Open Access Article
Open Access ArticleLong term storage of miRNA at room and elevated temperatures in a silica sol–gel matrix†
Rajat Chauhana,
Theodore S. Kalbfleischa,
Chinmay S. Potnis b,
Meenakshi Bansalc,
Mark W. Linderd,
Robert S. Keynton*e and
Gautam Gupta
b,
Meenakshi Bansalc,
Mark W. Linderd,
Robert S. Keynton*e and
Gautam Gupta *a
*a
aDepartment of Chemical Engineering, University of Louisville, Louisville, Kentucky 40292, USA. E-mail: gautam.gupta@louisville.edu
bDepartment of Chemistry, University of Louisville, Louisville, Kentucky 40292, USA
cDepartment of Chemistry, Thomas More University, Crestview Hills, KY 41017, USA
dDepartment of Pathology and Laboratory Medicine, University of Louisville, Louisville, Kentucky 40292, USA
eWilliam States Lee College of Engineering, University of North Carolina, Charlotte, 28223, USA
First published on 23rd September 2021
Abstract
Storage of biospecimens in their near native environment at room temperature can have a transformative global impact, however, this remains an arduous challenge to date due to the rapid degradation of biospecimens over time. Currently, most isolated biospecimens are refrigerated for short-term storage and frozen (−20 °C, −80 °C, liquid nitrogen) for long-term storage. Recent advances in room temperature storage of purified biomolecules utilize anhydrobiosis. However, a near aqueous storage solution that can preserve the biospecimen nearly “as is” has not yet been achieved by any current technology. Here, we demonstrate an aqueous silica sol–gel matrix for optimized storage of biospecimens. Our technique is facile, reproducible, and has previously demonstrated stabilization of DNA and proteins, within a few minutes using a standard benchtop microwave. Herein, we demonstrate complete integrity of miRNA 21, a highly sensitive molecule at 4, 25, and 40 °C over a period of ∼3 months. In contrast, the control samples completely degrade in less than 1 week. We attribute excellent stability to entrapment of miRNA within silica-gel matrices.
1. Introduction
This work builds upon our previous efforts in stabilizing nucleic acids (i.e., DNA) and proteins, utilizing the silica sol–gel technology.1,2 The silica sol–gel has previously demonstrated: (1) single-step preservation of E. coli DNA at ambient conditions with no significant nucleolytic and/or oxidative degradation and (2) single-step stabilization of hemoglobin under ambient and refrigeration conditions.1,2 MicroRNAs are an important class of biomarker for autoimmune diseases and human carcinogenesis.3 However, the rapid degradation of miRNA presents a significant challenge to standardization and reproducibility of studies to validate their utility as routine biomarkers.4 Herein, we have utilized the sol–gel technology to stabilize microRNA 21 over a range of temperatures, with single-step recovery.The National Cancer Institute (NCI) has identified room temperature biospecimen storage as a critical requirement of best practices for maximizing the quality of biospecimens against sources of pre-analytical error (collection, process and storage).5 A powerful yet conventional infrastructure of cryo or refrigeration-based storage techniques is presently controlling the ‘biospecimen pre-analytical variables’ (BPV), in a pursuit of addressing ∼100% preservation of biospecimen-integrity and reproducibility in the downstream analyses.5 However, these conventional techniques require significant infrastructure and the expense associated with liquid nitrogen storage make it impractical for many laboratories and in field operations.6–8 Especially RNA samples are stored at sub-zero temperatures [−20, −80] °C to avoid loss of total RNA integrity and unpredictability of miRNA concentration profiles during their downstream assays.9–11 Furthermore, the studies generally indicate that sample degradation increases with storage time, and the freeze–thaw cycles negatively impact biospecimens as the formation of ice crystals results in physical shearing. Clearly, there is dearth of techniques that can store biospecimens at room temperature. Recent advances in room temperature storage include commercial products such as Biomatrica (San Diego, CA, USA), GenTegra (IntegenX, Pleasanton, CA, USA) and Imagene (Evry Cedex, France) that employ Anhydrobiosis (“life without water”) however are constrained with multi-step rehydration and drying cycles of their ultra-dry bio specimens, prior to downstream processing.12–15
The potential for using hybrid functional materials to stabilize biological samples has been recognized by several scientists.16 Encapsulation in silica sol gels, in particular, has been explored due to the relative simplicity and biocompatibility of the gels, and the ability to control its relative water content and salinity.16–20 Despite interesting results over the last 3 decades, traditional sol–gel preparations are inherently complex, time-consuming and require the use of acids or bases as a catalyst, along with alcohols as co-solvents; thus, they may be deleterious to biological samples, and therefore practical solutions compatible with current clinical practices have not been achieved. Another critical aspect of sol–gel immobilization is that the conventional techniques utilize extremely high concentration of silica precursors, that results in intact gels/glasses, however the recovery of biospecimen in solution remains extremely challenging and downstream processing is not feasible.16 Although a higher degree of biospecimen-encapsulation is ubiquitous in higher concentration silica precursors, the biospecimen's integrity is gradually deteriorated among higher concentration sol–gel samples. Such deterioration or lack of ∼100% biospecimen recovery is attributed to the significant non-covalent interactions (i.e., electrostatic, van der walls, electronic) between sol–gel and biospecimen during their long-term storage.17–24
An ideal host matrix for entrapping i.e. encapsulating and immobilizing biospecimen should therefore be (i) neutral aqueous solution with minimal chemical interactions with the biospecimen (ii) feasible to achieve with high reproducibility in any environment (iii) demonstrate intact biospecimen over long-term room temperature storage (iv) should possibly prevent the denaturation by proteases and nucleases that can arise form contamination (v) easily amenable for biospecimen down-stream processing without any separations, and finally (vi) the process could be performed with minimal technical expertise. None of the current techniques can address all these critical requirements.
Here, we demonstrate a simple room temperature storage breakthrough technology using capture and release gels for optimized storage (CaRGOS) for biospecimens. In this article, we investigated the preservation of miRNA 21, which is a potential biomarker of cancer diagnosis.10,11 The CaRGOS formulations are prepared utilizing a deliberately ultra-low concentration of tetramethoxysilane/water suspension that is hydrolyzed in a standard microwave typically for 30 s. Biomolecule (DNA, RNA, protein) of interest can be added to the hydrolyzed silica at room temperature, resulting in its stabilization. Specifically, in our study, we have addressed the room temperature integrity and preservation challenges using a representative highly sensitive bioanalyte miRNA 21. We have demonstrated a single step ∼100% recovery of miRNA 21 at room and elevated temperature using aqueous formulations of CaRGOS with extremely low silica concentrations (0.5%). The aqueous formulations of the CaRGOS are significantly versatile for downstream processing when compared to conventional sol–gel matrices with immobilized biomolecular entities. These often require physical or chemical methods to overcome the non-covalent interactions, with a strong likelihood of disrupting biological activity before downstream usage.16,25,26 Moreover, our technique is completely compatible with a host of proteins as well as other nucleotides such as DNA.
Stabilization of biomolecular entities within the silica matrices typically entails their immobilization. However, the highly aqueous CaRGOS formulations lack the concentration-range requisite for such immobilization. Therefore, to validate the mode of stabilization of biospecimen in highly aqueous formulations of CaRGOS matrices, we have investigated the inherently dual nature of silica precursors: immobilization and nuclease-inhibition. In our highly stable CaRGOS formulations, a remarkable resistance to nuclease digestion was observed by demonstrating ≈100% integrity of yeast RNA in the presence of RNase A. Also, in this paper, we have shown that a ≈69 nm hydrodynamic-sized aqueous formulations of CaRGOS efficiently preserve miRNA 21 up to 82 days at above-freezing temperatures (4, 25, 40) °C with ∼100% single-step recovery.
2. Experimental
2.1 Materials
Taqman microRNA assay (hsa-miR-21; catalog: 4427975), TaqMan® MicroRNA Reverse Transcription Kit (catalog # 4366596), Tris EDTA buffer, Bovine pancreatic RNase A, yeast RNA MW 5000–8000, ethidium bromide, sterile 15.0 ml centrifuge tubes, tetramethyl orthosilicate (TMOS) and sodium chloride were purchased from Sigma Aldrich (Saint Louis, MO, USA). Nuclease free water was purchased from New England BioLabs (MA, USA). miRNA21 (5′-CAA CAC CAG UCG AUG GGC UGU-3′) was purchased from IDT technologies. qPCR tubes were purchased from USA scientific (California, USA) and 96 well plates were purchased from Thermo Fisher Scientific (MA, USA). The reverse transcription thermal cycle was performed at Eppendorf thermocycler (NY, USA). The Dynamic Light Scattering measurements (DLS) were acquired on a Zetasizer (Zetasizer Nano ZS90, Malvern Instruments Ltd, Westborough, MA, USA). The zeta potential measurements were acquired on latter samples using a NanoBrook Zeta PALS Zeta Potential Analyzer (Brookhaven Instruments, Holtsville, NY, USA). Fluorescence measurements were acquired on Molecular Devices SpectraMax M2 plate reader (San Jose, California, USA) and modulus fluorimeter's green module with emission range: 580–640 nm (Sunnyvale, California, USA). FT-IR spectra were measured with the FT-IR spectrometer (PerkinElmer Spectrum 100) with universal ATR (attenuated total reflectance) sample accessory. Raman spectra were acquired on Reva Educational Raman platform (Hellma, Plainview, NY).2.2 Degradation of yeast RNA with enhancement of RNase A concentration
The degradation of yeast RNA with respect to change in the concentration of bovine pancreatic RNase A was monitored at pH 7.5 (0.05 M Tris buffer) containing 0.1 M NaCl. The yeast RNA and EtBr solutions were mixed and incubated for 30 min. A 2.7 ml of pH 7.46 ‘CaRGOS Buffer’ [1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio of (A) CaRGOS (1.5 wt/v%); (B) 0.05 M Tris buffer with 0.1 M NaCl/pH 7.5 and (C) nuclease free water; pH 7.46] or ‘Control Buffer’ [0.05 M Tris buffer with 0.1 M NaCl; pH 7.5] were mixed with 0.2 ml (1 mg ml−1 RNA with 0.077 mM EtBr) and incubated for 100 s. These samples (with or w/o CaRGOS) were added into a respective well in a 96-well reaction plate and mixed gently to bring solution to the bottom of the wells. To the 96-well plates, 1–120 μl of 2.0 μM RNase A were added with the final volume to 200.0 μl and the change in fluorescence intensity monitored.
1 volume ratio of (A) CaRGOS (1.5 wt/v%); (B) 0.05 M Tris buffer with 0.1 M NaCl/pH 7.5 and (C) nuclease free water; pH 7.46] or ‘Control Buffer’ [0.05 M Tris buffer with 0.1 M NaCl; pH 7.5] were mixed with 0.2 ml (1 mg ml−1 RNA with 0.077 mM EtBr) and incubated for 100 s. These samples (with or w/o CaRGOS) were added into a respective well in a 96-well reaction plate and mixed gently to bring solution to the bottom of the wells. To the 96-well plates, 1–120 μl of 2.0 μM RNase A were added with the final volume to 200.0 μl and the change in fluorescence intensity monitored.
2.3 Reverse transcription
Reverse transcription (RT) master mix was prepared using the TaqMan MicroRNA Reverse Transcription Kit components before preparing the reaction (Table S4†). RT components were thawed on ice and 5X RT primers were vortexed. The 10 μl of Master mix – 5X RT Primer was pipetted into a respective well in a 96-well reaction plate using 200 μl 96-well plate. The 5.0 μl of miRNA samples (with or w/o CaRGOS) were added into a respective well in a 96-well reaction plate, cap-sealed and mixed gently to bring solution to the bottom of the wells. The 96-well plates were further incubated on ice for 5 minutes and transferred to Eppendorf thermocycler at 85 °C for 65 minutes.2.4 Real-time qPCR amplification
The 8.67 μl of master mix made for each miRNA 21 (with or w/o CaRGOS) was pipetted into a 100 μl PCR 96-well reaction plate respective well, (Table S5†). The 1.33 μl of RT product was transferred into respective 96-well reaction plate well, cap-sealed and gently mixed to bring solution to the bottom of the tube before real time qPCR amplification.3. Results and discussions
3.1 Synthesis and spectroscopic characterization of CaRGOS
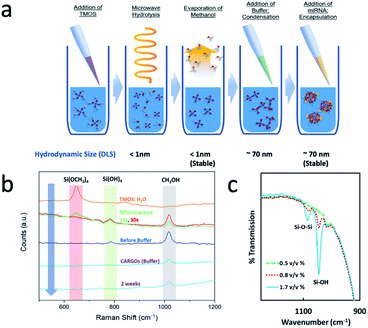
Fig. 1a shows a step by step schematic of the formulation of CaRGOS. Typically, tetramethoxy silane in a desired concentration is mixed with deionized water. A standard microwave oven is used to impart mixing, induce hydrolysis (15–30 s) and simultaneous sterilization that results in the formation of Si(OH)4 without the use of additional chemicals. miRNA, TE buffer and RNase free water is then added and the condensation reaction (formation of Si–O–Si) continues, resulting in the stabilization of miRNA. miRNA does not require any separation and is measured simply by taking an aliquot on a desired day, which is followed by RT PCR studies to establish the quality and quantification of RNA. The process is extremely versatile, involves no use of acids nor alcohols, is compatible with a standard microwave and requires minimum expertise. Raman and IR spectroscopy along with dynamic light scattering (DLS) were utilized to understand the evolution of hydrolysis and condensation reactions in the silica precursor solutions. Fig. 1b shows the step by step Raman spectra of aqueous formulations over a period of time. The theoretical wavenumbers (cm−1) of TMOS precursor (Si(OCH3)4), intermediates [(Si(OCH3)3OH, Si(OCH3)2(OH)2), Si(OCH3)(OH)3)], silicic acid (Si(OH)4 and methanol (CH3OH) are expected at 640–650 cm−1, 673–725 cm−1, 750–780 cm−1 and 1020 cm−1 respectively.27 Experimentally, we observe a wavenumber at 646 cm−1 for 1.25% TMOS/water solution prior to microwave exposure. After 15 s of exposure, this peak gradually decreases, and TMOS intermediate/methanol wavenumbers are observed at 750–780 cm−1 and 1020 cm−1 respectively. After 30 s of exposure to microwaves, the TMOS wavenumber (cm−1) completely disappears indicating complete hydrolysis and an increase in Si(OH)4/dimer and methanol wavenumbers at 780 cm−1 and 1020 cm−1. The efficiency of the hydrolysis was computed utilizing the Raman peak of methanol aqueous solutions (Table S1†). The hydrolyzed precursor exhibits stability for up to 3 months. Buffer is subsequently added to the CaRGOS solution, and a decrease in methanol peak is observed due to subsequent dilution. In addition, the peak at 780 cm−1 completely disappeared indicating a change in the structure of the Si(OH)4/dimer. In the final solution, the methanol concentration was estimated to be around 80 μM (Table S2 and Fig. S1†).The disappearance of silicic acid/dimer peak was carefully studied using dynamic light scattering. Prior to the addition of buffer, around 1 nm hydrodynamic diameter is observed, indicating a possible absence of colloidal particles and therefore indicating presence of only silicic acid [Si(OH)4] or some dimers in highly aqueous CaRGOS. Upon the addition of buffer, CaRGOS formulations display a transition from a ∼1 nm hydrodynamic-sized dispersion to highly monodisperse and ∼69 nm hydrodynamic-sized dispersion (Table S3†). This is possibly due to a decrease in electrostatic repulsions between negatively charged silica precursors in the presence of a saline environment. The addition of miRNA shows a negligible change in zeta potential, polydispersity index and hydrodynamic diameter (Table S3†). Since the concentrations of our silica precursor are extremely low, the IR signatures of low concentration gels are similar to pure water. Therefore, the concentration of silica precursor was gradually increased to observe the characteristic peaks of silica. Fig. 1c shows the IR spectra of miRNA-CaRGOS with variable silica concentration demonstrating increases in bands of 1085 cm−1 (Si–O–Si asymmetric vibration) and 1045 cm−1 (Si–OH asymmetric vibration). After completing spectroscopic investigation of hydrolysis and condensation of silica precursor, we investigated the compatibility of our CaRGOS process with miRNA 21 measurements by RT-PCR.
3.2 Investigation of compatibility of CaRGOS with miRNA
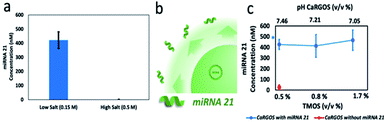
An ideal storage solution should have near physiological conditions and the key litmus test is to evaluate the compatibility of the CaRGOS with a sensitive biospecimen. It should be noted that the ionic strength, pH of the CaRGOS, and the concentration of silica precursor can drastically impact the stability of biomolecules. E.g. ionic strength of the solution can directly impact the electrostatic repulsions between negatively charged CaRGOS in a buffer environment and consequently affect the size, stability and monodispersity of the silica precursors (Table S3†). Also, salinity can dictate the nature of non-covalent interactions between biomolecules and CaRGOS matrices. Similarly, pH of the solution can dictate the extent of condensation reaction as well as the stability of biomolecules. Therefore, a systematic study was performed by varying these conditions and simultaneously monitoring the miRNA concentrations in each case respectively.Fig. 2a shows the miRNA concentration with 0.5% CaRGOS in Tris EDTA buffer with either 0.15 M NaCl or 0.5 M NaCl, while keeping fixed the rest of the CaRGOS storage parameters [10 mM Tris–HCl (pH 7.5), 1 mM EDTA, and ∼500 nM miRNA 21 ]. Measurement of miRNA by PCR amplification, utilizes sequence-specific primers and is dependent on the structural integrity of the nucleic acid. Thus, this technique allows for the simultaneous analysis of both the concentration and structural integrity of the miRNA under the various CaRGOS storage conditions. Reverse transcription (RT) was performed on the miRNA 21 sample aliquots (with and w/o CaRGOS) using TaqMan MicroRNA Reverse Transcription Kit and thermal cycler, followed by real-time quantitative polymerase chain reaction (qPCR) amplification with an applied 0.1 CT threshold value (Tables S4 and S5†). The miRNA 21 concentration (nM) of CaRGOS aqueous formulations were evaluated by measuring mean CT values using standard calibration curves (Fig. S2 and S3†). A strong correlation between miRNA 21 concentration (nM) of CaRGOS aqueous formulations and mean CT values was observed with R2 = 0.99 respectively (Fig. S2†). In response to salt stress, the miRNA concentration levels in low salt buffer were 426.63 ± 46.33 nM (CT: 11.5 ± 6.0), whereas in high salt buffer the miRNA was not measurable (CT: 31.1 ± 1.2) [CT ≥ 30 are equivalent to nuclease free water]. These conditions are similar to biological pH and ionic strength with an extremely low concentration (0.5% TMOS) of silica. Under these conditions, the nucleic acids have a negative charge, and the silica colloids also exhibit a negative charge as well. Therefore, electrostatic repulsive forces dominate and the miRNA21 is stabilized. Fig. 2b shows a schematic of miRNA 21 in CaRGOS. We anticipate that at such low concentrations of silica that are slightly viscous in nature, the miRNA 21 will be repelled around silica colloids and will remain stable. These repulsive interactions also allow for ease of measurement in the presence of CaRGOS without requirement of a separation step.
Once, the concentration of salt was optimized, we varied the concentration of silica precursor, Fig. 2c. As the concentration of hydrolyzed silica (silicic acid) is increased, a slight decrease in pH is observed for the CaRGOS with buffer and biospecimen. Once prepared, the pH of the CaRGOS formulations is stable, reproducible [SD < ±0.2 pH units] and not altered by air exposure. A ∼500 nM payload of miRNA 21 was added to each CaRGOS formulation. Fig. 2c shows the miRNA concentration of miRNA measured within 1 h from addition to CaRGOS formulations with variable TMOS concentrations.
3.3 Long-term evaluation of miRNA concentration in CaRGOS
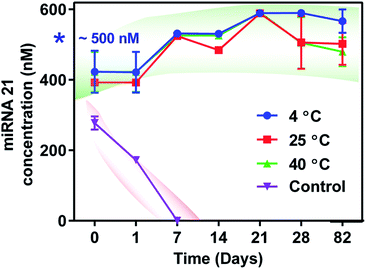
Based on the observation of better reproducibility and no significant difference in miRNA measurements between 0.5% vs. 1.7% TMOS, the 0.5% TMOS formulation was utilized for long-term miRNA 21 storage experiments. At room temperature, the miRNA 21 concentration (nM) in the mixtures without CaRGOS sol–gel matrices were (277.6 ± 18.8) nM within 3 h of addition, (172.0 ± 0.2) nM within 24 hours and was not measurable on day 7, Fig. 3. We utilized three temperature conditions (a) refrigeration (4 °C) (b) ambient temperature (25 °C) and (c) 40 °C respectively. Fig. 3 shows the quantitative RT-PCR analysis of 0.5 v/v% CaRGOS/miRNA 21 mixtures at 4, 25 and 40 °C over a period of 82 days. In contrast to the absence of CaRGOS, we observed ∼100% recovery of miRNA 21 (∼500 nM) in the CaRGOS mixtures at each temperature over the 82 day period: (426.63 ± 46.33) nM on day 0, (392.35 ± 8.28) nM on day 1, (524.53 ± 6.54) nM on day 7, (484.32 ± 2.46) nM on day 14, (588.94 ± 0.54) nM on day 21, (505.95 ± 75.00) nM on day 28 and (500.19 ± 59.92) nM on day 82 inferring a thermal stable miRNA 21 within CaRGOS formulations. The low concentration of miRNA measured on day 0 and day 1 is attributed to possible interference of PCR amplification by the methanol by product of TMOS generation.2 We observed similar results for our DNA preservation studies.2 This interference is avoided by preferential evaporation of methanol due to its higher vapor pressure. The mode of miRNA stability in lower concentrations CaRGOS sol–gel matrices might be attributed to the combined effect of some miRNA immobilization, caused by sterically-restricted synergistic interactions with CaRGOS network and local solvent/pH microenvironment [Tris EDTA buffer/pH > 7] of these aqueous CaRGOS formulations.16,28,29 However, such long-term stability cannot simply arise from repulsive interaction, and we anticipate some other factors play a critical role. A key possibility is that the CaRGOS can interact with RNase (which can arise from compromised sterility) and therefore can prevent the denaturation of miRNA by RNase. E.g. previously, Buijs et al. had reported an electrostatic adsorption induced destabilization of proteins (e.g. RNase A/Lysozyme) on 11 nm sized silica particles.30 Also the non-covalent interaction between biological entities and silica nanomaterials are well-known to electrostatically destabilizes nucleases (i.e. RNase) and protease activity, as well as providing stability to biospecimens (e.g. lipids, proteins, nucleic acids) in their immobilization matrices.16,19,22,23,25,263.4 Evaluation of stability in the presence of RNase
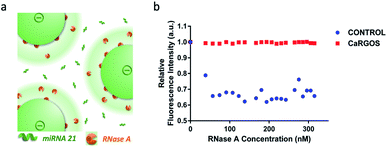
At pH 7.4, RNase A has asymmetrically stronger positive charge density across the longest axis of the molecule (PDB 2AAS).22,31,32 Also, RNase A's active site has been reported to reside in this electropositive region. Therefore, the potential for electrostatic interaction between the positive domain of RNase A and negatively charged CaRGOS as shown in Fig. 4a suggests that CaROGS may inhibit the activity of RNase's.To validate this hypothesis, we examined CaRGOS capability to prevent degradation of RNA in presence of pancreatic RNase A.33 As mentioned in the Experimental section and shown in the Fig. 4b and S4,† the fluorescence emission intensities of yeast RNA intercalated ethidium bromide [EtBr: (em: 600 nm; ex: 510 nm)] solutions in CaRGOS and control buffers were monitored in the presence of incrementally increasing RNase A concentrations (range: 0–320) nM. Normalized with EtBr's fluorescence emission at 0 nM RNase A concentration, a ∼40% relative fluorescence quenching was observed in control buffer solutions, (without CaRGOS) indicating degradation of yeast RNA with incremental increase in RNase A concentrations.33 However, unaltered fluorescence intensity was observed in CaRGOS buffer solutions within (0–320) nM range of RNase A concentrations, Fig. 4b. Such unaltered fluorescence emissions were attributed to the electrostatic adsorption induced RNase A inhibition with CaRGOS.22,23,30–32,34
CaRGOS demonstrates larger hydrodynamic size of ∼69 nm and displayed high stability in their buffer dispersions with zeta potential ∼ −21 mV respectively. Therefore, CaRGOS is an excellent candidate for preventing RNA degradation from environmental RNase contamination (e.g., bacteria, fungi), during transportation/storage and downstream processing.7,8 Here, we have accordingly elucidated the mechanism of RNA storage at room/elevated temperatures demonstrating (a) RNA immobilization within CaRGOS and (b) RNase inhibition respectively.
4. Conclusions
Novel, facile and efficient ultra-low concentration aqueous CaRGOS have been formulated using a bench top microwave that allows for the long-term stabilization of biospecimens. Spectroscopic studies indicate complete hydrolysis of silica precursor that can be utilized to stabilize any biospecimen of interest. We demonstrate stability of a highly sensitive cancer biomarker miRNA 21 at ambient and elevated temperatures (4, 25 and 40 °C) for nearly 3 months, with ∼100% single-step recovery without the need of any purification protocol. The mode of stabilization is attributed to the RNase (sterile-compromised contaminated environment) inhibition by optimized aqueous formulations of CaRGOS [low ionic strength, optimum pH (∼7.46), large hydrodynamic sized and high surface potential] as well as miRNA immobilization caused by localized interaction of CaRGOS matrix. CaRGOS formulations are completely compatible with current clinical practices and require no technical expertise and we anticipate their utility in storage of several relevant biospecimens as well as in enhancing the shelf life of several drugs including monoclonal antibodies.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors acknowledge financial support from the Office of the Executive Vice President for Research and Innovation at the University of Louisville (UofL). Author also acknowledge financial support from National Cancer Institute (NCI) exploratory/development grant [ID: Grant number: 1R21CA251042-01]. Gupta acknowledge the startup funds from University of Louisville. We acknowledge Price Institute of Surgical Research, Hiram C. Polk Jr MD Dept of Surgery, School of Medicine (UofL) for their qRT-PCR technical support and Nanotherapeutics laboratory, Bioengineering Department (UofL) for sterilized biological hood/accessories support.References
- J. Boylan, R. Chauhan, K. Koneru, M. Bansal, T. Kalbfleisch, C. S. Potnis, K. Hartline, R. S. Keynton and G. Gupta, RSC Adv., 2021, 11, 13034–13039 RSC.
- C. W. Narvaez Villarrubia, K. C. Tumas, R. Chauhan, T. MacDonald, A. M. Dattelbaum, K. Omberg and G. Gupta, Emergent Mater., 2021, 1–8, DOI:10.1007/s42247-021-00208-3.
- H. Lan, H. Lu, X. Wang and H. Jin, BioMed Res. Int., 2015, 2015, 125094 Search PubMed.
- G. Cirmena, M. Dameri, F. Ravera, P. Fregatti, A. Ballestrero and G. Zoppoli, Cancers, 2021, 13, 3460 CrossRef PubMed.
- K. B. Engel, J. Vaught and H. M. Moore, Biopreserv. Biobanking, 2014, 12, 148–150 CrossRef CAS PubMed.
- J. J. Lou, L. Mirsadraei, D. E. Sanchez, R. W. Wilson, M. Shabihkhani, G. M. Lucey, B. Wei, E. J. Singer, S. Mareninov and W. H. Yong, Clin. Biochem., 2014, 47, 267–273 CrossRef CAS PubMed.
- A.-L. Fabre, M. Colotte, A. Luis, S. Tuffet and J. Bonnet, Eur. J. Hum. Genet., 2013, 22, 379 Search PubMed.
- G. L. Mutter, D. Zahrieh, C. Liu, D. Neuberg, D. Finkelstein, H. E. Baker and J. A. Warrington, BMC Genomics, 2004, 5, 88 CrossRef PubMed.
- D. Ibberson, V. Benes, M. U. Muckenthaler and M. Castoldi, BMC Biotechnol., 2009, 9, 102 CrossRef PubMed.
- J. Zhu, L. Chen, L. Zou, P. Yang, R. Wu, Y. Mao, H. Zhou, R. Li, K. Wang, W. Wang, D. Hua and X. Zhang, Hum. Immunol., 2014, 75, 348–353 CrossRef CAS PubMed.
- A. H. Harrill, S. D. McCullough, C. E. Wood, J. J. Kahle and B. N. Chorley, Toxicol. Sci., 2016, 152, 264–272 CrossRef CAS PubMed.
- H. S. Stevenson, Y. Wang, R. Muller and D. C. Edelman, Biopreserv. Biobanking, 2015, 13, 114–122 CrossRef CAS PubMed.
- H. Martinez, G. Beaudry, J. Veer, M. Robitaille, D. Wong, B. Iverson and R. Nuñez, Ambient temperature storage of RNA in GenTegra™ for use in RT-qPCR, 2010 Search PubMed.
- A. G. Kansagara, H. E. McMahon and M. E. Hogan, Nat. Methods, 2008, 5, iv–v CrossRef.
- X. Liu, Q. Li, X. Wang, X. Zhou, X. He, Q. Liao, F. Zhu, L. Cheng and Y. Zhang, Biopreserv. Biobanking, 2015, 13, 49–55 CrossRef CAS PubMed.
- W. Xiaolin, A. Nada Ben, S. A. Gisela, V. T. Maria, H. Christophe, F. D. Martin and C. Thibaud, Curr. Top. Med. Chem., 2015, 15, 223–244 CrossRef PubMed.
- Z. Chen, S. Han, M. Shi, G. Liu, Z. Chen, J. Chang, C. Wu and Y. Xiao, Appl. Mater. Today, 2018, 10, 184–193 CrossRef.
- H. Meng, M. Liong, T. Xia, Z. Li, Z. Ji, J. I. Zink and A. E. Nel, ACS Nano, 2010, 4, 4539–4550 CrossRef CAS PubMed.
- D. M. Schlipf, S. E. Rankin and B. L. Knutson, ACS Appl. Mater. Interfaces, 2013, 5, 10111–10117 CrossRef CAS PubMed.
- J. Shen, Q. He, Y. Gao, J. Shi and Y. Li, Nanoscale, 2011, 3, 4314–4322 RSC.
- S. Peng, B. Bie, Y. Sun, M. Liu, H. Cong, W. Zhou, Y. Xia, H. Tang, H. Deng and X. Zhou, Nat. Commun., 2018, 9, 1293 CrossRef PubMed.
- W. Shang, J. H. Nuffer, J. S. Dordick and R. W. Siegel, Nano Lett., 2007, 7, 1991–1995 CrossRef CAS PubMed.
- A. A. Vertegel, R. W. Siegel and J. S. Dordick, Langmuir, 2004, 20, 6800–6807 CrossRef CAS PubMed.
- B. G. Cha, J. H. Jeong and J. Kim, ACS Cent. Sci., 2018, 4, 484–492 CrossRef CAS PubMed.
- C. J. Lee, J. H. Jung and T. S. Seo, Anal. Chem., 2012, 84, 4928–4934 CrossRef CAS PubMed.
- V. B. Kandimalla, V. S. Tripathi and H. Ju, Crit. Rev. Anal. Chem., 2006, 36, 73–106 CrossRef CAS.
- T. W. Zerda and G. Hoang, J. Non-Cryst. Solids, 1989, 109, 9–17 CrossRef CAS.
- C. Carrasquilla, P. S. Lau, Y. Li and J. D. Brennan, J. Am. Chem. Soc., 2012, 134, 10998–11005 CrossRef CAS PubMed.
- S. Perumal, S. k. Ramadass and B. Madhan, Eur. J. Pharm. Sci., 2014, 52, 26–33 CrossRef CAS PubMed.
- H. Larsericsdotter, S. Oscarsson and J. Buijs, J. Colloid Interface Sci., 2001, 237, 98–103 CrossRef CAS PubMed.
- C. S. Lee and G. Belfort, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 8392–8396 CrossRef CAS PubMed.
- J. Santoro, C. González, M. Bruix, J. L. Neira, J. L. Nieto, J. Herranz and M. Rico, J. Mol. Biol., 1993, 229, 722–734 CrossRef CAS PubMed.
- D. R. Tripathy, A. K. Dinda and S. Dasgupta, Anal. Biochem., 2013, 437, 126–129 CrossRef CAS PubMed.
- P. Roach, D. Farrar and C. C. Perry, J. Am. Chem. Soc., 2006, 128, 3939–3945 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04719a |
| This journal is © The Royal Society of Chemistry 2021 |