Open Access Article
Open Access ArticleCold–hot nature identification based on GC similarity analysis of Chinese herbal medicine ingredients
Guohui Wei ab,
Xianjun Fua,
Xueying Heb,
Peng Qiub,
Lu Yueb,
Rong Rong*a and
Zhenguo Wang*a
ab,
Xianjun Fua,
Xueying Heb,
Peng Qiub,
Lu Yueb,
Rong Rong*a and
Zhenguo Wang*a
aKey Laboratory of Theory of TCM, Ministry of Education of China, Shandong University of Traditional Chinese Medicine, Jinan 250355, China
bCollege of Intelligence and Information Engineering, Shandong University of Traditional Chinese Medicine, Jinan 250355, China. E-mail: rosierong@163.com; zhenguow@126.com
First published on 27th July 2021
Abstract
The theory of cold–hot nature of Chinese herbal medicines (CHMs) is the core theory of CHM. It has been found that the volatile oil ingredients in CHMs are closely related to their cold–hot nature. Guided by the scientific hypothesis that “CHMs with similar component substances should have similar medicinal natures”, exploration of the intelligent identification of the cold–hot nature of CHMs based on the similarity of their volatile oil ingredients has become a research focus. Gas chromatography (GC) chemical fingerprints have been widely used in the separation of volatile oil ingredients to analyze the cold–hot nature of CHMs. To verify the above hypothesis, in this work, we study the quantification of the similarity of the volatile oil ingredients of CHMs to their fingerprint similarity and explore the relationship between the volatile oil ingredients of CHMs and their cold–hot nature. In this study, we utilize GC technology to analyze the chemical ingredients of 61 CHMs that have a clear cold–hot nature (including 30 ‘cold’ CHMs and 31 ‘hot’ CHMs). Using the constructed fingerprint dataset of CHMs, a distance metric learning algorithm is applied to measure the similarity of the GC fingerprints. Furthermore, an improved k-nearest neighbor (kNN) algorithm is proposed to build a predictive identification model to identify the cold–hot nature of CHMs. The experimental results prove our inference that CHMs with similar component substances should have similar medicinal natures. Compared with existing classical models, the proposed identification scheme has better predictive performance. The proposed prediction model is proved to be effective and feasible.
1. Introduction
As one of the core elements of traditional Chinese medicine (TCM), the nature theory of Chinese herbal medicines (CHMs) has attracted the attention of scholars and research institutions for many years. The nature of CHMs can be classified into four types, i.e., cool, cold, hot, and warm, of which the cold or hot nature is an important part of TCM nature theory.1,2 The principle of “treating a hot syndrome with a cold-nature medicine and treating a cold syndrome with a hot-nature medicine” indicates that cold–hot medicinal nature theory is an important basis for TCM treatment in regulating the balance between Yin and Yang in the human body, and the application of the cold–hot medicine nature theory is effective for clinical treatment using TCM.3Nature studies from different perspectives have been proposed to reveal the scientific significance of CHM nature. Some current investigations have focused on analyzing the cold–hot nature of CHMs based on their component substances. Chemical component elements were included to construct a three-element mathematical analysis model for difference analysis for the biological characterization of cold–hot nature.4 Some research has analyzed the cold–hot nature of CHMs using animal behavioral methods. One study found that cold-nature medicine can regulate the body temperature of rats with yeast-induced fever.5 Another study found that cold–hot nature was closely related to energy metabolism rates.6 Some research has analyzed the cold–hot nature of CHMs using bioinformatics methods.7,8 Liang et al. used a molecular network to analyze the cold–hot nature of CHMs.7 They found that inflammation and immunity regulation were more related to CHMs with the hot nature, and cold-nature CHMs possessed the tendency to impact cell growth, proliferation and development. Our group concluded that compounds associated with a cold nature had a sedative function, associated with “mental and behavioral disorder” diseases, while compounds associated with a hot nature had cardio-protection function, associated with “endocrine, nutritional and metabolic diseases” and “diseases of the circulatory system”.8
The bioactivity of a CHM1 is determined by its composition, and the bioactivities of CHMs are the key to identifying their medicinal nature. Thus, the material composition of CHMs indirectly determines their nature. Current investigations of medicinal nature are focused on revealing the connection between the nature of a CHM and its material composition. Studies of the chemical basis of CHM nature have shown that hot CHMs contained volatile oil components, while cold CHMs contained glycosides.9 The medicinal nature of CHMs is closely related to their chemical components. For example, CHMs containing aromatic components in their volatile oil often have a hot nature.10 Chemical fingerprinting techniques have been used to analyze the chemical components of CHMs.11,12 Therefore, researchers have explored identification of the cold–hot nature of CHMs using their chemical fingerprints.13
Generally, discrimination of the cold–hot nature of CHMs consists of two parts: feature representation and nature classification. Feature representation uses the original effects of the CHMs, metabolomics methods, molecular descriptors or fingerprint technology to extract the characteristics of the CHMs. Nature classification requires the use of classical machine learning classifiers or constructed classifiers to discriminate the cold–hot nature of CHMs. The original effects of CHMs are an effective characteristic expression. Xue's research group explored the original efficacy features of CHMs and used classical classifiers (such as an artificial neural network) to classify the nature of unknown CHMs.11,12 Metabolomics methods are also applied to represent CHMs. Nie et al. studied the metabolomics features of CHMs and constructed a random forest model to discriminate the nature of unknown CHMs.14 Molecular descriptor technology is an important method for analyzing cold–hot nature. Long et al. analyzed the molecular descriptors of compounds of 284 CHMs with clear medicinal natures, and explored a combination system for predicting the cold–hot nature of other CHMs.15 Other methods, such as proton nuclear magnetic resonance spectroscopy (1H-NMR), are used to investigate the features of CHMs. Li et al. studied the characteristics of CHMs using 1H-NMR and applied pattern recognition techniques to analyze the unknown nature of CHMs.16 Chemical fingerprints can be used to characterize the overall composition of CHMs. Our group studied multi-solvent ultraviolet fingerprints for cold–hot nature identification.13
As mentioned above, the discrimination of the cold–hot nature of CHMs has been studied extensively. However, chemical fingerprint technology has not been studied in depth. Our previous research focused on the cold–hot nature discrimination of CHMs based on UV spectra.13 However, according to the existing research results, studies on the chemical basis of CHM nature have shown that hot CHMs contain volatile oil components, while cold CHMs contain glycosides.9,10 The volatile oil information of CHMs can be extracted using gas chromatography. It is possible to obtain a better discrimination rate for cold–hot nature using gas chromatography of CHMs. Furthermore, most investigations used existing classical algorithms to construct prediction models, which would result in poor classification. Designing a classifier based on the characteristics of the fingerprint data may improve the discrimination performance. In this work, gas chromatography technology is applied to extract the characteristic information of CHM ingredients. Using the chemical fingerprint data of CHMs from our research group, a distance metric learning algorithm is constructed to measure the similarity of gas fingerprints, and a prediction model is built to identify the cold–hot nature of CHMs.
2. Materials and methods
2.1 TCM dataset
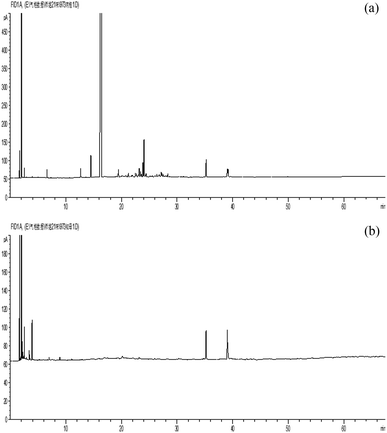
61 representative CHMs are analyzed in this study, of which 30 CHMs are ‘cold’ medicines and the others are ‘hot’ medicines.13 All CHMs have been marked in the classical texts Chinese Materia Medica and Shen Nong's Herbal Classic. GC technology with the steam distillation method was used to obtain the characteristic information of the CHM ingredients. The instrument used in the experiments was a GC6890N GC with a chromatographic data processing system; the reagents were distilled water, anhydrous Na2SO4, and ethyl acetate. Our group recorded the fingerprint information of a total of 61 CHMs for identification of their nature. Fig. 1 shows the gas chromatography fingerprints of cortex Cinnamomi and rhizoma Anemarrhenae.2.2 Gas chromatography fingerprint similarity
In this paper, we study the relationship between the cold–hot nature and material composition of CHMs. A GC reflects the volatile oil ingredients of a CHM. Therefore, we have conducted an exploration to reveal the material basis of cold–hot nature on the basis of gas chromatography fingerprints. According to the current research, the material composition of CHMs is the material basis of their cold–hot nature.8,13 If the volatile oil ingredients of CHMs are similar, we can assume that their medicinal nature is similar. Hence, CHMs with similar gas chromatography fingerprints should have the same medicinal nature.The similarity of the chromatographic fingerprints of CHMs has been widely used in the quality evaluation of CHMs.17 In this study, the similarity of chromatographic fingerprints is applied to evaluate the cold–hot nature of CHMs. We define the similarity of chromatographic fingerprints as the fingerprint similarity and semantic relevance. Fingerprint similarity is the feature similarity of the CHM ingredients, which means that the fingerprints of the two CHMs are similar. Semantic relevance depends on the cold or hot classification of CHMs, which means that if two CHMs have the same label, they are semantically similar.18 We want to learn a Mahalanobis distance to measure the similarity of chromatographic fingerprints, which preserves fingerprint similarity and semantic relevance. The smaller the Mahalanobis distance is, the higher the similarity of the chromatographic fingerprints is.
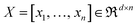 , with
, with 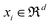 being the ith CHM chromatographic fingerprint in the original space and n being the total number of CHMs. The Mahalanobis distance between xi and xj is denoted as dM(xi,xj), which is defined as:19
being the ith CHM chromatographic fingerprint in the original space and n being the total number of CHMs. The Mahalanobis distance between xi and xj is denoted as dM(xi,xj), which is defined as:19
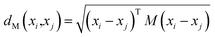 | (1) |
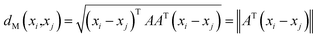 | (2) |
From eqn (2), solving this Mahalanobis distance is actually equivalent to learning a transformation of Euclidean distance between CHM fingerprints in the input space. In this study, we learn transformation matrix A from the fingerprint similarity and semantic relevance. With the transformation matrix A, the Mahalanobis distance between xi and xj can be learned according to eqn (2).
Considering a given X = [x1, …, xn]T ∈ Rn×d, for each xi ∈ X, its local patch is Xi = [xi, xi1, xi2, …, xik]T ∈ R(k+1)×d, which is calculated by the k-nearest neighbors of xi according to the Euclidean distance. For each patch Xi, we have a transformation model gi: Xi → Yi, Yi = [yi,yi1, …, yik]T ∈ R(k+1)×l. To learn a transformation model gi, we would like to minimize the error between the new feature representation Yi and the linear mapping of a patch Xi, and then align all the patches.23 The local patch errors can be minimized as:
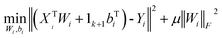 | (3) |
It is assumed that the samples are centered, i.e., XTi1k+1 = 0. To obtain the optimal solution to eqn (3), we set the derivatives of the objective function with respect to Wi and bi to zero. The solution is:
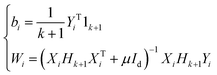 | (4) |
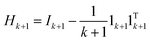 is the local centering matrix. By substituting Wi and bi into eqn (3) using eqn (4), eqn (4) is then rewritten as:
is the local centering matrix. By substituting Wi and bi into eqn (3) using eqn (4), eqn (4) is then rewritten as:
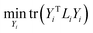 | (5) |
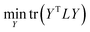 | (6) |
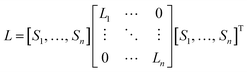 | (7) |
Define Y ∈ Rn×l is a representation of dataset X in feature space. Based on the assumption of linearization that Y = XTA, we can obtain the errors of the global patches:
| min tr(ATXLXTA) | (8) |
According to the definition of semantic relevance, it represents the separability of the cold–hot nature, which requires an increase in the class separability when the size of the within-class scatter matrix decreases or the size of the between-class scatter matrix increases. This can be modelled by the differential scatter discriminant criterion (DSDC) formula,24 which is defined as:
| A = argmax(tr(ATSBA) − λ tr(ATSWA)) | (9) |
The variation is defined as:
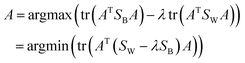 | (10) |
The similarity of CHM ingredients includes the fingerprint similarity and the semantic relevance. Therefore, we integrate eqn (8) from fingerprint similarity and eqn (10) from semantic relevance to construct the similarity metric model. The similarity metric model is as follows:
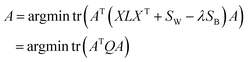 | (11) |
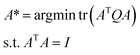 | (12) |
In this case, the solution of optimal projections A* can be obtained through eigenvalue decomposition on matrix Q, and A* can be constructed by the u eigenvectors of M associated with the u smallest eigenvalues.
2.3 A kNN (k-nearest neighbor) scheme for cold–hot nature identification
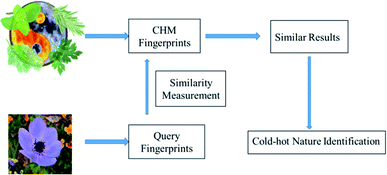
Using the learned Mahalanobis distance, an improved similarity-based kNN scheme is developed to identify cold–hot nature. The improved kNN scheme description is shown in Fig. 2. For a CHM with an unknown nature, we first determine the ingredients of the CHM by gas chromatography, and then calculate the similarity between the gas chromatography fingerprint of the CHM and those of CHM datasets with a clear nature. The similarity-based kNN algorithm using the Mahalanobis distance (SNNM) is used to search for r similar CHM fingerprints with the smallest distances. The r ‘most similar’ CHMs are ranked based on increasing Mahalanobis distance metrics to the query CHM. Finally, a cold nature probability is calculated to represent the degree of coldness of the CHM, which is the ratio of the number of cold CHMs to the total number of CHMs searched. The formula is defined as eqn (13):
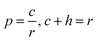 | (13) |
2.4 SNNM scheme for cold–hot nature identification
The cold–hot nature identification algorithm:Given a CHM fingerprint dataset X = [x1, x2, …, xn] ∈ Rd×n, and a number of classes C.
2.5 Performance assessment
In this subsection, to evaluate the effectiveness and feasibility of the proposed SNNM scheme, numerous experiments are constructed for cold–hot nature identification. We compare the identification performance of our scheme with that of the state-of-the-art schemes in terms of extrapolation evaluation and stability evaluation, including information-theoretic metric learning (ITML),25 large margin nearest neighbor (LMNN),20 retrieval system (RS),26 Pearson correlation coefficient (PCC) and ELM.27 ITML and LMNN are classical distance metric learning models. RS and PCC have been used for nature identification of CHMs. ELM has been applied for the nature identification of chemical compounds in CHMs. All performance evaluation experiments are based on an existing CHM fingerprint dataset. The application allows a researcher to test a CHM of unknown nature by searching for similar CHM fingerprints with a clear nature. In this study, we first determined the CHM ingredients using gas chromatography. Secondly, we proposed a SNNM scheme for the nature identification of CHMs. Finally, we designed extensive experiments to demonstrate the feasibility of our proposed scheme.In our experiments, the extrapolation evaluation represents the extent to which cold CHMs can be calculated on the basis of the CHMs that are retrieved based on similarity in the search. We divided the CHM fingerprint dataset into training fingerprints and test fingerprints and computed the probability of each test CHM belonging to the cold CHM group. By varying the threshold of the cold probability, a receiver operating characteristic (ROC) curve was calculated. The area under the ROC curve (AUC) and identification accuracy (ACC) were applied to assess the performance of our scheme. The ACC formula is as follows: ACC = n/r, where n is the number of correctly identified CHMs, and r is the total number of identified CHMs. The second evaluation method, stability evaluation, reflects the proportion of retrieved CHMs that are medically relevant to the query CHMs, which can be calculated by the leave-one-CHM-out method for the whole CHM fingerprint dataset.26 In this evaluation scheme, the cold probability of each CHM can be calculated from the r ‘most similar’ CHMs in the remaining 60 test samples. Finally, the probabilities for the 61 CHMs are obtained. We calculate the stability evaluation as eqn (14):
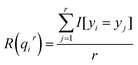 | (14) |
3. Results
3.1 Parameter configurations
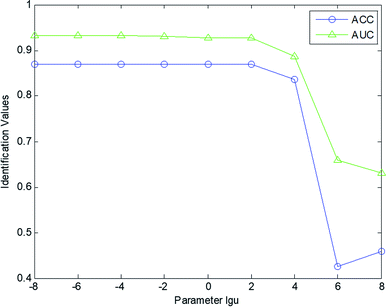
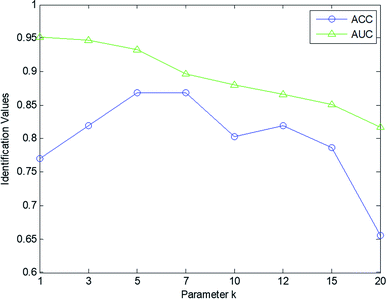
In this section, several parameters were set for nature identification. The parameter analyses were performed based on the experiments conducted on the CHM fingerprint database. In our experiments, we analyzed several factors, namely, μ in eqn (3) for patch building; the tradeoff parameter λ in eqn (9); and the number of retrieved CHMs k in the kNN scheme.In this work, the stability evaluation was applied to set the parameters of our identification model. AUC and ACC values were calculated to evaluate the performance of our identification scheme with different parameters (μ, λ, and k). The AUC and ACC values were computed as a function of the set parameters to obtain more comprehensive curves for evaluating the performance of our scheme. Fig. 3 shows the AUC and ACC value curves for the nature identification of the gas chromatography fingerprints using different μ values in the range [10−8, 10−6, 10−4, 10−2, 1, 102, 104, 106, 108]. As shown in Fig. 3, our scheme is not suitable for a large μ. When μ ≥ 104, the identification performance of our model drops. Based on comprehensive analysis of the ACC and AUC curves, our scheme reaches the optimal value when μ = 10−4. In this experiment, the tradeoff parameter λ was set as 1, and the number of retrieved CHMs k was set as 5.
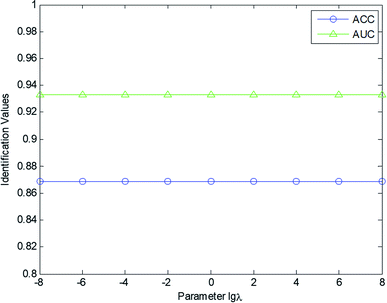
In this study, the effect of the tradeoff parameter λ in eqn (9) was investigated to evaluate the identification performance of the cold–hot nature. The value of the parameter λ was set within the range [10−8, 10−6, 10−4, 10−2, 1, 102, 104, 106, 108]. Fig. 4 displays the AUC and ACC value curves with different λ values. According to Fig. 4, our scheme and fingerprint data are not sensitive to the parameter λ. No matter what the parameter value is, the performance of the model remains at a certain level. Setting λ = 1, the AUC and ACC values of our model are 0.8689 and 0.9333, respectively. In this experiment, the parameter μ was set as 10−4, and the number of retrieved CHMs k was set as 5.
Furthermore, the parameter k in the kNN scheme was varied to evaluate the predictive performance of our scheme. The value of k was set within the range [1, 3, 5, 7, 12, 15, 20]. Fig. 5 shows the AUC and ACC value curves with different k values. From this figure, we can see that the performance of our scheme tends to decline with increasing k value. Based on comprehensive analysis of the ACC and AUC curves, our scheme reaches the optimal value when k = 5. In this experiment, the parameter μ was set as 10−4, and the tradeoff parameter λ was set as 1.
3.2 Model performance assessment
To demonstrate the feasibility and the efficiency of our proposed SNNM scheme for predicting the cold–hot nature of CHMs, this study compares the prediction performance of our model with those of some classical distance metric learning algorithms (i.e., ITML and LMNN) or classifiers used in CHM nature identification (RS, PCC, and ELM). The Pearson correlation coefficient (PCC) is applied as a comparative reference to measure the similarity of gas chromatography. Table 1 displays a comparison between the performance of the extrapolation evaluation of SNNM and other algorithms. For the extrapolation evaluation, 40 CHMs were randomly selected as the training dataset, among which the number of cold and hot CHMs was about 20 each; the remaining CHMs served as the test dataset. The parameters of our model for the extrapolation evaluation were λ = 1, μ = 10−4, k = 7. The parameters of the compared models were optimized. Based on the results for the classification of cold–hot nature, we drew the following conclusions. First, the identification performance of our SNNM scheme was the best, and was better than that of the RS algorithm. The RS algorithm only considers the semantic relevance between fingerprints. Therefore, fingerprint similarity is also important for similarity metric of CHM ingredients. Secondly, distance metric learning algorithms have better classification accuracy than ELM and PCC. This demonstrates that it is feasibility for nature prediction with similarity metric of CHM ingredients, and also illustrates that CHMs with similar gas chromatography have similar medicinal nature. Thirdly, ELM with gas chromatography is poor in identifying cold–hot nature. Finally, according to the extrapolation evaluation, our scheme is efficiency.| Classifier | AUC | ACC |
|---|---|---|
| ITML | 0.872 | 0.823 |
| LMNN | 0.855 | 0.786 |
| ELM | 0.587 | 0.525 |
| RS | 0.882 | 0.824 |
| PCC | 0.834 | 0.754 |
| SNNM | 0.891 | 0.852 |
Table 2 shows a performance comparison between the stability evaluation of SNNM and the other algorithms. Based on the cold–hot nature prediction results, we can draw similar conclusions to those obtained from Table 1. First, our SNNM scheme performs best in the identification of cold–hot nature. Secondly, distance metric learning algorithms outperform ELM and PCC in identifying the cold–hot nature. Thirdly, the stability assessment of our scheme is the best. Finally, Tables 1 and 2 comprehensively verify the feasibility and effectiveness of our scheme.
| Classifier | AUC | ACC |
|---|---|---|
| ITML | 0.896 | 0.869 |
| LMNN | 0.894 | 0.869 |
| ELM | 0.683 | 0.623 |
| RS | 0.872 | 0.820 |
| PCC | 0.603 | 0.557 |
| SNNM | 0.9333 | 0.869 |
3.3 Nature identification examples
The leave-one-CHM-out method was applied to calculate examples of the nature identification. The nature identification examples of the two CHMs are displayed in Table 3. The query CHMs (the second row) and their top k = 7 similar reference CHMs are shown in this table. The similar reference CHMs were calculated using SNNM and arranged in order of monotonically increasing Mahalanobis distance.| Prediction example | CHMs with a cold nature | CHMs with a hot nature |
|---|---|---|
| Query CHM | Anemarrhena asphodeloides Bunge (cold) | Euodiae fructus (hot) |
| Similar reference CHMs | Phellodendri chinensis cortex (cold) | Notopterygii rhizoma et radix (hot) |
| Isatidis folium (cold) | Corydalis rhizoma (hot) | |
| Lophatheri herba (cold) | Aconiti lateralis radix praeparata (hot) | |
| Stephaniae tetrandrae radix (cold) | Alpiniae katsumadai semen (hot) | |
| Puerariae lobatae radix (cold) | Psoraleae fructus (hot) | |
| Gardeniae fructus (cold) | Nardostachyos radix et rhizoma (hot) | |
| Notopterygii rhizoma et radix (hot) | Aucklandiae radix (hot) |
A cold CHM (Anemarrhena asphodeloides Bunge) and a hot CHM (Euodiae fructus) were selected as examples to explain the principle of cold–hot nature identification. In the second column, the query CHM is Anemarrhena asphodeloides Bunge, which is a CHM with a cold nature. The reference CHMs similar to it are six CHMs with a cold nature, and one CHM with a hot nature. The calculated cold nature probability is 85.7%, which means that the query CHMs are more likely to have a cold nature. In the third column, the query CHM is Euodiae fructus. The calculated reference CHMs are all hot-nature CHMs. Its cold nature probability is 0, meaning that the query CHM is most likely to have a hot nature. The identification examples indicate that similar gas fingerprints can represent the same CHM nature.
3.4 Overall prediction performance
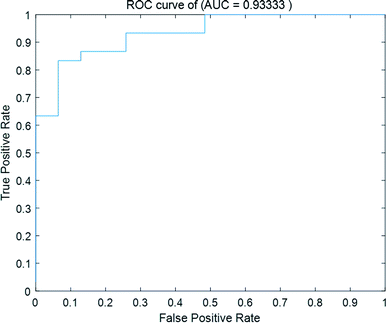
In this paper, we performed a holistic evaluation of the proposed SNNM method. A confusion matrix, recall, precision and F-score were introduced as evaluation indexes. The identification confusion matrix of 61 CHMs is displayed in Table 4. The identification accuracy of hot-nature CHMs is 87.1% (27/31), while the prediction accuracy of cold-nature CHMs is 86.7% (26/30). The total identification accuracy is 86.9% (53/61). The recall, precision and F-score of nature identification of the 61 CHMs are listed in Table 5. Based on comprehensive analysis of Tables 4 and 5, we can conclude that our method has good prediction performance for cold–hot nature using gas chromatography. The ingredients of the volatile oils of CHMs are closely related to their cold–hot nature. Fig. 6 displays the ROC curve of the cold–hot nature identification.| Ground truth | Identification | |
|---|---|---|
| Cold | Hot | |
| Cold | 26 | 4 |
| Hot | 4 | 27 |
| Cold | Hot | |
|---|---|---|
| Recall | 86.7% | 87.1% |
| Precision | 86.7% | 87.1% |
| F-Score | 86.7% | 87.1% |
4. Discussion
CHM gas chromatography fingerprints pose a challenge to existing distance metric methods. Traditional similarity measurement methods, such as Pearson correlation, suffer from high-dimensional problems. Distance metric learning methods, such as ITML and LMNN, only consider the semantic relevance of the components without including the fingerprint similarity. However, semantic relevance alone does not reflect all the similarities of CHMs. Herein, we introduce the fingerprint similarity and semantic relevance to represent the similarity of the CHM ingredients. The model is more consistent with the characteristics of the fingerprint data. We find that fingerprint similarity is very important in similarity measurement, which can effectively improve the identification accuracy.Volatile ingredients are an important part of CHMs and play an important role in the therapeutic effect of traditional Chinese medicine. In this study, the volatile ingredients of CHMs are extracted via gas chromatography. It is found that the volatile ingredients are closely related to the cold–hot nature of the CHMs. According to our previous studies on the nature identification of CHMs, gas chromatography has a better nature identification rate than UV spectroscopy. This indirectly proved the correlation between the volatile oils and the cold–hot nature of CHMs.
Classical classification approaches, such as ELM, are general classifiers that do not consider the characteristics of the data. These methods suffer from the problem of small samples and high dimensionality, resulting in low classification accuracy. Compared to classical classification approaches, our scheme not only considers the class separability of the samples, but also introduces the fingerprint similarity. Therefore, our scheme achieves better identification performance.
However, our research still has some limitations. First, this study only analyzes volatile oil ingredients via gas chromatography. Other ingredients in the CHMs are not taken into account in this study. In the future, we want to explore cold–hot nature identification of CHMs based on total component information. Second, we studied the similarity of the gas chromatography data using a distance metric. The fingerprints have the characteristics of high dimension and small sample. Based on these characteristics, the design of the forecasting model will be the focus in the future. Thirdly, this study focuses on modeling a similar scheme for cold–hot nature identification. The GC characteristics have not been thoroughly mined. In the future, we will combine more effective fingerprint information to represent CHM ingredients for cold–hot nature identification.
5. Conclusions
In this study, a SNNM scheme is proposed to predict the cold–hot nature of CHMs. Gas chromatography is used to analyze the volatile oil ingredients of CHMs. It is found that the volatile oil ingredients of CHMs are closely related to their cold–hot nature. We demonstrate that if the ingredients of CHMs are similar, their nature is similar. Based on the gas chromatography fingerprints of the CHMs, effective experiments demonstrate that our scheme has better performance than classical classifiers in identifying the cold–hot nature of the CHMs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Basic Research Development Program (973 Program) (No. 2007CB512600), National Natural Science Foundation of China (No. 81473369), Key Research and Development Plan of Shandong Province (No. 2016CYJS08A01-1), and Shandong Province TCM Science and Technology Development Plan Project (2019-0037).Notes and references
- J. Gao and C. Chen, Acta Univ. Tradit. Med. Sin. Pharmacol. Shanghai, 2007, 21, 16–18 Search PubMed.
- B. Ouyang, Z. G. Wang and P. Wang, J. Beijing Univ. Tradit. Chin. Med., 2006, 29, 592–594 Search PubMed.
- L. J. Wu, Y. Y. Wang, Y. D. Li, X. G. Zhang, S. Li and Z. Q. Zhang, IEE Proc.: Syst. Biol., 2007, 1, 51–60 Search PubMed.
- R. Jin, B. Zhang, X.-Q. Liu, S.-M. Liu, X. Liu, L.-Z. Li, Q. Zhang and C.-M. Xue, J. Chin. Integr. Med., 2011, 9, 715–724 CrossRef PubMed.
- H.-Y. Wan, X.-Y. Kong, X.-M. Li, H.-W. Zhu, X.-H. Su and N. Lin, China J. Chin. Mater. Med., 2014, 39, 3813–3818 Search PubMed.
- Y. Zhao, L. Jia, J. Wang, W. Zou, H. Yang and X. Xiao, Pharm. Biol., 2016, 54, 1298–1302 CrossRef CAS PubMed.
- F. Liang, L. Li, M. Wang, X. Niu, J. Zhan, X. He, C. Yu, M. Jiang and A. Lu, J. Ethnopharmacol., 2013, 148, 770–779 CrossRef CAS PubMed.
- X. Fu, L. H. Mervin, X. Li, H. Yu and J. Li, J. Chem. Inf. Model., 2017, 57, 468–483 CrossRef CAS PubMed.
- G. J. Xu, J. H. Hu and W. Yang, J. Nanjing Pharmaceut. College, 1961, 6, 92–100 Search PubMed.
- Q. D. Jiang, W. G. Yang, H. Cai, M. Ma, H. Zhang, P. Liu, J. Chen and J. Duan, China J. Chin. Mater. Med., 2016, 41, 2500–2505 Search PubMed.
- W.-H. Liu, Y. Li, Y.-J. Ji, P. Wang, Y.-Q. Zhang and F.-Z. Xue, J. Shandong Univ., Health Sci., 2012, 50, 151–154 Search PubMed.
- X.-X. Zhang, Y. Li, Y.-J. Ji, P. Wang, Y.-Q. Zhang and F.-Z. Xue, J. Shandong Univ., Health Sci., 2012, 50, 143–146 Search PubMed.
- G. H. Wei, X. J. Fu and Z. G. Wang, J. Chem. Inf. Model., 2019, 59, 5065–5073 CrossRef CAS PubMed.
- B. Nie, Z.-L. Hao, B. Gui, Z. Wang, J.-Q. Du, G.-L. Wang and X. Zhang, Journal of Jiangxi University of Traditional Chinese Medicine, 2015, 27, 82–86 Search PubMed.
- W. Long, P. Liu, J. Xiang, X. Pi, J. Zhang and Z. Zou, Comput. Methods Programs Biomed., 2011, 101, 253–264 CrossRef PubMed.
- H. Li, Q. Xu, J. Zhang and H. Xu, 2017 International Conference on Medical Science and Human Health (MSHH 2017), 2017, pp. 174–179 Search PubMed.
- M. J. Li, Y. R. Sha, X. M. Luo, P. Y. Gong and J. Gu, Chin. Tradit. Herb. Drugs, 2021, 52, 1–6 Search PubMed.
- Y. Liu, J. Rong, M. Lily, S. Rahul, G. Adam, B. Zheng, S. C. H. Hoi and S. Mahadev, IEEE Trans. Pattern Anal. Mach. Intell., 2010, 32, 30–44 Search PubMed.
- K. Q. Weinberger, J. Blitzer and L. K. Saul, J. Mach. Learn. Res., 2009, 10, 207–244 Search PubMed.
- G. Wei, H. Ma, W. Qian and M. Qiu, Curr. Med. Imaging Rev., 2017, 13, 210–216 CrossRef.
- G. Wei, H. Ma, W. Qian and M. Qiu, Med. Phys., 2016, 43, 6259–6269 CrossRef PubMed.
- T. Zhang, D. Tao, X. Li and J. Yang, IEEE Trans. Knowl. Data Eng., 2009, 21, 1299–1313 Search PubMed.
- D. Tao, X. Li, X. Wu and S. J. Maybank, IEEE Trans. Pattern Anal. Mach. Intell., 2007, 29, 1700–1715 Search PubMed.
- J. V. Davis, B. Kulis, P. Jain, S. Sra and I. S. Dhillon, The International Conference on Machine Learning (Corvallis, Oregon, USA), 2007, pp. 209–216 Search PubMed.
- G. H. Wei, X. J. Fu and Z. G. Wang, TMR Mod. Herb. Med., 2019, 2, 183–191 CAS.
- E. Malar, A. Kandaswamy, D. Chakravarthy and A. G. Dharan, Comput. Biol. Med., 2012, 42, 898–905 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |