Open Access Article
Open Access ArticleMultifunctional electrochemical biosensor with “tetrahedral tripods” assisted multiple tandem hairpins assembly for ultra-sensitive detection of target DNA†
Yuqi Huanga,
Shuhui Zhaoa,
Wenxiu Zhanga,
Qiuyue Duana,
Qi Yana,
Hu Fub,
Liang Zhonga and
Gang Yi *a
*a
aKey Laboratory of Medical Diagnostics of Ministry of Education, Department of Laboratory Medicine, Chongqing Medical University, Chongqing 400016, PR China. E-mail: yigang666@cqmu.edu.cn
bClinical Laboratory of Chengdu First People's Hospital, Chengdu 610000, PR China
First published on 4th June 2021
Abstract
Nucleic acids are genetic materials in the human body that play important roles in storing, copying, and transmitting genetic information. Abnormal nucleic acid sequences, base mutations, and genetic changes often lead to cancer and other diseases. Meanwhile, methylated DNA is one of the main epigenetic modifications, which is considered to be an excellent biomarker in the early detection, prognosis, and treatment of cancers. Therefore, a multifunctional electrochemical biosensor was constructed with sturdy tetrahedral tripods, which assisted multiple tandem hairpins through base complementary pairing and effective ultra-sensitive detection of targets (DNA, microRNA, and methylated DNA). In the experiments, experimental conditions were optimized, and different DNA concentrations in serum were detected to verify the sensitivity of the biosensor and the feasibility of this protocol. In addition, microRNA and DNA methylation were detected through different designs of tetrahedral tripods (TTs) that capture probes to prove the superiority of this scheme. A sturdy pyramid structure of TTs extremely enhanced the capture efficiency of targets. The targets triggered the one-step isothermal multi-tandem amplification reaction by incubating multiple hairpin assemblies. To our knowledge, a combination of two parts, which greatly reduced background interference and decreased non-specific substance interference, has appeared for the first time in this paper. Moreover, the load area of electrochemical substances was significantly increased than that in previous studies. This greatly increased the detection range and detection limit of targets. The electrochemical signal responses were generated in freely diffusing hexaammineruthenium(III) chloride (RuHex). RuHex could adhere to the DNA phosphate backbone by a powerful electrostatic attraction, causing increased current responses.
1. Introduction
Nucleic acids, as the major genetic materials in the human body, determine the structure of biomolecules, proteins, and cellular components and play vital roles in storing, regulating, and transmitting genetic information in the body. Structural abnormalities, gene-based mutations, and chromosomal abnormalities of nucleic acids have considerably increased cancers and diseases.1,2 DNA methylation is one of the main epigenetic modifications, where a methyl group is bound to 5-methylcytosine via catalysis by DNA methyltransferases (DNMTs). Methylated DNA generally occurs at the cytosine-phosphate-guanine site (CpG),3–5 and the epigenetic changes are indispensable for the early detection, prognosis, and treatment of cancers. Therefore, more sensitive and precise molecular detection methods have become important in various fields for the early detection of key molecular biomarkers that help avoid cancers and other malignancies and to meet research and clinical demands.Traditional nucleic acid detection methods include PCR,6–9 Southern blot,10,11 Northern blot,12 and microarrays.13,14 These methods have the advantages of high sensitivity for early detection and treatment. However, these methods could cause high false positives. Moreover, the tedious experimental procedures and complicated reaction systems lead to poor stability, which greatly hinders their broad clinical application. Thus, these methods are not applicable for long-term research.15,16 At present, many rapid and efficient nucleic acid detection methods, such as photoelectrochemical biosensors,17–19 fluorescent biosensors,20–22 surface-enhanced Raman scattering (SERS),23–25 and colorimetry26–28 are also applied to detect nucleic acids. Nevertheless, after many researches, detailed exploration, and verification experiments, electrochemical biosensors with ultra-sensitive detection, simpler operation, and lower cost are widely established to detect important molecular markers in clinical and other research fields.29–31 Target amplification strategies from various groups are based on target-induced hybridization chain reaction (HCR),32–35 molecular labeling reaction,36,37 rolling circle amplification reaction,38,39 and strand displacement amplification reaction,40,41 which are well-known effective methods to amplify the bio-signals of targets.
Taking advantage of electrochemical methods, the electrochemical biosensor based on tetrahedral tripods (TTs) was introduced, which was assisted by a one-step strategy of isothermal mixing of hairpin assemblies to form multiple tandem hairpins assembly (MTHsA) for the rapid detection of targets. The combination of the two parts has appeared for the first time in this paper, which greatly reduced background interference and decreased non-specific substance interference. Moreover, the load area of electrochemical substances was significantly increased than that in previous studies, which greatly increased the detection range and limit of the targets. In this scheme, TTs were quickly synthesized on an electrode in vitro using four single-stranded DNAs, modified on the electrode surface. The TTs based on the strict base complementary pairing principle were 55 bases in length, which included three parts that were complementarily paired and hybridized with other parts42 and had a sturdy spatial structure. The extremely high stability could effectively promote the capture efficiency of the targets.43–47 Meanwhile, MTHsA in the presence of targets automatically triggered the cascade HCR.48,49 HCR was first proposed by Dirks and Pierce in 2004,50 which mainly elaborated the effective self-assembly of a multiple hairpin structure. HCR was based on the principle of base complementary pairing to produce long tandem DNA double strands and was extensively used in various signal amplification fields for nucleic acid detection.51–54 Based on the HCR of two hairpins in previously published methods, the cascade hybridization of the four hairpins incubated on the electrode could generate multi-branched tandem hybrid chains, which could increase the carrying capacity of DPV signal substances and improve the DPV detection signal. The experiment was extremely simple and the result of the RuHex DPV signal detection was not only stable and reproducible, but RuHex could also be diffused freely while existing in the solution.55–57 RuHex could undergo fast and powerful static adsorption onto a DNA phosphate backbone, resulting in current increases. Moreover, the corresponding electrochemical signals reflected the amount of DNA molecules absorbed on the electrode surface. Compared with the long chain formed by the cyclic hybridization of multiple chains, the DNA nano-biological scaffold was composed of a TT structure carrying a small amount of electroactive substances and could be disregarded.
The newly developed multifunctional electrochemical biosensor could constructively test different targets, including DNA, microRNA, and methylated DNA on devising versatile toehold capture probes. Methylation of DNA mostly occurs at the cytosine phosphate guanine site (CpG); the target DNA was methylated at the 5′-CCmGG-3′ site in the second 5′C, and the unmethylated DNA could be identified and digested with Hpa II methylation restriction endonuclease.54,58 Therefore, the program could be used in clinical applications through accurately identifying methylated and unmethylated sites, which could play an important auxiliary role in early clinical diagnosis.
2. Experiment section
2.1. Materials and reagents
The DNA and microRNA sequences involved in all experiments were purified by high performance liquid chromatography (HPLC) (listed in Table S1†) and chemically synthesized by Sangon Biotech Inc. (Shanghai, China). The RNA enzyme-free water and RNase inhibitor were purchased from Sangon Biotech Inc. (Shanghai, China). Tris (2-carbox-yethy) phosphine hydrochloride (TCEP) was purchased from Aladdin (Shanghai, China), while 6-mercapto-1-hexanol (MCH) and hexaammineruthenium(III) chlorid [(Ru(NH3)6)3+, RuHex] were purchased from Sigma-Aldrich (St. Louis, USA). Hpa II methylation restriction endonuclease and 10× CutSmart®buffer were purchased from New England Biolabs (Beijing, China). The ultrapure water used in the experiment came from the Millipore water purification system (18.2 MΩ, MilliQ, Millipore).All buffers included the following: 1× TE buffer (pH = 8.0) as a DNA sequence preparation buffer containing 10 mM Tris·HCl and 1 mM EDTA; 1× TM buffer (pH = 8.0) as the TT preparation buffer containing 20 mM Tris and 50 mM MgCl2·6H2O; 1× SPSC buffer (pH = 7.5) used as the hybridization buffer containing 1 M NaCl and 50 mM NaH2PO4; DNA hybridization buffer containing 0.1 M PB, 1 M NaCl, and 20 mM MgCl2·6H2O; PB buffer containing A buffer (0.2 M Na2HPO4) and B buffer (0.2 M NaH2PO4); 1× PBS buffer (0.01 M, pH = 7.4) used as the washing buffer containing 137 mM NaCl, 10 mM Na2HPO4, 2.65 mM KCl, and 1.75 mM KH2PO4; 10 mM Tris–HCl buffer (pH 7.4) used as the differential pulse voltammetry (DPV) solution; and 5 mM (Fe(CN)6)3−/4− with 0.1 M KCl solution used as the cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) solutions.
Reagents for gel electrophoresis containing 5× TBE buffer (445 mM Tris, 445 mM Boric acid, and 10 mM EDTA; pH = 8.0–8.6) and 30% Acryl/Bis (29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were purchased from Sangon Biotech Inc. (Shanghai, China). Ammonium persulfate (APS) and N,N,N′,N′-tetramethylethylenediamine (TEMED) were purchased from Sigma-Aldrich (St. Louis, USA). GoldView I was purchased from Solarbio Technology Co. Ltd. (Beijing, China). The 1000 bp DNA marker was purchased from TaKaRa (Dalian, China).
1) were purchased from Sangon Biotech Inc. (Shanghai, China). Ammonium persulfate (APS) and N,N,N′,N′-tetramethylethylenediamine (TEMED) were purchased from Sigma-Aldrich (St. Louis, USA). GoldView I was purchased from Solarbio Technology Co. Ltd. (Beijing, China). The 1000 bp DNA marker was purchased from TaKaRa (Dalian, China).
2.2. Apparatus and characterization
Cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS), and differential pulse voltammetry (DPV) were all performed on the CHI660D electrochemical workstation (Shanghai Chen Hua Instruments, Shanghai, China). The traditional three-electrode system used included: Ag/AgCl electrode as the reference electrode, platinum wire electrode as the auxiliary electrode, and gold electrode (disk diameter = 3 mm) as the working electrode (GaossUnion Technology Co., Ltd, Wuhan, China). The construction of TTs was carried out with a Bio-Rad T100 thermal cycler (Bio-Rad, USA). The gel electrophoresis experiment was performed on an electrophoresis analyzer (Bio-Rad, USA) and the gel was recorded on an imaging system (Bio-Rad, USA). MTHsA was characterized by the SPM-9700HT atomic force microscope (AFM) (Shimadzu, Kyoto, Japan).2.3. Construction of tetrahedral tripods (TTs)
Four DNA single strands were used for TT synthesis and the modification process was mainly derived from the published research methods with slightly improved measurements.59 The four TT strands (S1–S4) were first centrifuged at 4 °C and dissolved in a TE buffer to obtain the 100 μM stock solution, which was stored at −20 °C. Then, 1 μL of four strands from TTs (S1–S4, listed in Table S1†) were equally added to a homogeneous solution of 86 μL TM buffer and 10 μL TCEP (30 mM in ultrapure water) to make a total volume of 100 μL. The DNA sample products were placed at room temperature for 1 h to be completely reduced. The DNA sample mixture was then heated at 95 °C for 5 min and the temperature was quickly dropped to 4 °C within 30 s using a Bio-Rad T100 thermal cycler. The reaction product was taken out, placed on ice for 30 min, and finally diluted to 0.5 μM in the TM buffer and stored in a refrigerator at 4 °C for further use.2.4. Preparation of MTHsA
All the important DNA hairpin sequences required for MTHsA are reasonably listed in Table S1.† The related procedures and DNA sequences were mainly derived from published papers but with slightly improved measurements.60 Before the next process, all single strands of hairpins needed were annealed to form a better hairpin structure. First, 10 μM of every single-stranded DNA was diluted with 1× TE buffer, heated and denatured at 95 °C for 5 min, and slowly cooled to 25 °C overnight to form a hairpin DNA. Subsequently, hairpin H1 (1 μM), H2 (1 μM), H3 (1 μM), and H4 (1 μM) were stably mixed in a SPSC buffer at 4 °C for further experiments.2.5. Preparation of the electrochemical biosensor
First, the new bare gold electrodes were polished with 0.3 and 0.05 μm alumina powder for 3 min to form a clean “mirror,” and then ultrasonically cleaned in ultrapure water, ethanol, and ultrapure water for 5 min to remove excess aluminum powder. Then, the electrodes were immersed in a freshly prepared piranha solution (98% H2SO4 and 30% H2O2 at a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 volume ratio) for 15 min, following which they were rinsed thoroughly with ultrapure water and dried in air. Subsequently, the gold electrodes were electrochemically activated with 0.5 M H2SO4 in the potential range of −0.2 V to 1.6 V until a steady state reproducible CV was obtained. The cleaned gold electrodes were then dried and prepared for the next experiments.
1 volume ratio) for 15 min, following which they were rinsed thoroughly with ultrapure water and dried in air. Subsequently, the gold electrodes were electrochemically activated with 0.5 M H2SO4 in the potential range of −0.2 V to 1.6 V until a steady state reproducible CV was obtained. The cleaned gold electrodes were then dried and prepared for the next experiments.
2.6. Modification and characterization of the fabricated electrochemical biosensor
All measurements were carried out using a conventional three-electrode system with an Ag/AgCl (3 M KCl) reference electrode, a Pt counter electrode, and a modified gold electrode as the working electrode. First, 10 μL of the fully prepared TTs was modified on the electrodes at room temperature overnight. The next day, the electrodes were rinsed with 1× PBS and ultrapure water, respectively, three times and the electrodes were blocked with 6 μL of 1 mM MCH at 4 °C. After rinsing with ultrapure water, 10 μL of the target was dropped on the electrode and incubated for 2 h at 37 °C. The target DNA was diluted in DNA hybridization buffer. The miRNA was first dissolved in an RNA enzyme-free water and then diluted in a hybridization buffer with 1 μL RNase inhibitor, washed with 1× PBS and ultrapure water, respectively, 3 times, and dried in air. Considering it as the most important part of the MTHsA, 10 μL of the prepared mixture of multiple hairpin chains was directly incubated on the electrode surface at 37 °C for 5 h. Then, the electrodes were thoroughly rinsed with 1× PBS and ultrapure water, and tested in the corresponding working buffers.Cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) were carried out in 0.1 M KCl solution containing 5 mM (Fe(CN)6)3−/4−, and the scanning rate in CV was 100 mV s−1 from −0.1 V to 0.5 V. EIS was measured in the above solution with a frequency range from 0.1 Hz to 10 kHz and an amplitude of 5 mV. Differential pulse voltammetry (DPV) was carried out in a 10 mM Tris–HCl buffer (pH 7.4) containing RuHeX from −0.5 V to 0.1 V at the rate of 50 mV s−1, pulse width of 0.05 s, and pulse period of 0.5 s.
2.7. Verification of methylation digestion reaction
First, 10 μL methylated DNA or unmethylated DNA fully scattered in a DNA hybridization buffer was incubated at 37 °C for 2 h. Then, the electrode was rinsed thoroughly with 1× PBS and ultrapure water, respectively. The methylation restriction enzyme digestion reaction was performed in 50 U mL−1 Hpa II enzyme at 37 °C for 2 h, which was in 1× CutSmart®buffer (50 mM KAc, 20 mM Tris-Ac, 10 mM Mg(Ac)2, 100 μg ml−1 BSA, pH = 7.9). The electrode was again thoroughly washed with 1× PBS and ultrapure water, respectively, and dried in air for further modification process.542.8. Native polyacrylamide gel electrophoresis of tetrahedral tripods (TTs)
An 8% natural polyacrylamide gel was employed to verify the successful preparation of the synthesized TTs in one step. A running electrophoresis was carried out in a 1× TBE buffer at 100 V for 40 min; each sample was added to a gel pore containing 10 μL reaction product and 2 μL 6× loading buffer as follows: S1, S2, S3, S4, S123, S124, S134, S234, and S1234, wherein each single strand in the reaction was diluted to 0.5 μM in the TM buffer. After the electrophoresis was completed, the gel was mixed and stained well with Goldviw I nuclear staining dyes, avoiding light and at room temperature for 30 min, and the electrophoresis gel bands were imaged clearly on the ChemiDoc XRS system.3. Results and discussion
3.1. Principle of the fabricated electrochemical biosensor
As illustrated in Scheme 1, TTs were synthesized in one step by four single DNA strands and stably modified on the electrode surface overnight at room temperature. Three vertexes of TTs were labeled with the thiol group at the 5′ end, which were modified on the electrode surface, and the other apex end including toehold was better designed to capture targets. Then, MCH was covered on the electrode surface to block other unbound active sites. In the presence of targets, targets were recognized and captured on the electrode. Targets have two recognition regions, and one could bind to TTs and one part to hairpin H1 to form multiple tandem hairpin assemblies (MTHsA). MicroRNA was tested in the experiment. Due to the short microRNA sequence, we designed the TTs' protruding end capture probe as a hairpin, with only the microRNA present, and the hairpin could be opened to perform the subsequent MTHsA reaction. Thereafter, the prepared mixture of H1, H2, H3, and H4 was incubated on the electrode surface at a constant temperature to produce MTHsA. At the beginning of the entire reaction, the target as a catalyst would strictly follow the principle of base pair complementarity and be hybridized with the long end of the hairpin H1 stem to open H1. The opened H1 would also bind to the long end of the hairpin H2 stem, causing H2 to spontaneously open, and a tandem DNA double helix with unlimited repeating units would be produced. Subsequently, the other domains of hairpin H2 tails were also initiators that could hybridize with hairpin H3 to open the folded stems. The opened hairpin H3 could then simultaneously bind with hairpin H4, while H3 and H4 hybrid chains also could produce a long double helix through the MTHsA of the two DNA chains. Along with the opening of H4, the required dendritic multi-branched cascade of long double-stranded chains of H1/H2/H3/H4 on the electrode was well produced. After that, the prepared modified electrodes were immersed in 10 mM Tri-HCl solution containing 50 μM RuHex for the next electrochemical measurements. The RuHex cation has been widely explored as an electrochemical indicator, which can be combined with the DNA phosphate backbone through electrostatic interaction. A small reduction peak could be observed without the targets. Otherwise, when the methylated DNA or unmethylated DNA was incubated on the electrode with Hpa II endonuclease, it could target the double stranded DNA (dsDNA) sequences 5′-CCGG-3′ and cleave dsDNA (unmethylated DNA) between the cytosine. The methylated DNA could not be cleaved by Hpa II, while the MTHsA reaction was well processed, which could induce a stronger electrochemical signal.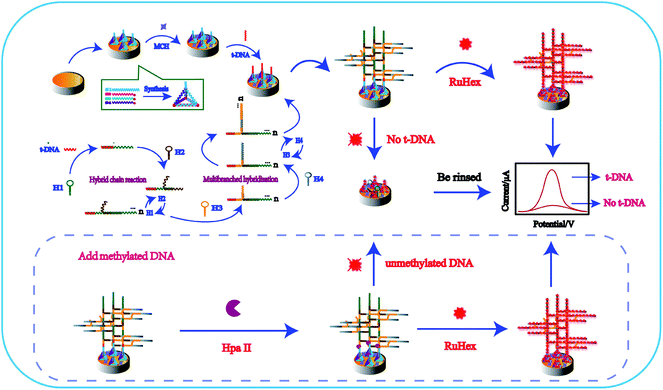 | ||
| Scheme 1 Schematic of the fabricated electrochemical biosensor. TTs assisted multiple tandem hairpins assembly for the ultra-sensitive detection of target DNA. | ||
3.2. Characterization of the constructed tetrahedral tripods (TTs)
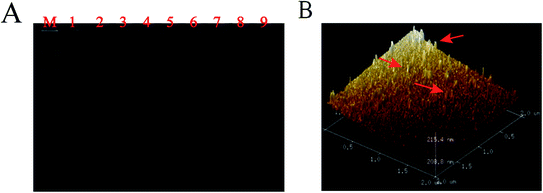
The key component in the experiment was to verify the feasibility of the TT capture probes.61 An 8% polyacrylamide gel electrophoresis (PAGE) was used to evaluate the successful synthesis of four well-designed DNA single-strands without interference bands, as shown in Fig. 1A, wherein rows 1–4 were single-stranded S1–S4 and rows 5–8 showed that the molecular bands of the three DNA strands were completely synthesized, which was compared with TTs in row 9 to verify the successful assembly of TTs into a larger molecular weight. AFM was employed to characterize TTs; the obvious three-dimensional structure indicated that the TTs were effectively synthesized.3.3. Characterization of the biosensor fabrication process
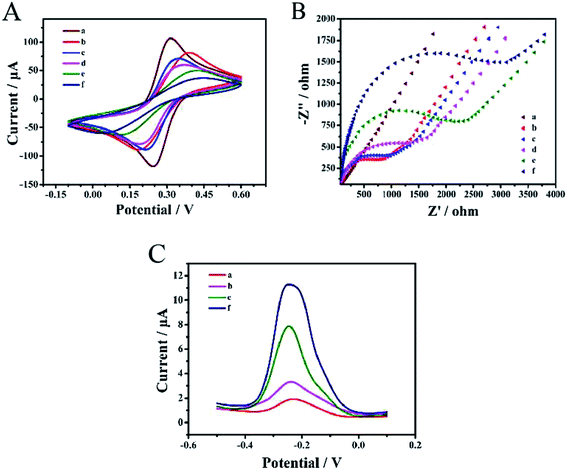
CV and EIS electrochemical analysis for each modification process in 0.1 M KCl solution containing 5 mM (Fe(CN)6)3−/4−, is depicted in Fig. 2A and Fig. 2B. DPV signal responses were recorded (Fig. 2C) for each modification process in 10 mM Tris–HCl with 50 μM RuHex.The bare gold electrode exhibited a pair of obvious redox peaks of (Fe(CN)6)3−/4− and a low DPV signal response; the straight line (curve a) reflected that the electroactive ions were transported easily on the electrode surface. The redox peak currents of (Fe(CN)6)3−/4− decreased and the electron-transfer resistance (Ret) increased in the modified electrode (curve b) with TTs, indicating that TTs were successfully modified on the gold electrode surface. When MCH was added on the electrode (curve c), although the current decreased, the blocking effect of MCH was not obvious. The stable three-dimensional TT space structure greatly reduced the interference of non-specific substances and thus reduced the background signal. Moreover, the target DNA was dropped on the electrode and captured by TTs (curve d), causing Ret to rise again. To compare the hybridization effects between two DNA strands and four DNA strands, only the mixture of H1 and H2 was incubated on the electrode and tested (curve e). The Ret and DPV signal response increased remarkably upon the hybridization of H1 and H2 (ref. 56 and 62) but when four DNA hairpins were assembled on the electrode surface successfully (curve f), the Ret increased and the current response decreased, which effectively confirmed the successful hybridization of multiple hairpin tandem strands and thus was more conducive to the following test.
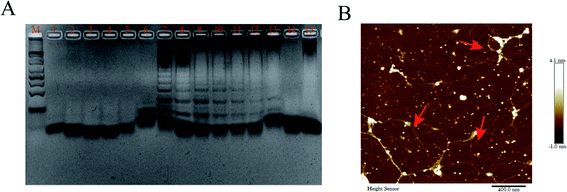
To prove the feasibility of MTHsA, 3.5% agarose gel electrophoresis and AFM imaging experiments were used (Fig. 3). The four DNA hairpins in the experiment that coexisted with the target DNA were incubated at 37 °C for 5 h; the different brighter bands suggested the successful MTHsA reaction of the hairpin probes hybridized to different concentrations of target DNA. Finally, the DPV changes of the stepwise modification process were analyzed in 10 mM Tris–HCl containing 50 μM RuHex and the results are shown in Fig. 2C. To further evaluate the feasibility of the multifunctional electrochemical biosensor, miRNA and methylated DNA were verified in the electrochemical experiment. When the target miRNA was present, there was a correspondingly high electrical signal as shown in Fig. S1.† Moreover, methylated DNA assisted by Hpa II enzyme digestion and all the proposed processes of the stepwise modification were successfully carried out. Hpa II enzyme could recognize unmethylated 5′-CCGG-3′ specific site and cleave it. After the DNA double strand was digested, the subsequent MTHsA reaction could not be performed, no electrical signal was generated; the CV and EIS signal characterizations of the gradually constructed biosensor were recorded (Fig. S2†).
3.4. Optimization of experimental parameters
To obtain the best experimental parameters, the crucial reaction parameters were assessed well, as illustrated in Fig. S3A.† RuHex in Tris–HCl, as an important electroactive substance, was highly sensitive to target amplification, and the DPV responses increased with RuHex concentration until it reached 50 μM. Thus, 50 μM RuHex was found to be the optimum detection concentration and was chosen for further experiments. This is because the saturated concentration of MTHsA determined the carrying capacity of RuHex. To improve the amount of RuHex adsorbed on the DNA phosphate backbone, the hybridization time of MTHsA was one of the important signal amplification parameters in preparing biosensors. As shown in Fig. S3B,† DPV responses increased with the hybridization time of MTHsA until the current responses saturated at 240 min. The reason was that the hybridization efficiency of a certain concentration of multiple hairpins was restricted and the reaction was saturated on the electrode. Thus, 240 min was chosen as the optimal incubation time for the subsequent experiments. In addition, the hybridization time of the target DNA played a key role in effective DNA detection (Fig. S3C†). As the DNA hybridization time increased, the DPV responses gradually increased until 120 min and then an increase in hybridization time caused the responses to decrease instead. Thus, it was obvious that each target probe with a sufficiently high concentration would fully meet the demand for a certain concentration of DNA detection. Subsequently, the TT concentration modified on the electrode surface for signal amplification was also of great significance for target DNA capture and signal amplification. As shown in Fig. S3D,† the DPV signals increased with the enhancement of TT concentration, and reached a plateau when it reached 0.5 μM. Once the electrode surface had been saturated with TTs, the DPV signals were then slightly decreased. Therefore, 0.5 μM TTs was selected for the next experimental operation. Moreover, the Hpa II enzyme digestion time was a key factor for the characterization and verification of methylated and unmethylated DNA when the endonuclease concentration was high enough to reach 50 U mL−1, as demonstrated in Fig. S4.† The DPV signals toward unmethylated DNA decreased with the increase in enzyme digestion time. It could be noted that the DNA was digested fully when the digestion time exceeded 120 min, and its current signal could be approximated to the blank signal value.3.5. Specificity, stability, and TT sensitivity of the constructed biosensor
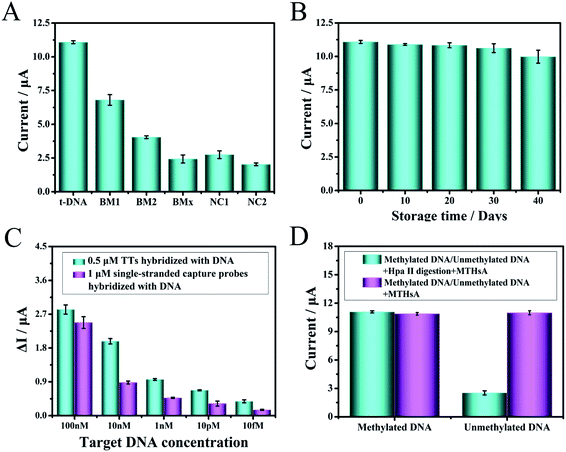
Under optimal experimental conditions, a series of mutant target molecules and normal target DNA combined with MTHsA reaction were evaluated to study the specificity of target DNA detection, which were as follows: single-base mismatched DNA (BM1), two-base mismatched DNA (BM2), multi-base mismatched DNA (BMx), non-complementary base mismatch-A (NC1), and non-complementary base mismatch-B (NC2). All the mutant sequences are shown in Table S1,† and the DPV current changes of the mutant target (Mt) molecules were fairly lower than the currents in the presence of the target DNA (Fig. 4A). The difference between non-complementary NC1 and NC2 was that half of the DNA sequences would hybridize with the toehold of TTs or one domain of hairpin H1. This specificity assay confirmed that when there were slight changes in the target nucleic acids, the detection results were different, and the efficiency of capture of the target analyte decreased with an increase in the number of base mismatches, e.g., the capture of a similar gene from the same family with a single-nucleotide polymorphism or a series of base mismatches could be effectively identified. The result clearly illustrated that the fabricated electrochemical platform had high sequence specificity.The assembled TTs and the prepared H1/H2/H3/H4 mixture were stored at 4 °C for days to verify the stability of the experiment. As shown in Fig. 4B, the experimental verification was to observe the DPV current changes from 0–40 days. A DPV current loss of 10% at 40 days of the original DPV signal was observed, and the stability reflected the superior electrochemical biosensing strategy design. Moreover, the sensitivity of TTs as probe to capture the target DNA was confirmed (Fig. 4C). A 0.5 μM TT and 1 μM single-stranded capture probes were incubated and modified on the electrode surface to capture and hybridize with several groups of the target DNA with different concentrations. The ΔI represented the signal change of DPV and defined as ΔI = I2 − I1, where I2 represents the DPV current response value of TTs or single-stranded capture probes in the presence of target DNA and I1 represents blank signal value without target captured. The ΔI of the target DNA captured after incubation with TTs was higher than that with the single-stranded capture probes despite the concentration of the single-stranded capture probes being higher. The results proved the superiority of TTs as a capture probe, which was important to detect even trace amounts of target DNA in human serum. Moreover, the comparison of the Hpa II enzymatic cleavage reaction of MTHsA between methylated and unmethylated DNA is shown in Fig. 4D.
3.6. Analytical performance of the constructed electrochemical biosensor
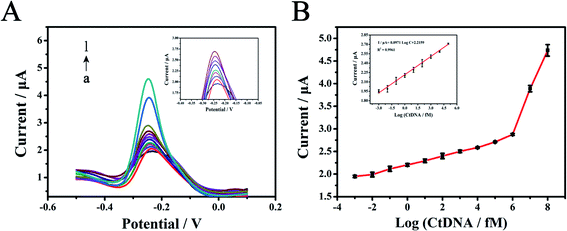
After exploring and determining various optimal experimental conditions, the electrochemical biosensor based on the TTs-assisted MTHsA could achieve ultra-sensitive detection of target DNA. As depicted in Fig. 5A, the DPV peak current response increased with the increase in the concentration of target DNA. The detection concentration could range from 1 aM to 100 nM and the DPV peak current response had a linear relationship with the target DNA logarithmic concentration from 1 aM to 100 pM with a correlation coefficient of 0.9961; the regression equation was I/μA = 2.2159 + 0.0971 lgC (fM) (Fig. 5B). The detection limit of the electrochemical biosensor was estimated to be 0.59 aM, which was calculated according to the following rule: 3 times the standard deviation of the 3 blank groups plus the average value. Compared with the previously reported submission, the electrochemical protocol and method in this experiment exhibited higher sensitivity and the entire reaction process was simplified via the highly efficient design of TTs-assisted MTHsA (Table S3†).3.7. Detection of targets spiked in human serum samples
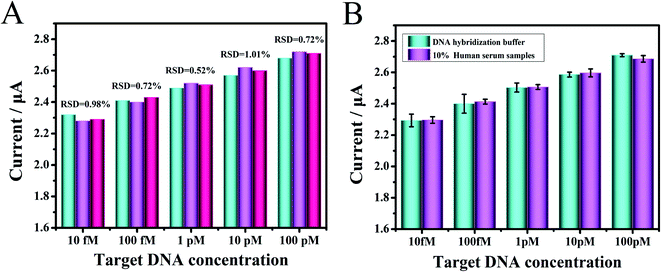
To verify the clinical applicability of the fabricated biosensor in biological conditions, the proposed biosensor's characteristics were assessed by testing target DNA and miRNA spiked in 10% human serum samples. Human serum samples spiked with five groups of different concentrations (10 fM to 100 pM) of chemically synthesized DNA, which were firstly dissolved to 100 μM from a freeze-dried powder, were diluted and tested in the experiment. As depicted in Fig. 6A, the reproducibility was assessed through five human serum sample measurements with different target DNA concentrations, and the results listed in Table S2† ranged from 95.1% to 105.54%, the relative standard deviation (RSD) value was as low as 0.52%, demonstrating that the biosensor had an excellent reproducibility. The DPV signal responses of the target DNA were simultaneously compared in DNA hybridization buffer and 10% human serum (Fig. 6B). Moreover, the reproducibility was assessed through five human serum sample (10 fM to 100 nM) measurements with different target miRNA, and the results are listed in Fig. S5.† Since miRNA was not stable, it could be seen that the detection value was lower than that of DNA. These results demonstrated that the biosensor could have potential clinical applications.4. Conclusion
In summary, we proposed a multifunctional electrochemical biosensor for the ultra-sensitive detection of targets by constructing a MTHsA reaction assisted by tetrahedral tripods (TTs). Thiol-modified TTs anchored on gold electrode surface provided a sturdy scaffold for site-specific anchoring of versatile biomolecular probes with high reproducibility. We tested miRNA and methylated DNA to verify the superiority of this protocol; as a simple design principle, the feasibility was promising, the cost was relatively lower, and the experiment operation was simpler. Under optimal conditions, the effective design of versatile TT toehold probes could excellently enhance target availability, specificity, and sensitivity, which decreased to 0.59 aM and improved sensitivity by several orders of magnitude. In the electrochemical measurement, the DPV signal responses were greatly amplified by the MTHsA reaction; the enzyme-free and label-free one-step isothermal multiple HCR could bring about the stable and repeated detection of targets, which could absorb a large amount of RuHex and greatly reduce background signal interference and realize the sensitive detection of DNA in human serum. In view of this, the multifunctional biosensor has great potential in biomedical research, drug therapy, and early clinical diagnosis of genetic diseases and basic research.Author contributions
Yuqi Huang: conceptualization, methodology. Shuhui Zhao: validation, formal analysis. Wenxiu Zhang: investigation, resources. Qiuyue Duan: data curation, visualization. Qi Yan: writing–original draft. Hu Fu: writing–review & editing. Liang Zhong: supervision. Gang Yi: project administration, funding acquisition.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.Acknowledgements
This work was supported by the Key Project of Science and Technology Research Program of Chongqing Education Commission (grant No. KJZD-K202000404), and the Science and Technology Research Program of Chongqing Yuzhong District Science Technology Commission (grant No. 20180127).References
- K. Matsuda, PCR-Based Detection Methods for Single-Nucleotide Polymorphism or Mutation: Real-Time PCR and Its Substantial Contribution Toward Technological Refinement, Elsevier Inc., 1st edn, 2017 Search PubMed.
- S. Minchin and J. Lodge, Understanding biochemistry: structure and function of nucleic acids, Essays Biochem., 2019, 63, 433–456 CrossRef CAS PubMed.
- J. Huang and L. Wang, Cell-Free DNA Methylation Profiling Analysis — Technologies and Bioinformatics, Cancers, 2019, 11, 1741 CrossRef CAS PubMed.
- L. Vrba and B. W. Futscher, A suite of DNA methylation markers that can detect most common human cancers, Taylor & Francis, 2018 Search PubMed.
- A. Ali, I. Sina, L. G. Carrascosa and M. Trau, Opinion DNA Methylation-Based Point-of-Care Cancer Detection: Challenges and Possibilities, Trends Mol. Med., 2019, 1–12 Search PubMed.
- M. R. Green and J. Sambrook, Analysis and Normalization of Real-Time Polymerase Chain Reaction (PCR) Experimental Data, Cold Spring Harbor Protocols, 2018, 10, 769–778 Search PubMed.
- M. C. Lebeau, G. A. Bolado, W. Wahli and S. Catsicas, PCR driven DNA-DNA competitive hybridization: a new method for sensitive differential cloning, Nucleic Acids Res., 1991, 17, 4778 CrossRef PubMed.
- A. Lishanski, N. Kurn and E. Ullman, Branch migration inhibition in PCR-amplified DNA: homogeneous mutation detection, Nucleic Acids Res., 2000, 28, 42–50 CrossRef PubMed.
- J. Wilhelm, A. Pingoud and M. Hahn, Validation of an algorithm for automatic quantification of nucleic acid copy numbers by real-time polymerase chain reaction, Anal. Biochem., 2003, 317, 218–225 CrossRef CAS PubMed.
- S. Larseni, R. E. Rygaard, S. Asnies and M. Spang-thomsen, Northern and Southern blot analysis of human RNA and DNA in autopsy material, Apmis, 1992, 100, 498–502 CrossRef PubMed.
- M. H. Schiffman, H. M. Bauer., A. T. Lorincz., M. M. Manos., J. C. Byrne., A. G. Glass., D. M. Cadell and P. M. Howley, Comparison of Southern blot hybridization and polymerase chain reaction methods for the detection of human papillomavirus DNA, J. Clin. Microbiol., 1991, 29, 573 CrossRef CAS PubMed.
- M. L. Quintana, R. Rauhut, W. Lendeckel and T. Tuschl, Identification of novel genes coding for small expressed RNAs, Science, 2001, 294, 853–858 CrossRef PubMed.
- T. R. Hughes, S. L. Hiley, A. L. Saltzman, T. Babak and B. J. Blencowe, 14 Microarray Analysis of RNA Processing and Modification, Methods Enzymol., 2006, 410, 300–316 CAS.
- J. Peplies, C. Lachmund, F. O. Glöckner and W. Manz, A DNA microarray platform based on direct detection of rRNA for characterization of freshwater sediment-related prokaryotic communities, Appl. Environ. Microbiol., 2006, 72, 4829–4838 CrossRef CAS PubMed.
- P. Jolly, P. Estrela and M. Ladomery, Oligonucleotide-based systems: DNA, microRNAs, DNA/RNA aptamers, Essays In, Biochem, 2016, 60, 27–35 Search PubMed.
- L. K. Ali, S. Mitsuharu, T. Isabelle, M. Emmanuelle, B. Eloy Bernal, C. Bourget, M. Lionel., L. Jean and T. Alain, Aryldiazomethanes for universal labeling of nucleic acids and analysis on DNA chips, Bioconjugate Chem., 2003, 14, 1298–1306 CrossRef PubMed.
- C. Li, H. Wang, J. Shen and B. Tang, Cyclometalated iridium complex-based label-free photoelectrochemical biosensor for dna detection by hybridization chain reaction amplification, Anal. Chem., 2015, 87, 4283–4291 CrossRef CAS PubMed.
- M. Wang, H. Yin, Y. Zhou, C. Sui, Y. Wang, X. Meng, G. I. N. Waterhouse and S. Ai, Photoelectrochemical biosensor for microRNA detection based on a MoS 2/g-C 3 N 4/black TiO 2 heterojunction with Histostar@AuNPs for signal amplification, Biosens. Bioelectron., 2019, 128, 137–143 CrossRef CAS PubMed.
- H. Wang, M. Li, Y. Zheng, T. Hu, Y. Chai and R. Yuan, Biosensors and Bioelectronics An ultrasensitive photoelectrochemical biosensor based on Ru (dcbpy) 2 dppz 2 +/Rose Bengal dyes co-sensitized fullerene for DNA detection, Biosens. Bioelectron., 2018, 120, 71–76 CrossRef CAS PubMed.
- N. Yotapan, D. Nim-anussornkul and T. Vilaivan, Pyrrolidinyl peptide nucleic acid terminally labeled with fluorophore and end-stacking quencher as a probe for highly specific DNA sequence discrimination q, Tetrahedron, 2016, 72, 7992–7999 CrossRef CAS.
- H. Wu, H. Wang, Y. Liu, J. Wu and P. Zou, Fluorometric determination of microRNA by using target-triggered cascade signal amplification and DNA-templated silver nanoclusters, Microchim. Acta, 2019, 186, 669 CrossRef CAS PubMed.
- H. Zhang, X. Liu, C. Zhang, Y. Xu, J. Su and X. Lu, et al., A DNA tetrahedral structure-mediated ultrasensitive fluorescent microarray platform for nucleic acid test, Sens. Actuators, B, 2020, 321, 128538 CrossRef CAS.
- J. Zhang, Y. Yang, X. Jiang, C. Dong, C. Song, C. Han and L. Wang, Ultrasensitive SERS detection of nucleic acids via simultaneous amplification of target-triggered enzyme-free recycling and multiple-reporter, Biosens. Bioelectron., 2019, 141, 111402 CrossRef CAS PubMed.
- W. Zhou, Y. F. Tian, B. C. Yin and B. C. Ye, Simultaneous SERS Detection of Multiplexed MicroRNA Biomarkers Simultaneous SERS Detection of Multiplexed MicroRNA Biomarkers, Anal. Chem., 2017, 89, 6120–6128 CrossRef CAS PubMed.
- A. Kowalczyk, J. Krajczewski, A. Kowalik, J. L. Weyher, I. Dziecielewski and M. Chlopek, et al., New strategy for the gene mutation identification using surface enhanced Raman spectroscopy (SERS), Biosens. Bioelectron., 2019, 132, 326–332 CrossRef CAS PubMed.
- H. He, J. Dai, Y. Meng, Z. Duan, C. Zhou and B. Zheng, et al., Self-assembly of DNA nanoparticles through multiple catalyzed hairpin assembly for enzyme-free nucleic acid amplified detection, Talanta, 2018, 179, 641–645 CrossRef CAS PubMed.
- S. Wei, G. Chen, X. Jia, X. Mao, T. Chen, D. Mao, W. Zhang and W. Xiong, Exponential amplification reaction and triplex DNA mediated aggregation of gold nanoparticles for sensitive colorimetric detection of microRNA, Anal. Chim. Acta, 2020, 1095, 179–184 CrossRef CAS PubMed.
- S. Li, X. Shang, J. Liu, Y. Wang, Y. Guo and J. You, A universal colorimetry for nucleic acids and aptamer-specific ligands detection based on DNA hybridization amplification, Anal. Biochem., 2017, 528, 47–52 CrossRef CAS PubMed.
- S. Campuzano, M. Pedrero and J. M. Pingarron, Electrochemical Nucleic Acid-Based Biosensing of Drugs of Abuse and Pharmaceuticals, Curr. Med. Chem., 2018, 25, 4102–4118 CrossRef CAS PubMed.
- S. Liu, Y. Lin, T. Liu, C. Cheng, W. Wei, L. Wang and F. Li, Biosensors and Bioelectronics Enzyme-free and label-free ultrasensitive electrochemical detection of DNA and adenosine triphosphate by dendritic DNA concatamer-based signal amplification, Biosens. Bioelectron., 2014, 56, 12–18 CrossRef CAS PubMed.
- J. Zhuang, L. Fu, M. Xu, H. Yang, G. Chen and D. Tang, Sensitive electrochemical monitoring of nucleic acids coupling DNA nanostructures with hybridization chain reaction, Anal. Chim. Acta, 2013, 783, 17–23 CrossRef CAS PubMed.
- D. Yang, Y. Tang and P. Miao, Trends in Analytical Chemistry Hybridization chain reaction directed DNA superstructures assembly for biosensing applications, Trends Anal. Chem., 2017, 94, 1–13 CrossRef CAS.
- L. Song, Y. Zhang, J. Li, Q. Gao, H. Qi and C. Zhang, Non-Covalent Fluorescent Labeling of Hairpin DNA Probe Coupled with Hybridization Chain Reaction for Sensitive DNA Detection, Appl. Spectrosc., 2016, 70, 688–694 CrossRef CAS PubMed.
- Z. Shi, X. Zhang, R. Cheng, B. Li and Y. Jin, Sensitive detection of intracellular RNA of human telomerase by using graphene oxide as a carrier to deliver the assembly element of hybridization chain reaction, Analyst, 2016, 141, 2727–2732 RSC.
- X. Chen, J. Huang, S. Zhang, F. Mo, S. Su, Y. Li, L. Fang, J. Deng, H. Huang, Z. Luo and J. Zheng, Electrochemical Biosensor for DNA Methylation Detection through Hybridization Chain-Amplified Reaction Coupled with a Tetrahedral DNA Nanostructure, ACS Appl. Mater. Interfaces, 2019, 11, 3745–3752 CrossRef CAS PubMed.
- X. Su, H. Teh, X. Lieu and Z. Gao, Enzyme-Based Colorimetric Detection of Nucleic Acids Using Peptide Nucleic Acid-Immobilized Microwell Plates, Anal. Chem., 2007, 79, 7192–7197 CrossRef CAS PubMed.
- Y. C. Su, H. Y. Chen, N. C. Ko, C. C. Hwang, M. H. Wu and L. F. Wang, et al., Effective and site-specific phosphoramidation reaction for universally labeling nucleic acids, Anal. Biochem., 2014, 449, 118–128 CrossRef CAS PubMed.
- C. Ding, N. Wang, J. Zhang and Z. Wang, Rolling circle amplification combined with nanoparticle aggregates for highly sensitive identification of DNA and cancer cells, Biosens, Bioelectron, 2013, 42, 486–491 CrossRef CAS PubMed.
- W. Tian, P. Li, W. He, C. Liu and Z. Li, Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs, Biosens. Bioelectron., 2019, 128, 17–22 CrossRef CAS PubMed.
- P. Chen, P. Wu, Y. Zhang, J. Chen, X. Jiang and C. Zheng, et al., Strand Displacement-Induced Enzyme-Free Amplification for Label-Free and Separation-Free Ultrasensitive Atomic Fluorescence Spectrometric Detection of Nucleic Acids and Proteins, Anal. Chem., 2016, 88, 12386–12392 CrossRef CAS PubMed.
- Z. Chen, Y. Liu, C. Xin, J. Zhao and S. Liu, A cascade autocatalytic strand displacement amplification and hybridization chain reaction event for label-free and ultrasensitive electrochemical nucleic acid biosensing, Biosens. Bioelectron., 2018, 113, 1–8 CrossRef CAS PubMed.
- S. Li, T. Tian, T. Zhang, X. Cai and Y. Lin, Advances in biological applications of self-assembled DNA tetrahedral nanostructures, Mater. Today, 2019, 24, 57–68 CrossRef CAS.
- Z. Ge, M. Lin, P. Wang, H. Pei, J. Yan and J. Shi, et al., Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor, Anal. Chem., 2014, 86, 2124–2130 CrossRef CAS PubMed.
- M. Lin, J. Wang, G. Zhou, J. Wang, N. Wu, J. Lu, J. Gao, X. Chen, J. Shi, X. Zuo and C. Fan, Programmable Engineering of a Biosensing Interface with Tetrahedral DNA Nanostructures for Ultrasensitive DNA Detection, Angewandte, 2015, 2151–2155 CrossRef CAS PubMed.
- H. Pei, N. Lu, Y. Wen, S. Song, Y. Liu and H. Yan, et al., A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing, Adv. Mater., 2010, 22, 4754–4758 CrossRef CAS PubMed.
- X. Wang, J. Wu, W. Mao, X. He, L. Ruan, J. Zhu, P. Shu, Z. Zhang, B. Jiang and X. Zhang, A tetrahedral DNA nanostructure-decorated electrochemical platform for simple and ultrasensitive EGFR genotyping of plasma ctDNA, Analyst, 2020, 145, 4671–4679 RSC.
- X. Wang, F. Chen, D. Zhang, Y. Zhao, J. Wei, L. Wang, S. Song, C. Fan and Y. Zhao, Single copy-sensitive electrochemical assay for circulating methylated DNA in clinical samples with ultrahigh specificity based on a sequential discrimination-amplification strategy, Chem. Sci., 2017, 8, 4764–4770 RSC.
- Y. Li, C. Z. Huang and Y. F. Li, Ultrasensitive Electrochemiluminescence Detection of MicroRNA via One-Step Introduction of a Target-Triggered Branched Hybridization Chain Reaction Circuit, Anal. Chem., 2019, 91, 9308–9314 CrossRef CAS PubMed.
- W. Yang, X. Zhou, J. Zhao and W. Xu, A cascade amplification strategy of catalytic hairpin assembly and hybridization chain reaction for the sensitive fluorescent assay of the model protein carcinoembryonic antigen, Microchim. Acta, 2018, 185, 100 CrossRef PubMed.
- R. M. Dirks and N. A. Pierce, Triggered amplification by hybridization chain reaction, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- S. Bi, M. Chen, X. Jia, Y. Dong and Z. Wang, Branched Hybridization Chain Reaction for Triggered Signal Amplification and Concatenated Logic Circuits, Angew. Chem., 2015, 54, 8144–8148 CrossRef CAS PubMed.
- J. Huang, Y. Wu, Y. Chen, Z. Zhu, X. Yang and C. J. Yang, et al., Pyrene-excimer probes based on the hybridization chain reaction for the detection of nucleic acids in complex biological fluids, Angew. Chem., 2011, 50, 401–404 CrossRef CAS PubMed.
- Y. Tang, X. L. Zhang, L. J. Tang, R. Q. Yu and J. H. Jiang, In Situ Imaging of Individual mRNA Mutation in Single Cells Using Ligation-Mediated Branched Hybridization Chain Reaction (Ligation-bHCR), Anal. Chem., 2017, 89, 3445–3451 CrossRef CAS PubMed.
- X. Chen, J. Huang, S. Zhang, F. Mo, S. Su, Y. Li, L. Fang, J. Deng, H. Huang, Z. Luo and J. Zheng, Electrochemical Biosensor for DNA Methylation Detection through Hybridization Chain-Amplified Reaction Coupled with a Tetrahedral DNA Nanostructure, ACS Appl. Mater. Interfaces, 2019, 11, 3745–3752 CrossRef CAS PubMed.
- S. Dai, W. Lu, Y. Wang and B. Yao, Universal DNA biosensing based on instantaneously electrostatic attraction between hexaammineruthenium (III) and DNA molecules, Biosens. Bioelectron., 2019, 127, 101–107 CrossRef CAS PubMed.
- Q. Guo, Y. Yu, H. Zhang, C. Cai and Q. Shen, Electrochemical Sensing of Exosomal MicroRNA Based on Hybridization Chain Reaction Signal Amplification with Reduced False-Positive Signals, Anal. Chem., 2020, 92, 5302–5310 CrossRef CAS PubMed.
- S. Sato and S. Takenaka, PCR-free telomerase assay using chronocoulometry coupled with hexaammineruthenium(III) chloride, Anal. Chem., 2012, 84, 1772–1775 CrossRef CAS PubMed.
- J. Huang, X. Y. Li, Y. C. Du, L. N. Zhang, K. K. Liu, L. N. Zhu and D. M. Kong, Sensitive fluorescent detection of DNA methyltransferase using nicking endonuclease-mediated multiple primers-like rolling circle amplification, Biosens. Bioelectron., 2017, 91, 417–423 CrossRef CAS PubMed.
- M. Lin, P. Song, G. Zhou, X. Zuo, A. Aldalbahi and X. Lou, et al., Electrochemical detection of nucleic acids, proteins, small molecules and cells using a DNA-nanostructure-based universal biosensing platform, Nat. Protoc., 2016, 11, 1244–1263 CrossRef CAS PubMed.
- L. Liu, J.-W. Liu, H. Wu, X.-N. Wang, R.-Q. Yu and J.-H. Jiang, Branched Hybridization Chain Reaction Circuit for Ultrasensitive Localizable Imaging of mRNA in Living Cells, Anal. Chem., 2018, 90, 1502–1505 CrossRef CAS PubMed.
- H. Liu, J. Luo, L. Fang, H. Huang, J. Deng and J. Huang, et al., An electrochemical strategy with tetrahedron rolling circle amplification for ultrasensitive detection of DNA methylation, Biosens. Bioelectron., 2018, 121, 47–53 CrossRef CAS PubMed.
- L. P. Jia, L. J. Wang, R. N. Ma, L. Shang, W. Zhang and Q. W. Xue, et al., An electrochemical aptasensor for the highly sensitive detection of 8-hydroxy-2'-deoxyguanosine based on the hybridization chain reaction, Talanta, 2018, 179, 414–419 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02424h |
| This journal is © The Royal Society of Chemistry 2021 |