23.47 ± 0.85 |
| 72 |
26.62 ± 1.43 |
20.39 ± 0.85 |
11.85 ± 0.95 |
14.56 ± 0.87 |
| 96 |
14.74 ± 0.93 |
11.80 ± 0.78 |
7.43 ± 0.74 |
9.31 ± 0.79 |
| 2 |
24 |
92.86 ± 1.97 |
36.94 ± 1.12 |
21.85 ± 0.86 |
29.84 ± 0.86 |
| 48 |
53.22 ± 1.73 |
25.13 ± 1.01 |
14.58 ± 0.76 |
14.97 ± 0.74 |
| 72 |
29.20 ± 1.47 |
19.96 ± 1.00 |
11.59 ± 0.77 |
11.57 ± 0.75 |
| 96 |
17.74 ± 1.25 |
9.86 ± 0.86 |
1.86 ± 0.53 |
7.75 ± 0.70 |
| Anba |
48 |
>1000 |
975.4 ± 1.76 |
>1000 |
>1,000,0 |
| Ambt |
48 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>1000 |
| Clba |
48 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
>1000 |
>1000 |
| Cisplatin |
48 |
37.34 ± 1.57 |
36.50 ± 1.76 |
37.37 ± 2.01 |
43.81 ± 1.62 |
3.4. Cell cycle arrest
The distribution of HepG2 cells across various stages of the cell cycle was explored by flow cytometry and PI staining. The proportions of HepG2 cells at G0/G1 were 55.38% at 24 h (Fig. 6) and 58.68% at 48 h (Fig. 7). After incubation with complexes 1 or 2 to the time point, the proportion of G0/G1 phase increased, whereas the proportion of G2/M and S phase declined. Compared with the control, [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] triggered G0/G1 stage arrest in HepG2 cells, and the proportion of G0/G1 phase was concentration- and time-reliance, with [Cu(ambt)2(clba)4] showing a stronger activity than [Cu(ambt)2(cnba)4].
3.5. Cell apoptosis assay
With untreated HepG2 cells served as control, flow cytometry investigated the apoptosis in HepG2 cells (Fig. 8 and 9). After exposing the copper(II) complexes with 5 μM and 20 μM to the cells for 24 hours, the ratios of apoptotic or necrosis cells were 5.4% and 13.9% for [Cu(ambt)2(cnba)4] while 8.3% and 19.7% for [Cu(ambt)2(clba)4]. The ratios of apoptotic or necrosis cells for the control was 2.9%. After 48 hour exposure to the same conditions, the ratios of apoptotic or necrosis cells were 11.3% and 35.9% for [Cu(ambt)2(cnba)4] while 10.3% and 40.9% for [Cu(ambt)2(clba)4]. The ratios of apoptotic or necrosis cells for the control was 3.0%. By comparing with the control, the ratio of living cells had declined while that of apoptotic or necrosis cells had grown in [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] treatments, and [Cu(ambt)2(clba)4] exhibited a stronger activity on apoptosis activity against HepG cells. The apoptotic or necrosis effect of the copper(II) complexes on the HepG2 cells was both concentration- and time-dependent.
 |
| | Fig. 8 HepG2 cells were inoculated with various levels of [Cu(ambt)2(cnba)4] for different durations. | |
 |
| | Fig. 9 HepG2 cells were inoculated with various levels of [Cu(ambt)2(clba)4] for different durations). | |
4. Conclusions
Herein, two new copper dinuclear complexes were synthesized with ambt as the main ligand. Both complexes had copper binuclear coordination structures. [Cu(ambt)2(cnba)4] was triclinic system with P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group while [Cu(ambt)2(clba)4] was monoclinic system with C2/c space group. Both [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] bound intercalatively to DNA and [Cu(ambt)2(clba)4] showed greater binding constant than [Cu(ambt)2(cnba)4]. The fluorescence quenching of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] to CT-DNA was static quenching. Compared with ligands and cisplatin, the complexes had stronger anti-proliferative activity against HepG2, A549, and HeLa tumor cells. The inhibitory ability of [Cu(ambt)2(clba)4] on the three kinds of tumor cells was stronger. This is congruent with the binding power of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] with DNA, indicating that the antitumor movement of the complexes may be linked to their binding ability with DNA. [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] inhibit tumor cells by inducing apoptosis or necrosis in HepG2 cells, and arresting HepG2 cell cycle in G0/G1 phase. This work may aid in the development of Cu(II) complexes as potential antitumor reagents in the future.
space group while [Cu(ambt)2(clba)4] was monoclinic system with C2/c space group. Both [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] bound intercalatively to DNA and [Cu(ambt)2(clba)4] showed greater binding constant than [Cu(ambt)2(cnba)4]. The fluorescence quenching of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] to CT-DNA was static quenching. Compared with ligands and cisplatin, the complexes had stronger anti-proliferative activity against HepG2, A549, and HeLa tumor cells. The inhibitory ability of [Cu(ambt)2(clba)4] on the three kinds of tumor cells was stronger. This is congruent with the binding power of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] with DNA, indicating that the antitumor movement of the complexes may be linked to their binding ability with DNA. [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] inhibit tumor cells by inducing apoptosis or necrosis in HepG2 cells, and arresting HepG2 cell cycle in G0/G1 phase. This work may aid in the development of Cu(II) complexes as potential antitumor reagents in the future.
Author contributions
Zhenfang Zeng: designed the methodology and drafted the original manuscript. Jiehui Cai: performed the experiments. Qiuping Huang: conceptualized the whole research. Yanying Weng: performed data curation. Qiuchan Huang: drafted and edited the manuscript. Youhuan Wei: drafted and reviewed the manuscript as well as sourced for funding.
Conflicts of interest
The authors report no conflicts of interest.
Acknowledgements
The work was funded by the Middle-aged and Young Teachers' Basic Ability Promotion Project of Guangxi (No. 2017KY0846 and 2019KY0769), the Scientific Research Project of Guangxi Normal University for Nationalities (No. 1011/3270020101, 327003070414, 327003070416, 327002060107, 31700702, 2019YB018, and 2020GP008).
References
- L. Qi, Q. Luo, Y. Zhang, F. Jia and F. Wang, Chem. Res. Toxicol., 2019, 32, 1469–1486 Search PubMed.
- S. Hosseinzadeh, M. E. Moghadam, S. Sheshmani and A. S. Shahvelayati, J. Biomol. Struct. Dyn., 2019, 2215–2228 Search PubMed.
- B. Q. Zou, X. L. Huang, Q. P. Qin, Z. F. Wang and H. Liang, Polyhedron, 2020, 181, 114482 CrossRef CAS.
- J. Alday, A. M. Mazzeo and S. Suarez, Inorg. Chim. Acta, 2020, 510, 119696 CrossRef CAS.
- S. Pereira, Am. J. Bioeth., 2019, 19, 75–76 CrossRef PubMed.
- H. H. Lee, R. Kalhor, N. Goela, J. Bolot and G. M. Church, Nat. Commun., 2019, 10, 2383–2394 CrossRef PubMed.
- S. S. A. Fathima, R. Paulpandiyan and E. R. Nagarajan, J. Mol. Struct., 2019, 1178, 179–191 CrossRef.
- S. Elleuchi, I. O. D. Luzuriaga, N. Sanchez-Gonzalez, X. Lopez and A. Gil, Inorg. Chem., 2020, 59, 12711–12721 CrossRef CAS PubMed.
- P. J. Jarman, F. Noakes, S. Fairbanks, K. Smitten, I. K. Griffiths, H. K. Saeed, J. A. Thomas and C. Smythe, J. Am. Chem. Soc., 2019, 141, 2925–2937 CrossRef CAS PubMed.
- Z. Yaghobi, Z. R. Ranjbar and S. Gharbi, Polyhedron, 2019, 164, 176–184 CrossRef CAS.
- G. Barone, A. Terenzi, A. Lauria, A. M. Almerico, J. M. Leal, N. Busto and B. Garcia, Coord. Chem. Rev., 2013, 257, 2848–2862 CrossRef CAS.
- L. Antonino, B. Riccardo, T. Alessio, S. Angelo, G. Francesco, L. Alessandro, A. A. Maria and B. Giampaolo, Dalton Trans., 2014, 43, 6108–6119 RSC.
- C. Zhang, C. Huang, S. Wei, X. Cai and A. A. Pimerzine, Thermochim. Acta, 2020, 690, 178661–178667 CrossRef CAS.
- J. Wang, X. P. Zhou, Q. Lv and H. L. Yuan, J. Mol. Struct., 2021, 1231, 129964–129970 CrossRef CAS.
- C. J. Nunes, A. H. Otake, S. O. Bustos, R. B. Fazzi and A. Ferreira, Chem.–Biol. Interact., 2019, 311, 108789 CrossRef CAS PubMed.
- T. Shen, X. X. Hu, Y. Liu, Y. Zhang and X. B. Zhang, ACS Appl. Mater. Interfaces, 2020, 12, 5403–5412 CrossRef CAS PubMed.
- A. Zafar, S. Singh, S. Ahmad, S. Khan, M. I. Siddiqi and I. Naseem, Bioorg. Chem., 2019, 87, 276–290 CrossRef CAS PubMed.
- S. Karpagam, R. Kartikeyan, P. P. Nachiyar, M. Velusamy and M. Kannan, J. Coord. Chem., 2019, 3102–3127 CrossRef CAS.
- M. Ociepa, S. Bakalarz, M. Nentwig, O. Oeckler, M. Gajewska, E. Turlej, J. Wietrzyk and D. Michalska, Inorg. Chim. Acta, 2019, 490, 68–77 CrossRef.
- S. Jiang, H. Ni, F. Liu, S. Gu and Y. Gou, Inorg. Chim. Acta, 2019, 499, 119186–119195 CrossRef.
- G. L. Chang, Z. Li, M. J. Niu and S. N. Wang, J. Coord. Chem., 2019, 72, 2422–2436 CrossRef CAS.
- F. Y. Ju and S. Li, Russ. J. Gen. Chem., 2020, 90, 1083–1087 CrossRef CAS.
- X. Wang, M. Kong, D. Li, J. Fang, Z. Deng and H. Zhang, Crystengcomm, 2019, 21, 2144–2153 RSC.
- M. M. Zhu, J. Cui, Y. L. Zeng, N. Ren and J. J. Zhang, Polyhedron, 2019, 158, 485–493 CrossRef CAS.
- L. N. Zhu, Z. P. Deng, S. W. Ng, L. H. Huo and S. Gao, Dalton Trans., 2019, 48, 7589–7601 RSC.
- F. Y. Jie, C. Y. I. Ping, G. X. Mei, H. H. Bing and G. Z. Hui, J. Mol. Struct., 2019, 1195, 929–935 CrossRef.
- M. Bakir, M. W. Lawrence, P. Nelson and M. B. Yamin, J. Coord. Chem., 2019, 72, 2261–2278 CrossRef CAS.
- K. B. Manjunatha, R. Rajarao, P. Poornesh, B. J. Rudresha, G. Umesh and B. R. Bhat, Opt. Mater., 2019, 89, 494–497 CrossRef CAS.
- P. Thanakit, D. Limthin, P. Leepheng, S. Suramitr and D. Phromyothin, Ferroelectrics, 2019, 552, 108–120 CrossRef CAS.
- T. Madanhire, H. Davids, M. C. Pereira, E. C. Hosten and A. R. Abrahams, Polyhedron, 2020, 185, 114583 CrossRef CAS.
- W. H. El-Shwiniy, L. M. Abbass, S. A. Sadeek and W. A. Zordok, Russ. J. Gen. Chem., 2020, 90, 483–488 CrossRef CAS.
- N. Alvarez, F. Velluti, F. Guidali, G. Serra and M. H. T. vz, Inorg. Chim. Acta, 2020, 508, 119622 CrossRef CAS.
- H. Kotani, D. Hong, K. Satonaka, T. Ishizuka and T. Kojima, Inorg. Chem., 2019, 58, 3676–3682 CrossRef CAS PubMed.
- S. R. Shah, Z. Shah, M. Khiat, S. A. Halim, A. Khan, J. Hussain, R. Csuk, M. U. Anwar and A. Al-Harrasi, Appl. Organomet. Chem., 2020, 34, e5842 CAS.
- M. Kaufmann, C. Müller, A. A. Cullen, M. P. Brandon and M. T. Pryce, Inorg. Chem., 2020, 60, 760–773 CrossRef PubMed.
- S. Safaei, J. Wang and P. C. Junk, J. Solid State Chem., 2020, 294, 121762–1121773 CrossRef.
- A. M. Khedr, H. El-Ghamry, M. A. Kassem, F. A. Saad and N. El-Guesmi, Inorg. Chem. Commun., 2019, 108, 107496 CrossRef CAS.
- J. Zhao, X. Chen, J. Wu, M. Jian and Z. Chi, J. Mater. Chem. C, 2020, 8, 10390–10400 RSC.
- B. H. Pursuwani, B. S. Bhatt, F. U. Vaidya, C. Pathak and M. N. Patel, J. Biomol. Struct. Dyn., 2020, 39, 1–12 Search PubMed.
- N. Ning, D. Q. Zhao, H. Li, J. Nan, J. Y. Wen and H. Y. Liu, Molecules, 2015, 21, 54 CrossRef PubMed.
- H. W. Mo, Y. X. Liu, D. H. Cai, F. Shen and X. Y. Le, Chin. J. Inorg. Chem., 2019, 35, 477–484 CAS.
- V. N. Kadam, K. Saikrishnan and K. N. Ganesh, J. Phys. Chem. C, 2018, 122, 14004–14013 CrossRef CAS.
- A. Draksharapu, A. Boersma, M. Leising, A. Meetsma, W. R. Browne and G. Roelfes, Dalton Trans., 2015, 44, 3647–3655 RSC.
- C. L. Zhang, Y. X. Liu, X. M. Zhang, S. Chen, F. Shen, Y. H. Xiong, W. Liu, Z. W. Mao and X. Y. Le, Mater. Sci. Eng. C, 2018, 91, 414–425 CrossRef CAS PubMed.
- T. Sohrabi, M. Hosseinzadeh, S. Beigoli, M. R. Saberi and J. Chamani, J. Mol. Liq., 2018, 256, 127–138 CrossRef CAS.
- Y. Zhang, J. Y. Wen, M. H. Mahmood, X. L. Wang, B. B. Lv, X. Ying, H. Wang, L. N. Ji and H. Y. Liu, Luminescence, 2015, 30, 1045–1054 CrossRef CAS PubMed.
- G. Zhang and Y. Ma, Food Chem., 2013, 136, 446–449 Search PubMed.
- B. H. Pursuwani, B. S. Bhatt, F. U. Vaidya, C. Pathak and M. N. Patel, J. Fluoresc., 2021, 31, 349–362 CrossRef CAS PubMed.
- A. A. Hassan, A. A. Aly, N. K. Mohamed, K. M. E. Shaieb, M. M. Makhlouf, E. S. M. N. Abdelhafez, S. Brase, M. Nieger, K. N. Dalby and T. S. Kaoud, Bioorg. Chem., 2019, 85, 585–599 CrossRef CAS PubMed.
- N. Vamsikrishna, M. P. Kumar, G. Ramesh, N. Ganji, S. Daravath and Shivaraj, J. Chem. Sci., 2017, 129, 609–622 CrossRef CAS.
- B. Anupama, J. Mol. Struct., 2020, 1210, 127988 CrossRef CAS.
- Z. F. Zeng, Q. P. Huang, J. H. Cai, G. J. Zheng, Q. C. Huang, Z. L. Liu, Z. L. Chen and Y. H. Wei, Molecules, 2021, 26, 4028 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2086765 and 2086766. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ra05798g |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article ,
Jiehui Cai,
Fuyan Li,
Yanying Weng,
Qiuping Huang,
Honglan Yang,
Qiuchan Huang* and
Youhuan Wei
,
Jiehui Cai,
Fuyan Li,
Yanying Weng,
Qiuping Huang,
Honglan Yang,
Qiuchan Huang* and
Youhuan Wei
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) 3411, ν(
CH) 3411, ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH) 3149 and 2932, ν(C
NH) 3149 and 2932, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1661, 1535 and 780, ν(N
C) 1661, 1535 and 780, ν(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1474, ν(C–N) 1380 and 1218, ν(C–N–C) 1051, 745, and 665, and ν(C–O–C) 921, 830, and 554.
O) 1474, ν(C–N) 1380 and 1218, ν(C–N–C) 1051, 745, and 665, and ν(C–O–C) 921, 830, and 554.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH) 3413, ν(
CH) 3413, ν(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH) 3311 and 2935, ν(C
NH) 3311 and 2935, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1646, 1597 and 833, ν(N
C) 1646, 1597 and 833, ν(N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) 1521, ν(C–N) 1416 and 1340, 733, ν(C–N–C) 1221, 1058, and 749, and ν(C–O–C) 1100, 1029, and 544.
O) 1521, ν(C–N) 1416 and 1340, 733, ν(C–N–C) 1221, 1058, and 749, and ν(C–O–C) 1100, 1029, and 544.![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 217/5121[Rint = 0.0416]
217/5121[Rint = 0.0416]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 762/4857 [Rint = 0.0259]
762/4857 [Rint = 0.0259]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) voriginal CCK-8 solution = 10
voriginal CCK-8 solution = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were introduced into each well of the 96-well plates. To calculate the rate of cell proliferation, the OD value (λ = 450 nm) of each well was determined.
1) were introduced into each well of the 96-well plates. To calculate the rate of cell proliferation, the OD value (λ = 450 nm) of each well was determined.![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , however, the crystallization of [Cu(ambt)2(clba)4] was shown in the monoclinic crystal system with space group C2/c (Fig. 1). The Cu(II) ions in [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] were hexa-coordinated by one oxygen (O) atom from ambt, one Cu(II) atom from the other nuclear, and four oxygen (O) atoms from cnba and clba for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–O distances ranged from 1.9640 (13)–1.9873 (13) and 1.9558 (19)–1.989 (2) Å for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–Cu(II) distance was 2.7290 (4) and 2.6970 (6) for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The O/N–Cu–O/N/Cu bond angles ranged from 78.44(4)–172.83(4)° to 81.09(6)–173.01(6)° for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively (Table S1†).
, however, the crystallization of [Cu(ambt)2(clba)4] was shown in the monoclinic crystal system with space group C2/c (Fig. 1). The Cu(II) ions in [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] were hexa-coordinated by one oxygen (O) atom from ambt, one Cu(II) atom from the other nuclear, and four oxygen (O) atoms from cnba and clba for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–O distances ranged from 1.9640 (13)–1.9873 (13) and 1.9558 (19)–1.989 (2) Å for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The Cu(II)–Cu(II) distance was 2.7290 (4) and 2.6970 (6) for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively. The O/N–Cu–O/N/Cu bond angles ranged from 78.44(4)–172.83(4)° to 81.09(6)–173.01(6)° for [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4], respectively (Table S1†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka + n
Ka + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[C]
log[C]
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000

![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group while [Cu(ambt)2(clba)4] was monoclinic system with C2/c space group. Both [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] bound intercalatively to DNA and [Cu(ambt)2(clba)4] showed greater binding constant than [Cu(ambt)2(cnba)4]. The fluorescence quenching of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] to CT-DNA was static quenching. Compared with ligands and cisplatin, the complexes had stronger anti-proliferative activity against HepG2, A549, and HeLa tumor cells. The inhibitory ability of [Cu(ambt)2(clba)4] on the three kinds of tumor cells was stronger. This is congruent with the binding power of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] with DNA, indicating that the antitumor movement of the complexes may be linked to their binding ability with DNA. [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] inhibit tumor cells by inducing apoptosis or necrosis in HepG2 cells, and arresting HepG2 cell cycle in G0/G1 phase. This work may aid in the development of Cu(II) complexes as potential antitumor reagents in the future.
space group while [Cu(ambt)2(clba)4] was monoclinic system with C2/c space group. Both [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] bound intercalatively to DNA and [Cu(ambt)2(clba)4] showed greater binding constant than [Cu(ambt)2(cnba)4]. The fluorescence quenching of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] to CT-DNA was static quenching. Compared with ligands and cisplatin, the complexes had stronger anti-proliferative activity against HepG2, A549, and HeLa tumor cells. The inhibitory ability of [Cu(ambt)2(clba)4] on the three kinds of tumor cells was stronger. This is congruent with the binding power of [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] with DNA, indicating that the antitumor movement of the complexes may be linked to their binding ability with DNA. [Cu(ambt)2(cnba)4] and [Cu(ambt)2(clba)4] inhibit tumor cells by inducing apoptosis or necrosis in HepG2 cells, and arresting HepG2 cell cycle in G0/G1 phase. This work may aid in the development of Cu(II) complexes as potential antitumor reagents in the future.








