Open Access Article
Open Access ArticleThe sustainable cycle of a new cacao-based bioplastic: from manufacturing to exploitable biodegradation products†
Allan Calmont de Andrade Almeidaa,
João Guilherme de Moraes Pontes a,
Gabriel Rodrigues Alvarengaa,
Henrique Finocchio*b and
Taicia Pacheco Fill*a
a,
Gabriel Rodrigues Alvarengaa,
Henrique Finocchio*b and
Taicia Pacheco Fill*a
aUniversidade Estadual de Campinas (UNICAMP), Organic Chemistry, Laboratório de Biologia Química Microbiana (LABIOQUIMI), P. O. Box 6154, Campinas, SP 13083-970, Brazil. E-mail: taicia@unicamp.br
bAfinko Soluções em Polímeros, São Carlos, SP 13570-591, Brazil. E-mail: henrique@afinkopolimeros.com.br
First published on 8th September 2021
Abstract
The exponential growth of plastic consumption in the last decade became a large economic and ecological issue; therefore, strategies have been used to mitigate the environmental impacts, including the manufacture of biodegradable bio-based plastics and biodegradation strategies. Herein, a new bio-based plastic was developed consisting of a polymeric recyclable matrix (polyethylene or polypropylene) with a vegetal polymeric material from cocoa husk. Mechanical and rheological properties were evaluated and the new material showed interesting tensile strength compared to completely non-biodegradable plastics. The new polymeric material was submitted to biodegradation processes using different fungi species. The biodegradation caused by Colletotrichum gloeosporioides, Xylaria sp. and Fusarium graminearum in the new polymeric material was analyzed through scanning electron microscopy with energy-dispersive X-ray spectroscopy (SEM-EDS) and tensile tests. Furthermore, ultra performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) and mass spectrometry imaging (MSI) were applied to identify metabolites produced in consequence to the biodegradation process. Interestingly, some compounds produced present high economic value.
1. Introduction
Polymers are widely used, versatile and practical materials that are inserted in almost every aspect of our daily lives. However, the exponential growth of plastics in the last decade became a large economic and ecological issue.1–3 Besides all the environmental concerns, the production keeps getting exponentially higher worldwide, especially in the United States, China, India and Brazil.4–6Many strategies have been used to mitigate the environmental impact caused by plastics pollution, including recycling,7–9 incineration,7,10,11 landfilling;12,13 application in construction of roads and pavements,14–16 use in the composition of fuels17,18 and the development of biodegradable polymers with interesting and applicable mechanical properties.19–21 Among these strategies, the biodegradation of biodegradable polymers is the most eco-friendly.22,23 According to Alshehrei, biodegradation is any chemical or physical process caused by biological activity, which impacts in terms of weight loss, loss of mechanical properties or visual changes.24 The biodegradability is related to the ability of a material to decompose after the interaction with specific biological elements such as microorganisms. The molecular structure of the polymer is the main contributing factor for its biodegradability.25,26 Non-biodegradable plastics (based on fossil resources) may be degraded in more than 1000 years,27 while bioplastics and biodegradable plastics may be degraded within 6 to 13 months.28
According to the European Bioplastic, in 2018 bioplastics represented roughly one percent of the 335 million tons of plastic produced annually, implicating in just 935 thousand tons of biodegradable consumed plastics.29 The long-period-degradation of non-biodegradable plastics, as polyolefins, is due to the fact that most of the microorganisms failed to adhere to the polymer surface hindering the production of enzymes necessary to degrade the polymer carbon backbone with its high molecular weight long chains.30 The environmental conditions also play an important role in the degradation kinetics of polymers.30,31 On the other hand, bioplastics, mainly the biodegradable ones, have showed a great degradation potential,32 in terms of percentage of biodegradability: polylactide (PLA) range 60–100%,33–36 polyhydroxyalkanoates (PHA) range 64.3–100%,37–39 and polybutylene succinate (PBS) 90%.40 Furthermore, vegetal fillers can be incorporated to non-degradable plastics to improve the properties of the polymer matrix and potentially accelerates degradation compared to the pure non-biodegradable fossil based plastics. The polyethylene manufactured from ethanol was the fourth most produced bio-based plastic in the world. In 2019, it represented 11.8% of the global production capacity of bio-based plastics.31 Therefore, polymers composed by biopolyethylene with vegetable fillers are very promising.
Degradation of polymers through the use of microorganisms is extensively studied.41–43 Sangale et al., for instance, demonstrated that Aspergillus sydowii was able to partially degrade polyethylene and they described the highest reduction in tensile strength so far.23 In another study performed by Ojha et al., it was isolated from soil samples two species of fungi (Penicillium oxalicum and Penicillium chrysogenum) that exhibited good abilities to degrade plastics such as low density polyethylene (LDPE) and high density polyethylene (HDPE).44 However, there are no data available concerning the metabolites produced by these microorganisms in the biodegradation process or the biodegradation products.
Herein, we describe the biodegradation of a new manufactured cocoa bio-based plastic using microorganisms isolated as endophyte from cocoa fruit and fruit phytopathogens. The study evaluated the mechanical properties of the degraded bio-based plastics through tensile tests, scanning electron microscopy (SEM), and energy dispersive spectroscopy (EDS). In addition, we identified the secondary metabolites produced in consequence of the biodegradation process by the microorganisms using liquid chromatography-mass spectrometry (LC-MS/MS) and imaging mass spectrometry. Therefore, we evaluated the biodegradation potential and the ability of the microorganisms to produce interesting and biological active products that may have industrial applications.
2. Material and methods
2.1. Microorganisms used in the study
In total five fungal strains were isolated from the cocoa fruit (Theobroma cacao). The fruit was washed using 4% sodium hypochlorite and rinsed with water. Posteriorly, a piece (2 cm × 2 cm) of the cacao fruit was inoculated in potato dextrose agar (PDA) plates.45 Colletotrichum gloeosporioides tested in our studies was characterized by sequencing and phylogenetic analysis of gene fragments from the ribosomal operon and part of the β tubulin gene (Fig. S1 and S2 in ESI†). In addition to the five fungi isolated from cacao, we investigated the degradation potential of the fungus F. graminearum CECT (Colección Española de Cultivos Tipos) 20924 from Universitat de València.2.2. Manufacture of bio-based plastic and material analyses
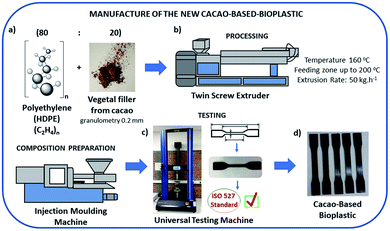
The bio-based plastics were manufactured using a matrix of polyethylene and vegetal filler from cocoa shells. The polymers used as synthetic matrix were provided by Braskem Company (HDPE HC7260LSL) and cocoa was acquired at the supermarket and prepared to be grounded with average grain size of 0.2 mm.The bio-based plastic was processed in a twin screw extruder with linear temperature from 160 °C in the feeding zone up to 200 °C in the die head and the extrusion rate fixed at 50 kg h−1. The composition prepared by extrusion was molded by injection using an injection-molding machine, Romi Pratica 130 model. The test pieces obtained were tested on an universal testing machine, Instron 2330, in accordance to ISO 527 standard (Fig. 1).
The nominal properties of the HDPE HC7260LSL that was used in this study correspond to a melt flow index equal to 0.72 g min−1, density of 0.96 g cm−3, tensile strength of 29 MPa, izod impact strength of 35 J m−1, roughness of 63 shore D, and thermal distortion temperature of 74 °C. Other characteristics related to granulometry of the cocoa bean are shown in Table S1 in ESI.†
2.3. Fungal biodegradation studies
The analysis of fungal degradation capabilities was done cultivating them with a test specimen of biopolymer (5A type follow the ISO 527 standard) in Czapeck culture medium. The biodegradation studies were performed according to the methodology described by Volova et al.46 using approximately 25 mL of Czapeck culture media (3.0 g L−1 NaNO3 Art Lab, 1.0 g L−1 KH2PO4 Ecibra, 0.50 g L−1 MgSO4 chemical kinetics, 0.050 g L−1 FeSO4·7H2O Vetec, 1.0 g L−1 KCl, 30.0 g L−1 glucose Synth, and 2.0% (m/m) agar–agar diluted in deionized water). The test specimen of plastic is deposited in the middle of the Petri dish measuring 90 mm × 15 mm and the inoculum is made. The plates were kept at 25 °C in a Tecnal TE-371 incubator. Ten samples of polymer were inoculated with the tested fungus and ten control samples were incubated during 45 days. Other ten samples of each group were incubated during 75 days.2.4. Scanning electron microscope with energy-dispersive X-ray spectroscopy analysis
After 45 days incubation, the degraded plastic specimen was removed and sterilized with ethanol 70% during 30 min. An environmental scanning electron microscope (ESEM; Quanta 250 FEG, FEI Co., OR, USA) equipped with energy-dispersive X-ray spectroscopy EDS (EDS; Oxford Xmax, 50 mm) operating in low-vacuum mode at 21 °C and 55–60% relative humidity was used to analyze images of cross-sectional and metal localization on the cross-section. ESEM was operated at 5 kV and used a secondary electron detector.2.5. Statistical analysis
Tensile strength, elongation, and Young's modulus were measured in quintuplicate for polymeric control and for each replicate of the polymers inoculated with the fungus species studied (C. gloeosporioides, F. graminearum, and Xylaria, sp.). These measurements were done in the period of 75 days after the biodegradation process. T-test was calculated in Microsoft Excel software. p-Values less than 0.05 were considered with statistical significance.2.6. Metabolites extraction and HRESIMS analyses
The culture medium containing the fungus was completely removed for solid liquid extraction (SLE) in 50 mL Falcon tubes using methanol, ethyl acetate and dichloromethane solution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (v/v).47 In the extraction process was used 30 mL of this solution per plate. The culture was then extracted in an ultrasonic bath (Branson 2800) for 40 min. The extracts were dried and analyzed by HRESIMS.
3 (v/v).47 In the extraction process was used 30 mL of this solution per plate. The culture was then extracted in an ultrasonic bath (Branson 2800) for 40 min. The extracts were dried and analyzed by HRESIMS.
Liquid chromatography analyses were performed on ultra-high performance liquid chromatography – UHPLC (UltiMate 3000) coupled to high-resolution Orbitrap Q-Exactive mass spectrometer (Thermo Fisher Scientific®) with heated ionization electrospray source (RESI-II) set at 3500 V. The m/z values range was scanned between 100 to 1000 in positive ion mode. The chromatographic separation was performed in a Thermo Scientific Accucore C18 (2.6 μm, 100 × 2.1 mm) column using water with 0.1% formic acid, v/v (phase A) and acetonitrile (phase B) as mobile phase and eluted in gradient mode at 45 °C with flow rate of 0.2 mL min−1.
Exact mass of metabolites were obtained using Thermo Xcalibur Roadmap™ software 3.0.63 version. The annotation of the metabolites was done using dictionary of natural products (DNP).48 Other databases used in metabolite annotation were the global natural product social molecular networking (GNPS),49 the human metabolome database (HMDB),50 and MassBank.
2.7. Mass spectrometry imaging (MSI) analyses
The MSI analysis was performed using a spectrometer from Thermo Scientific® Q-Exactive equipped with desorption electrospray ionization (DESI) source (Omni Spray 2D®-3201 model – Prosolia, Indianapolis, USA) and coupled to a mass analyzer of the type hybrid quadrupole-Orbitrap. It was used as mobile phase acetonitrile (LiChrosolv®) and methanol HPLC grade with a solvent flow rate of 1500 μL min−1 and resolution of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 (m/Δm at m/z 200). The analysis was carried out in negative and positive mode with m/z range between 100 to 1500 and spray voltage of 1.5 kV.
500 (m/Δm at m/z 200). The analysis was carried out in negative and positive mode with m/z range between 100 to 1500 and spray voltage of 1.5 kV.
The sample preparation was done with polymeric control and the polymer submitted to the biodegradation process with the different fungi tested. The samples were glued with tape on the glass plate and analyzed in the same scanning for comparative effect. The sample size used was 1.0 cm × 5.0 cm (height × width) for each polymeric region. Images were processed in BioMAP 3.8.0.4 (Novartis, Basel, Switzerland) software and exact mass was obtained using Thermo Xcalibur Roadmap software 3.0.63.
3. Results and discussion
3.1. Manufacture of the new bio-based plastic
Different compositions of polyethylene (PE) or polypropylene (PP) were tested with cocoa shells for the manufacture of the new bio-based plastic. The best rheological properties were obtained from the composition HDPE/cocoa (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) with rheological torque equivalent to 10.0 N m (Table S2 in ESI†) and melt flow index (MFI) of 5.4 g/10 min (at 190 °C) (Table S3 in ESI†), these characteristics are related to the flow behavior of polymer composites51 and polymer processability.52 Furthermore, mechanical properties (Young's modulus, tensile strength, and elongation) were also compared between each of the different compositions (PP100, PP80/20, PP80/20 + compatibilizing agent, PE100, PE80/20, PE80/20 + compatibilizing agent) and evaluated to select the optimal polymer composition (Table S4 in ESI†). The benefits of this new material are the lower amount of polyethylene with substitution of 20% cocoa fiber, which was being discarded by the Brazilian cocoa industry, in addition to the material resistance compatible with materials with 100% fossil origin. The applications of this new material are innumerable as sustainable utensils and packaging for products. This new bioplastic can be applied as materials of polyethylene molded parts such as chairs, toys, bottles, jars, and others. This new material was submitted to the biodegradation studies with different fungi species.
20) with rheological torque equivalent to 10.0 N m (Table S2 in ESI†) and melt flow index (MFI) of 5.4 g/10 min (at 190 °C) (Table S3 in ESI†), these characteristics are related to the flow behavior of polymer composites51 and polymer processability.52 Furthermore, mechanical properties (Young's modulus, tensile strength, and elongation) were also compared between each of the different compositions (PP100, PP80/20, PP80/20 + compatibilizing agent, PE100, PE80/20, PE80/20 + compatibilizing agent) and evaluated to select the optimal polymer composition (Table S4 in ESI†). The benefits of this new material are the lower amount of polyethylene with substitution of 20% cocoa fiber, which was being discarded by the Brazilian cocoa industry, in addition to the material resistance compatible with materials with 100% fossil origin. The applications of this new material are innumerable as sustainable utensils and packaging for products. This new bioplastic can be applied as materials of polyethylene molded parts such as chairs, toys, bottles, jars, and others. This new material was submitted to the biodegradation studies with different fungi species.
Various microorganisms such as bacteria and fungi are responsible for the degradation of both natural and synthetic plastics.53 Endophytic microorganisms, in particular, that can be isolated from vegetable sources, are potentially interesting organisms to be used as biodegradation agents as described by Russell et al.54 The cocoa endophytic fungi are the organisms with the highest biodegradation potential, since they constantly require the cocoa nutrients for their own survival. There are few data related to the use of endophytic microorganisms isolated from plant sources for biodegradation purposes. In this sense, we evaluated the degradation ability of the fungi that are naturally associated to the cocoa.55,56 Fungi used in our biodegradation study are shown in Fig. S3 in ESI.†
Rubini et al. reported various species of endophytic fungi growing in cocoa beans tissues, among them are Colletotrichum gloeosporioides and Xylaria sp.57 that were also isolated and used in our biodegradation studies. In our isolation procedure, five fungi species were isolated as endophytes from cocoa fruit (Theobroma cacao) using potato dextrose agar (PDA) culture medium, however just two of the isolated fungi were selected for further studies – Xylaria sp. and Colletotrichum gloeosporioides, due to the accelerated growth in the bioplastic surface. Fusarium graminearum, a corn phytopathogen was included in this study, since Rubini et al. reported three different species of Fusarium spp. as endophytic fungi of cocoa57 and F. graminearum exhibited an accelerated growth in the bioplastic surface (Fig. S3 in ESI†).
3.2. Evaluation of the polymer degradation by fungi
Biodegradation studies allow new solutions for the long-life cycle of non-biodegradable polymers in nature. The option to use microorganisms is a sustainable alternative since it accelerates the material degradation and may allow the production of compounds with commercial applications. Degradation of plastics usually involves physical, chemical or biological process, or a combination of them.58 The matrix used in our studies are polyethylene or polypropylene, which are polyolefins, and do not have oxidized regions, therefore, microorganisms hardly adhere to their hydrophobic surfaces.59 The addition of cocoa fibers into the bio-plastic may facilitate the penetration and access of microorganisms to the polymer, thus contributing to the biodegradation process. According to Arutchelvi et al. the biodegradation mechanism follows the steps: (i) attachment of the microorganism to the polymer surface; (ii) growth of the microorganism making use of the polymer as carbon source; (iii) primary degradation of polymer; and (iv) ultimate degradation.59 These steps can be visualized in a schema reported by Müller, 2005.30We analyzed the microbial degradation potential of HDPE containing 20% cocoa fiber using parameters and methodologies previously described.23,60 We evaluated the potential degradation of the new polymeric material by SEM and EDS analyses, in addition to tensile strength modifications. Scanning electron microscopy have been previously applied to evaluate the degradation of oxo-biodegradable plastic by Pleurotus ostreatus.61
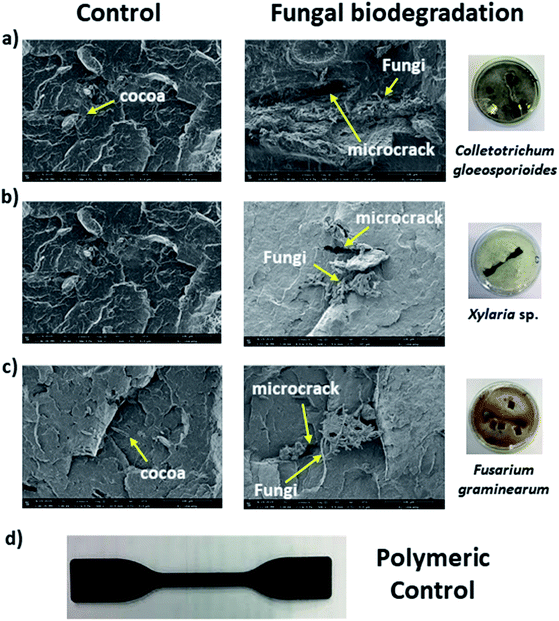
In our biodegradation experiments, hyphae of C. gloeosporioides covered the entire polymeric surface (Fig. 1). SEM cross section analyses (Fig. 1a) were performed and indicated the presence of the fungus inward the bioplastic as shown in the comparison between the polymeric control and the polymer submitted to the biodegradation studies for 75 days. We observed the presence of microcracks indicating material gap regions (Fig. 2), as was verified in other microbial biodegradation studies using SEM technique.62 These microcracks may imply a partial polymer degradation since the fungus hyphae could interfere with the mechanical parameters, and consequently create regions of stress concentration.63 SEM analyses were also used to monitor the biodegradation by Xylaria sp. and F. graminearum in this study (Fig. 1b and c).
Hyphae of the fungus Xylaria sp. was also identified inward the polymer after the biodegradation experiments for 75 days. We also observed the presence of microcracks as evidenced in Fig. 2b indicating a partial polymer degradation. The Young's modulus values however, did not significantly decrease in the tensile tests after 75 days experiment (Fig. 3a and b). Young's modulus is a mechanical property that measures the stiffness of a solid material. Defines the relationship between stress and strain in the elastic region of stress/strain curve.64
Finally, SEM analysis also indicated the presence of F. graminearum's hyphae inward the material and the detection of microcracks as showed in Fig. 2c in the right. In previous studies, Fusarium sp. AF4 was reported to be able to adhere to the surface of polyethylene.65 The data reported corroborate with our study.
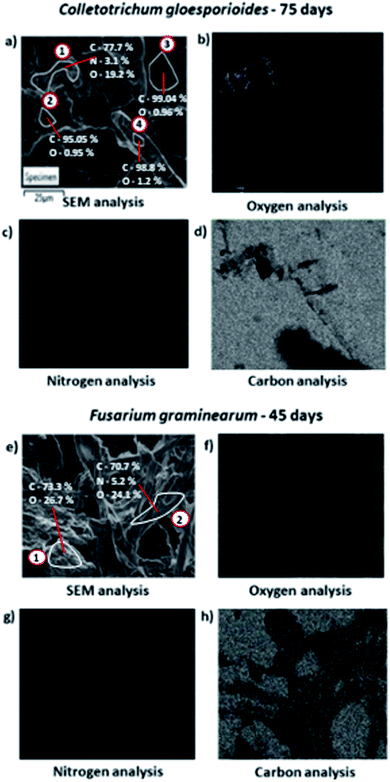
Based on the SEM data, partial biodegradation could be observed after 75 days experiment with all fungi evaluated. Energy dispersive spectroscopy (EDS) analysis (Fig. 3) was applied to confirm the presence of C. gloeosporioides (Fig. 3a–d) and F. graminearum (Fig. 3e–h) in the interior of the bioplastics.
Through EDS analysis applied into the polymer submitted to C. gloeosporioides biodegradation, it was detected in the region 1 (in the upper left corner of Fig. 3a) a rich area of oxygen (19.17%, Fig. 3b); nitrogen (3.09%, Fig. 3c), and the remaining of carbon (77.74%, Fig. 3d). While in the region 2 (in the right of Fig. 3a) it was just detected a small amount of oxygen (0.95%) and carbon was the element detected almost in the totality of the composition (99.05%), as well as region 3 with 99.04% of carbon and the region 4 with 98.78% of this element. These results indicate the presence of C. gloeosporioides in region 1, since this region shows oxygen and nitrogen compositions similar to the ones found in the fungal cells proportion.62 In the EDS analysis of F. graminearum, the oxygen and nitrogen were detected in the concentration of 24.07% (Fig. 3f), and 5.24% (Fig. 3g) respectively, also indicating the presence of F. graminearum inward the material. Furthermore, the detection of the hyphae is also an indicative of the presence of the fungus in the material.66
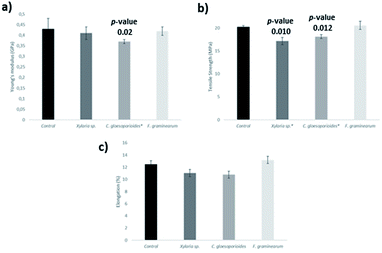
Another criterion used to compare the fungi biodegradation of the new cacao bioplastic was the tensile strength (TS) that is defined as the stress at the local maximum observed during a tensile test.67 The assays were performed based on the ISO 527.67 TS reduction has been previously reported as a parameter to measure plastic biodegradation.68 The polymeric control reference was used for comparison with each biodegraded polymer to evaluate the TS reduction in our analyses. Biodegradation experiments with the 20% cocoa polymer were conducted with each microorganism for 75 days and the specimens were removed and disinfected with 70% ethanol. Other parameters evaluated were Young's modulus (GPa) and elongation (%) that is defined as the strain at break (%)67 (Fig. 4).
C. gloeosporioides was able to decrease 9% of TS when compared with the polymeric control. F. graminearum did not change the TS value significantly, while Xylaria sp. showed a decrease of 15.2%, when incubated during 75 days. It is important to consider that Fig. 3 shows a relative measure, i.e., an average of values and the deviations may be related to the complexity of the polymeric matrix and also to heterogeneity of the material.
El-Shafei et al. tested the biodegradation of disposable polyethylene with Streptomyces sp., however only superficial change was observed when compared to the control sample.69 On the other hand, Sangale et al. observed a greater reduction (around 94%) of tensile strength in pure synthetic polyethylene with Aspergillus sydowii.23 So far, there are no data concerning biodegradation of polyethylene or bio-based polyethylene using C. gloeosporioides. Konduri et al. used Aspergillus oryzae for polymer biodegradation and they were able to observe 63% reduction in the TS of the synthetic polyethylene.70
The results of Young's modulus for the C. gloeosporioides showed that this fungus is able to reduce the stiffness of the plastic in 14.0% during 75 days incubation, however Xylaria sp. and F. graminearum did not reduce significantly the Young's modulus. The reduction of Young's modulus when C. gloeosporioides was applied as biodegradation agent may indicate that there was a partial degradation such as occurred in other polymeric studies reported in the literature.71,72
In terms of elongation, all the three fungi did not reduce significantly this parameter. Lee et al. registered a loss of the percentage of elongation when inoculated three different species of bacteria (Streptomyces viridosporus T7A, S. badius 252, and S. setonii 75Vi2), while the fungus Phanerochaete chrysosporium recorded a gain.73
The biodegradation process was also monitored through relative quantification of theobromine and caffeine from cocoa, since methylxanthine derivatives are compounds produced in the degradation process of cocoa shell.74 The relative quantification was performed based on the integrated area of the respective chromatographic peaks. LC-MS/MS with putative annotation of theobromine and caffeine, and table with integrated area values are shown in ESI (Fig. S12–S15 and Table S5†).
We observe a significant increase in the concentration of theobromine and caffeine after a period of 75 days of the biodegradation process. This increase was more pronounced when the bioplastic was biodegraded by C. gloeosporioides (p-value 0.0006) and F. graminearum (p-value 0.0025). However, the results of theobromine and caffeine after the biodegradation of Xylaria sp also shown significant difference (p-value 0.0266). These results indicate a partial biodegradation of the bioplastic.
3.3. Metabolites produced during the biodegradation process
LC-MS-based metabolomics is commonly used in the comparison of different metabolic profiles of biological sample groups.75 However, metabolomics studies of natural products that may be produced during the biodegradation process of bioplastics is still poorly explored. In this sense, this study sought to identify the metabolites that were produced during the fungal biodegradation process. Thus, the environmental issue of bioplastics biodegradation can be combined with the production of compounds of industrial interests.The secondary metabolites produced by Xylaria sp. in these study do not exhibit a significant increase in their concentration during the biodegradation process. Therefore, the data are not included in the discussion. Metabolites produced by C. gloeosporioides and F. graminearum are discussed in 3.3.1 and 3.3.2 Sections, respectively.
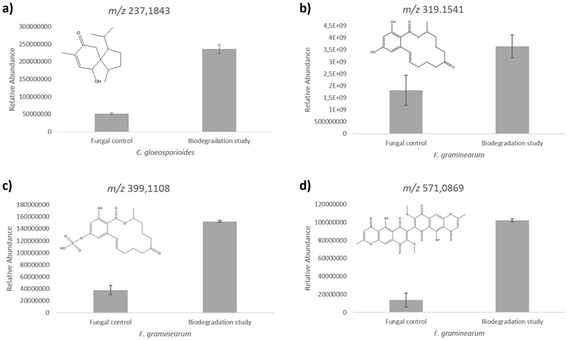
In our LC-MS/MS analysis we detected a compound at m/z 237.1852 (Δ = 0.14 ppm) in positive ionization mode, which was accumulated during the biodegradation process. André et al. reported the chemical structure of four acorane sesquiterpenes produced by C. gloeosporioides,77 among these compounds, 2-hydroxyacorenone showed m/z values very close to our experimental results. In addition, the MS/MS fragmentation pattern was compared to the predicted spectra from HMDB database, with similar fragmentation profile (Fig. S4 and S5†) that enable us to putatively identify this compound as produced in the biodegradaton studies. The metabolite 2-hydroxyacorenone was produced in higher levels during the biodegradation process. This increase in the production showed statistical significance (p-value < 0.05) in relation to the control group (Fig. 5a). This compound is a derivative of acorenone and exhibits antiglycation activity,77 which is interesting to pharmaceutical industries. The antiglycation activity reduces inflammatory reactions and is considered a strategy to slow down the development of diseases such as diabetes mellitus. The discovery and obtainment of compounds that exhibit antiglycation activity may be useful to the development of new drugs.78,79
In general, C. gloeosporioides biodegradation process presented the best results of Young's modulus and elongation in the mechanical tests. Furthermore, the presence of the fungus inside the material was detected through SEM-EDS analysis. This is an important indicative of a successful biodegradation. Among the three fungi that were analyzed herein, C. gloeosporioides showed the most interesting results to be considered with the higher potential to be used in the biodegradation of this new bioplastic. In addition, C. gloeosporioides produced metabolites that are known to exhibit anti-glycation properties.79 Therefore, C. gloeosporioides is a promising fungus to be used in the life cycle of this new bioplastic.
In our LC-MS/MS analysis, it was observed an increased level of zearalenone in the biodegradation experiments when compared to the fungus fermentation in artificial culture medium. Zearalenone was detected by LC-MS/MS in positive ionization mode at m/z 319.1541 (Δ = 0.06 ppm) and the confirmation of this compound was based on the same retention time and MS/MS fragmentation pattern comparison with the authentic standard (Fig. S6 and S7 in ESI†).
The levels of zearalenone produced by F. graminearum during the biodegradation process increased approximately two-fold in relation to the fungal control. The T-test calculation using relative peak areas of chromatograms showed the level alterations with a statistical significance of p-value 0.01 as shown in the Fig. 5b.
All the metabolites shown in Fig. 5 were detected before and after the biodegradation process. However, after a period of 75 days of biodegradation, the concentration of these metabolites was intensified (Fig. S4, S6, S8 and S10, in the ESI†). We believe that the presence of the bioplastic stimulates the production of those metabolites.
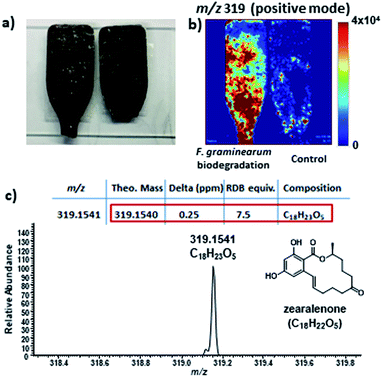
Zearalenone was also detected using desorption electrospray ionization mass spectrometry imaging (DESI-MSI) analysis. DESI-MSI is an analytical technique that allows to detect m/z values of compounds in different types of samples (polymers, plant tissues, culture medium, among others), without previous treatment, under ambient conditions.82,83 During the analysis, the mass spectra are acquired by scanning the surface of the evaluated sample. The images obtained indicate the spatial distribution of compounds in the sample surface.84,85
Through DESI-MSI was possible to confirm the increase in zearalenone levels during the F. graminearum biodegradation (Fig. 6) corroborating with the LC-MS/MS data. The zearalenone production may be related to a competition mechanism to colonize new substrates, since zearalenone and its derivatives can inhibit the growth of filamentous fungi on solid media at concentration lower than 10 μg mL−1.86
Zearalenone is a resorcylic acid lactone (RAL), and this class of compounds is generally used in different synthetic strategies. Therefore, zearalenone has a commercial importance as reagent in researches that aim to produce new analogues.87,88 Zearalenone can be converted to zeranol, an anabolic agent used in veterinary of many countries and it have been used as a growth additive in beef in the United States and Canada.89 The pathway to produce zearalenone using plastic biodegradation showed to be cheap and eco-friendly. It is important to highlight that this is the first report considering the production of active secondary metabolites as a result of bioplastic biodegradation for industrial applications. So far, biodegradation studies only focused in the physicochemical properties of the materials and in the classification of the biodegradation potential of the evaluated microorganisms.
Fusarium graminearum also produced high levels of a zearalenone derivative during the biodegradation of the bioplastic as shown in Fig. 5c. LC-MS/MS analyses detected zearalenone sulfate at m/z 399.1108 (Δ = 0.06 ppm) in positive ionization mode and its ammonium adduct [M + NH4]+ at m/z 416.1373 (Δ = −0.23 ppm). The protonated form was detected more intensively in the biodegradation experiments, according to Fig. 5, corroborating with the zearalenone data. Through LC-MS/MS spectra comparison and fragmentation pattern similarity between zearalenone and its analogue zearalenone sulfate (Fig. S8 and S9 in ESI†), the identity of the metabolite could be confirmed.
Another metabolite that showed a relative increase in the production during the biodegradation process was aurofusarin, a secondary natural product produced by Fusarium spp.90 This compound was detected in our LC-MS/MS analysis at m/z 571.0871 (Δ = −0.11 ppm) in positive ionization mode. The identity of aurofusarin was confirmed through putative assignment using MS/MS data available91 and comparison with MassBank database (Fig. S10 and S11 in ESI†).
Despite the high toxicity of aurofusarin when ingested,92,93 this compound is obtained from natural source and it is industrially used as a red pigment.92 Aurofusarin cannot be used as pigment for feed colorants, once toxicological effects of aurofusarin have been observed in the case of ingestion. However, aurofusarin can be used as pigment for non-food colorants such as inks and other commercial applications.92,93 Furthermore, antibacterial effects of aurofusarin have been studied against different bacteria species.94 The biodegradation of the bioplastic by F. graminearum caused an expressive increase (above 7-fold) of aurofusarin production in relation to the fungal control, which showed a statistical significance with p-value 0.004, as shown in Fig. 5d.
The fungal hyphae from F. graminearum detected inward the bioplastic through SEM-EDS analysis indicated the high potential of this fungus as biodegradation agent of the new material. Furthermore, it was detected through LC-MS/MS analysis a significative increase in the production of different natural products during the biodegradation process produced by this organism. One of these compounds was also detected on the surface of the degraded bioplastic through DESI-MSI analysis. The use of this technique opens new possibilities to monitor the production of compounds during the fungal colonization process of new substrates. Thus, the study of F. graminearum biodegradation contributes to accelerate the life cycle of the new material in a sustainable way and as a result, generate compounds with aggregate industrial values.
4. Conclusions
A new sustainable material was developed. The best composition in terms of mechanical and rheological properties were found with 80% of polyethylene and 20% cocoa bean husk powder from the residues of the Brazilian cocoa industry. This bio-based plastic was tested in terms of biodegradation with three fungi C. gloeosporioides, Xylaria sp. and F. graminearum. From a physical-chemical perspective, C. gloeosporioides and F. graminearum stood out due to the fact that both exhibited full adhesion capacity on the plastic surface and they were detected inside the material, close to the cocoa fiber region by SEM analysis. Xylaria sp. was able to reduce the TS, even though its presence was not observed in vicinity of the region of cocoa fiber. Through metabolomics studies, we identified one metabolite produced by C. gloeosporioides in the biodegradation experiment with the bioplastic, and three produced by F. graminearum that may have aggregate industrial value due to their biological activities in accordance to literature.77–79,88,89,92We believe that the use of polymers containing cocoa residues in its composition is advantageous from both ecological and industrial perspectives, since it adds new properties to the composite and allows endophytic fungi isolated from the fruit itself to biodegrade it. Furthermore, from the biodegradation experiments, secondary metabolites with high aggregate values may be produced.
Author contributions
TPF and HF designed the experiments. ACAA and GRA cultured the fungi. ACAA performed SEM-EDS analyses and did the sample preparation for LC-MS/MS analyses. ACAA and HF performed the mechanical tests. JGMP and ACAA performed DESI-MSI analyses. ACAA, JGMP, HF, and TPF interpreted the data and wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil (CAPES) – Finance Code 001 and Fundação de Amparo a Pesquisa no Estado de São Paulo [grant number FAPESP 2017/24462-4, 2018/03670-0 and 2019/06359-7, student fellowship 2018/20930-6] and L'Oréal Brazil, together with ABC and UNESCO in Brazil. We thank Miss Julia Florez Ablan for helping the mechanical tests and technical assistance.References
- E. J. North and R. U. Halden, Rev. Environ. Health, 2013, 28, 1–8 CAS.
- S. Pathak, C. L. R. Sneha and B. B. Mathew, J. Polym. Biopolym. Phys. Chem., 2014, 2, 84–90 CAS.
- Y. Chae and Y. J. An, Environ. Pollut., 2018, 240, 387–395 CrossRef CAS PubMed.
- World Wildlife Fund (WWF), https://www.worldwildlife.org/publications/solving-plastic-pollution-through-accountability, accessed 2021.
- J. M. Garcia and M. L. Robertson, Science, 2017, 358, 870–872 CrossRef CAS PubMed.
- G. P. Olivatto, M. C. T. Martins, C. C. Montagner, T. B. Henry and R. S. Carreira, Mar. Pollut. Bull., 2019, 139, 157–162 CrossRef CAS PubMed.
- W. R. Lea, J. Hazard. Mater., 1996, 47, 295–302 CrossRef CAS.
- K. Ragaert, L. Delva and K. V. Geem, Waste Manag., 2017, 69, 24–58 CrossRef CAS PubMed.
- R. Avolio, F. Spina, G. Gentile, M. Cocca, M. Avella, C. Carfagna, G. Tealdo and M. E. Errico, Polymers, 2019, 11, 208 CrossRef PubMed.
- J. Hopewell, R. Dvorak and E. Kosior, Philos. Trans. R. Soc., B, 2009, 364, 2115–2126 CrossRef CAS PubMed.
- R. Geyer, J. R. Jambeck and K. L. Law, Sci. Adv., 2017, 3, e1700782 CrossRef PubMed.
- V. T. Breslin, J. Environ. Polym. Degrad., 1993, 1, 127–141 CrossRef CAS.
- S. Seif, J. F. Provencher, S. Avery-Gomm, P. Y. Daoust, M. L. Mallory and P. A. Smith, Arch. Environ. Contam. Toxicol., 2018, 74, 349–360 CrossRef CAS PubMed.
- M. Batayneh, I. Marie and I. Asi, Waste Manag., 2007, 27, 1870–1876 CrossRef CAS PubMed.
- J. J. Jafar, KSCE Journal of Civil Engineering, 2016, 20, 243–249 CrossRef.
- A. Ossa, J. L. García and E. Botero, J. Cleaner Prod., 2016, 135, 379–386 CrossRef.
- M. Sarker, J. Energy Eng., 2011, 108, 35–43 CrossRef.
- B. K. Sharma, B. R. Moser, K. E. Vermillion, K. M. Doll and N. Rajagopalan, Fuel Process. Technol., 2014, 122, 79–90 CrossRef CAS.
- R. A. Gross and B. Kalra, Biodegradable polymers for the environment, Science, 2002, 297, 803–807 CrossRef CAS PubMed.
- T. Iwata, Angew. Chem., Int. Ed., 2015, 54, 3210–3215 CrossRef CAS PubMed.
- S. Lambert and M. Wagner, Chem. Soc. Rev., 2017, 46, 6855–6871 RSC.
- M. K. Sangale, M. Shahnawaz and A. B. Ade, J. Biorem. Biodegrad., 2012, 3, 1000164 Search PubMed.
- M. K. Sangale, M. Shahnawaz and A. B. Ade, Sci. Rep., 2019, 9, 5390 CrossRef PubMed.
- F. Alshehrei, J. Appl. Environ. Microbiol., 2017, 5, 8–19 CAS.
- P. Goswami and T. O'Haire, in Advances in Technical Nonwovens, ed. G. Kellie, Woodhead Publishing, 2016, ch. 1, pp. 1–17 Search PubMed.
- S. Kabasci, in Plastic Waste and Recycling: Environmental Impact, Societal Issues, Prevention, and Solutions, ed. M. Letcher, Academic Press, 2020, ch. 4, pp. 67–96 Search PubMed.
- D. K. Shaw and P. Sahni, IOSR J. Mech. Civ. Eng., 2014, 46–48 Search PubMed.
- A. Folino, A. Karageorgiou, P. S. Calabrò and D. Komilis, Sustainability, 2020, 12, 6030 CrossRef CAS.
- European Bioplastic, https://www.european-bioplastics.org/market, accessed 2021.
- J. R. Müller, Biopolymers, 2005, 365–374 Search PubMed.
- R. S. Devi, V. R. Kannan, K. Natarajan, D. Nivas, K. Kannan, S. Chandru and A. R. Antony, in Environmental waste management, ed. R. Chandra, CRC Press, Boca Raton, 1st edn, 2015, ch. 12, pp. 341–370 Search PubMed.
- S. M. Emadian, T. T. Onay and B. Demirel, Waste Manag., 2017, 59, 526–536 CrossRef CAS PubMed.
- G. Kale, R. Auras, S. P. Singh and R. Narayan, Polym. Test., 2007, 26, 1049–1061 CrossRef CAS.
- M. P. Arrieta, J. López, E. Rayón and A. Jiménez, Polym. Degrad. Stab., 2014, 108, 307–318 CrossRef CAS.
- M. Mihai, N. Legros and A. Alemdar, Polym. Eng. Sci., 2014, 54, 1325–1340 CrossRef CAS.
- R. Y. Tabasi and A. Ajji, Polym. Degrad. Stab., 2015, 120, 435–442 CrossRef CAS.
- N. Sridewi, K. Bhubalan and K. Sudesh, Polym. Degrad. Stab., 2006, 91, 2931–2940 CrossRef CAS.
- C. Thellen, M. Coyne, D. Froio, M. Auerbach, C. Wirsen and J. A. Ratto, J. Polym. Environ., 2008, 16, 1–11 CrossRef CAS.
- R. Jain and A. Tiwari, J. Environ. Health Sci. Eng., 2015, 13, 11 CrossRef PubMed.
- A. Anstey, S. Muniyasamy, M. M. Reddy, M. Misra and A. Mohanty, J. Polym. Environ., 2014, 22, 209–218 CrossRef CAS.
- T. Ishigaki, W. Sugano, A. Nakanishi, M. Tateda, M. Ike and M. Fujita, Chemosphere, 2004, 54, 225–233 CrossRef CAS PubMed.
- Y. Tokiwa, B. P. Calabia, C. U. Ugwu and S. Aiba, Int. J. Mol. Sci., 2009, 10, 3722–3742 CrossRef CAS PubMed.
- A. A. Shah, F. Hasan, A. Hameed and S. Ahmed, Biotechnol. Adv., 2008, 26, 246–265 CrossRef CAS PubMed.
- N. Ojha, N. Pradhan, S. Singh, A. Barla, A. Shrivastava, P. Khatua, V. Rai and S. Bose, Sci. Rep., 2017, 7, 39515 CrossRef CAS PubMed.
- O. Petrini, T. N. Sieber, L. Toti and O. Viret, Nat. Toxins, 1992, 1, 185–196 CrossRef CAS PubMed.
- T. G. Volova, A. N. Boyandin, A. D. Vasil, V. A. Karpov, I. V. Kozhevnikov, S. V. Prudnikova, V. P. Rudnev, B. B. Xuån, V. V. Dũng and I. I. Gitel’zon, Microbiol, 2011, 80, 252–260 CrossRef CAS.
- J. Smedsgaard, J. Chromatogr. A, 1997, 760, 264–270 CrossRef CAS PubMed.
- Dictionary of Natural Products (DNP), available at: http://dnp.chemnetbase.com, 2000 Search PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg and Y. Peng, et al., Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- D. S. Wishart, D. Tzur, C. Knox, R. Eisner, A. C. Guo and N. Young, et al., Nucleic Acids Res., 2007, 35, D521–D526 CrossRef CAS PubMed.
- B. Cheng, C. Zhou, W. Yu and X. Sun, Polym. Test., 2001, 20, 811–818 CrossRef CAS.
- T. Bremner, A. Rudin and D. G. Cook, J. Appl. Polym. Sci., 1990, 41, 1617–1627 CrossRef CAS.
- J. D. Gu, Int. Biodeterior. Biodegrad., 2003, 52, 69–91 CrossRef CAS.
- J. R. Russell, J. Huang, P. Anand, K. Kucera, A. G. Sandoval, K. W. Dantzler, D. Hickman, J. Jee, F. M. Kimovec, D. Koppstein, D. H. Marks, P. A. Mittermiller, S. J. Núñez, M. Santiago, M. A. Townes, M. Vishnevetsky, N. E. Williams, M. P. Vargas, L. A. Boulanger, C. Bascom-Slack and S. A. Strobel, Appl. Environ. Microbiol., 2011, 77, 6076–6084 CrossRef CAS PubMed.
- E. I. Rojas, S. A. Rehner, G. J. Samuels, S. A. Van Bael, E. A. Herre, P. Cannon, R. Chen, J. Pang, R. Wang, Y. Zhang, Y. Q. Peng and T. Sha, Mycologia, 2010, 102, 1318–1338 CrossRef PubMed.
- J. Crozier, S. E. Thomas, M. C. Aime, H. C. Evans and K. A. Holmes, Plant Pathol., 2006, 55, 783–791 CrossRef CAS.
- M. R. Rubini, R. T. Silva-Ribeiro, A. W. V. Pomella, C. S. Maki, W. L. Araújo, D. R. dos Santos and J. L. Azevedo, Int. J. Biol. Sci., 2005, 1, 24–33 CrossRef CAS PubMed.
- M. Mierzwa-Hersztek, K. Gondek and M. Kopeć, J. Polym. Environ., 2019, 27, 600–611 CrossRef CAS.
- J. Arutchelvi, M. Sudhakar, A. Arkatkar, M. Doble, S. Bhaduri and P. V. Uppara, Indian J. Biotechnol., 2008, 7, 9–22 CAS.
- O. Abdel-Kareem, Polymer science: research advances, practical applications and educational aspects, 2016, pp. 309–320 Search PubMed.
- J. M. Rodrigues da Luz, S. A. Paes, M. D. Nunes, M. C. S. da Silva and M. C. M. Kasuya, PLoS One, 2013, 8, e69386 CrossRef CAS PubMed.
- F. Awaja, S. Zhang, M. Tripathi, A. Nikiforov and N. Pugno, Prog. Mater. Sci., 2016, 83, 536–573 CrossRef CAS.
- F. Jbilou, C. Joly, S. Galland, L. Belard, V. Desjardin, R. Bayard, P. Dole and P. Degraeve, Polym. Test., 2013, 1565–1575 CrossRef CAS.
- L. C. Cossolino and A. H. A. Pereira, https://sonelastic.com/images/RT03-ATCP.pdf, accessed 2021.
- A. A. Shah, F. Hasan, A. Hameed and J. I. Akhter, Afr. J. Microbiol. Res., 2009, 3, 658–663 CAS.
- C. Pritsch, G. J. Muehlbauer, W. R. Bushnell, D. A. Somers and C. P. Vance, Mol. Plant-Microbe Interact., 2000, 13, 159–169 CrossRef CAS PubMed.
- American Society for Testing and Materials ASTM D790-17, www.astm.org, accessed 2021 Search PubMed.
- M. Karimi and D. Biria, Sci. Rep., 2019, 9, 2612 CrossRef CAS PubMed.
- H. A. El-Shafei, N. H. Abd El-Nasser, A. L. Kansoh and A. M. Ali, Polym. Degrad. Stab., 1998, 62, 361–365 CrossRef CAS.
- M. K. R. Konduri, K. S. Anupam, J. S. Vivek, D. B. R. Kumar and M. L. Narasu, Indian J. Biochem. Biophys., 2010, 6, 157–174 Search PubMed.
- L. Ding, R. L. Davidchack and J. Pan, J. Mech. Behav. Biomed. Mater., 2012, 5, 224–230 CrossRef CAS PubMed.
- M. I. Artsis, A. P. Bonartsev, A. L. Iordanskii, G. A. Bonartseva and G. E. Zaikov, Mol. Cryst. Liq. Cryst., 2010, 523, 21 CAS.
- B. Lee, A. L. Pometto, A. Fratzke and T. B. Bailey Jr, Appl. Environ. Microbiol., 1991, 57, 678–685 CrossRef CAS PubMed.
- S. Jokić, T. Gagić, Ž. Knez, D. Šubarić and M. Škerget, Molecules, 2018, 23, 1408 CrossRef PubMed.
- J. R. Belinato, J. M. Bazioli, A. Sussulini, F. Augusto and T. P. Fill, Quim. Nova, 2019, 42, 546–559 CAS.
- V. Gambarini, O. Pantos, J. M. Kingsbury, L. Weaver, K. M. Handley and G. Lear, mSystems, 2021, 6, e01112–e01120 CrossRef CAS PubMed.
- A. André, N. Wojtowicz, K. Touré, D. Stien and V. Eparvier, Tetrahedron Lett., 2017, 58, 1269–1272 CrossRef.
- W. J. Yeh, S. M. Hsia, W. H. Lee and C. H. Wu, J. Food Drug Anal., 2017, 25, 84–92 CrossRef CAS PubMed.
- A. A. Chinchansure, A. M. Korwar, M. J. Kulkarni and S. P. Joshi, RSC Adv., 2015, 5, 31113 RSC.
- K. D. Broders, P. E. Lipps, P. A. Paul and A. E. Dorrance, Plant Dis., 2007, 91, 1155–1160 CrossRef CAS PubMed.
- H. K. Kim, S. Lee, S. M. Jo, S. P. McCormick, R. A. E. Butchko, R. H. Proctor and S. H. Yun, PLoS One, 2013, 8, e68441 CrossRef CAS PubMed.
- Y. Liu, X. Ma, Z. Lin, M. He, G. Han, C. Yang, Z. Xing, S. Zhang and X. Zhang, Angew. Chem., 2010, 122, 4537–4539 CrossRef.
- J. H. Costa, J. M. Bazioli, E. V. Araújo, P. H. Vendramini, M. C. F. Porto, M. N. Eberlin, J. A. Souza-Neto and T. P. Fill, Fungal Biol., 2019, 123, 594–600 CrossRef CAS PubMed.
- E. Claude, E. A. Jones and S. D. Pringle, Methods Mol. Biol., 2017, 1618, 65–75 CrossRef CAS PubMed.
- J. G. M. Pontes, P. H. Vendramini, L. S. Fernandes, F. H. de Souza, E. J. Pilau, M. N. Eberlin, R. F. Magnani, N. A. Wulff and T. P. Fill, Sci. Rep., 2020, 10, 13457 CrossRef PubMed.
- J. Utermark and P. Karlovsky, Appl. Environ. Microbiol., 2007, 73, 637–642 CrossRef CAS PubMed.
- E. Vekiru, C. Hametner, R. Mitterbauer, J. Rechthaler, G. Adam, G. Schatzmayr, R. Krska and R. Schuhmacher, Appl. Environ. Microbiol., 2010, 76, 2353–2359 CrossRef CAS PubMed.
- N. Jana and S. Nanda, New J. Chem., 2018, 42, 17803–17873 RSC.
- D. Mukherjee, S. G. Royce, J. A. Alexander, B. Buckley, S. S. Isukapalli, E. V. Bandera, H. Zarbl and P. G. Georgopoulos, PLoS One, 2014, 9, e113632 CrossRef PubMed.
- K. Jarolim, K. Wolters, L. Woelflingseder, G. Pahlke, J. Beisl, H. Puntscher, D. Braun, M. Sulyok, B. Warth and D. Marko, Toxicol. Lett., 2018, 284, 170–183 CrossRef CAS PubMed.
- T. Neuhof, R. Köppen, M. Koch and I. Nehls, Eur. J. Mass Spectrom., 2008, 14, 329–333 CrossRef CAS PubMed.
- K. R. Westphal, R. D. Wollenberg, F. A. Herbst, J. L. Sørensen, T. E. Sondergaard and R. Wimmer, Toxins, 2018, 10, 485 CrossRef CAS PubMed.
- J.-E. Kim, K.-H. Han, J. Jin, H. Kim, J.-C. Kim, S.-H. Yun and Y.-W. Lee, Appl. Environ. Microbiol., 2005, 71, 1701–1708 CrossRef CAS PubMed.
- T. E. Sondergaard, M. Fredborg, A. M. O. Christensen, S. A. Damsgaard, N. F. Kramer, H. Giese and J. L. Sørensen, Toxins, 2016, 8, 355 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental and supplementary data. See DOI: 10.1039/d1ra04432j |
| This journal is © The Royal Society of Chemistry 2021 |