Open Access Article
Open Access ArticleA perspective on the isolation and characterization of extracellular vesicles from different biofluids
Reshma Banoa,
Farhan Ahmadb and
Mohd Mohsin *a
*a
aMetabolic Engineering Laboratory, Department of Biosciences, Jamia Millia Islamia (A Central University), New Delhi-110025, India. E-mail: mmohsin1@jmi.ac.in
bDepartment of Animal Biology, University of Hyderabad, Hyderabad, 500046, India
First published on 1st June 2021
Abstract
Extracellular vesicles (EVs) are small membrane-bound particles, which include exosomes, micro vesicles (MVs) and various-sized vesicles, released by healthy and diseased cells. EVs also include other vesicular structures, such as large apoptotic bodies (1–5 μm), as well as membrane particles (50–80 nm) originating from the plasma membrane. However, exosomes are nanosize (≈30–100 nm) extracellular vesicles of endocytic origin that are bud-off by most types of cells and circulate in bodily fluids. Extracellular nanovesicles contain a large variety of biomolecules, including miRNA, RNA, DNA, proteins, signaling peptides and lipids, that can have diagnostic and therapeutic value. The spectrum of the existing scientific interest in extracellular nanovesicles is comprehensive, which ranges from understanding their functions and pathways to their potential clinical usage. EVs can be obtained from different body fluids with minimally invasive techniques (e.g., urine, plasma, serum), so they are most useful in disease diagnosis. High yield and purity contribute to the accurate diagnosis of various diseases, but damaged EVs and impurities can cause misinterpreted results. Over the last decade, a plethora of approaches have been developed for examining EVs using optical and non-optical tools. However, EV isolation methods have different yields and purities. Moreover, the isolation method that is most appropriate to maximize EVs recovery depends on the different experimental situations. This review explores the emerging use of micro and nano-technologies to isolate and characterize exosomes and microvesicles (MVs) from different biological samples, and the application of these technologies for the monitoring and diagnosis of different pathological conditions.
1. Introduction
Many cell types actively secrete extracellular vesicles (EVs). EVs are released from cells, including tumor cells, immune cells, and others, into their surrounding environment by budding from the cells' plasma membrane or upon fusion of a multivesicular body (MVB) with the plasma membrane.1,2 EVs exhibit a wide diversity in terms of their diameter size [e.g., exosomes (30–100 nm), MVs (100–1000 nm), apoptotic bodies (1000–5000 nm)] with respect to the outer phospholipid membrane, and contain growth factors, receptors, cytokines, as well as lipids, RNAs and metabolites.3 EVs are biological nanoparticles found in extracellular fluids, such as saliva, milk, blood, and tears.4 Although research on EVs such as exosomes, microvesicles and others has been ongoing for more than 30 years,5 their significance in the discovery and development of disease-related biomarkers has come on the heels very recently.6,7 EVs perform the role of mediators of intercellular communication by conveying biological information between cells.Two major types of EVs (exosomes and MVs) have been focused in the current research field. The term ‘‘exosomes’’ is often used to entitle the EVs evaluated. Thus, the term exosome is commonly used to denote a diverse population of small EVs (sEVs) deprived of further demonstration of their intracellular origin. In fact, functions allocated to exosomes may reflect generic EV activity.8 Exosome secretion was reported for the first time in reticulocytes through their differentiation pathways.9
EVs also have been shown to participate in the propagation of cancer cells, and many groups have defined that tumor- and stroma-derived EVs are intricate in the different phases of the metastatic cascade.10,11 More recently, hematopoietic cells (including dendritic cells (DCs),12–14 B and T lymphocytes,15–18 and mast cells19,20) have been shown to release exosomes. Several studies have reported that non-hematopoietic cells, such as intestinal epithelial cells,21 neuroglial cells22 and tumor cells,23,24 also are competent to release exosomes.
The International Society of Extracellular Vesicles (ISEV), the Food and Drug Administration (FDA), the International Council for Harmonization (ICH) of Technical Requirements for Pharmaceuticals for Human Use, and the European Medicines Agency (EMA) offer extensive guidance for the enlargement and generation of novel biological medicines with regard to donor/patient care, product safety, and quality.25–28 EV-based therapeutics has opened up a whole new area of research opportunities for treating diseases related to inflammation. Researchers have been trying to categorize the roles of EVs, contained proteins and nucleic acids, and their release mechanisms. Over the studies of exosomes, they proposed that these could be deliberated as novel diagnostic markers and therapeutic targets for cancer. EVs are also produced by healthy cells and play important roles in physiological functions. It has been reported that many different types of cancer (such as prostate cancer,29 breast and ovarian cancer) patients contain higher concentrations of exosomes than healthy individuals.30–32 Therefore, in the recent year's standardized and accurate isolation, the quantification and exploration of cancer-specific exosomes have gained more attention.
Due to the intricate property of the sample matrix and physiochemical properties, the isolation of EVs poses significant challenges.33,34 Conventional methods, such as differential and density gradient centrifugation, chromatography and immunological-based separation, have been affianced to isolate exosomes.35–39 In recent time, the putative procedure for the isolation of exosomes includes ultracentrifugation, consistently in combination with sucrose density gradients or sucrose cushions to float the comparatively low-density exosomes,40 and using a novel class of synthetic peptide (Vn96) with specific affinity towards heat shock proteins (Hsps) and efficiently capture Hsp-containing exosomes from biological fluids.41 The recently released commercial kits co-isolate different types of EVs. At present, numerous commercial Exosome Isolation Kits are available. These kits elude the time-consuming variance steps by precipitating the vesicles with polyethylene glycol or similar components, while the isolation of non-vesicles has also been perceived.42 The size exclusion chromatography (SEC) method was also evaluated to isolate extracellular vesicles from plasma and cell culture.43 Although advances in nano-size particle detection approaches have been achieved, the routine detection and quantification of EVs are still challenging and clumsy,44,45 which is partly due to the lack of rapid, sensitive, reproducible and low-cost methodologies.
In recent years, various detection methodologies have been broadly developed to evaluate exosomes and MVs for both research and clinical purposes.46–48 For example, enzyme-linked immune-sorbent assay (ELISA),49 nanoparticle tracking analysis (NTA),50 tunable resistive pulse sensing (TRPS), flow cytometry35 and fluorescence-activated cell sorting (FACS),51 a modified ELISA assimilated with a lab-on-a-chip microfluidic device offer a modest, fast and robotic investigation of exosomes52 and some developing strategies based on microfluidics and electrochemical biosensors.53–57
The main aim of this review is to deliberate the current advances in EVs isolation and quantification methods that would be helpful in the future to identify various diseases, including cancer and neurodegenerative disorders and others in their early stages, and also discuss the limitation and some technical troubles of various methods used in the purification and detection of EVs.
2. Methods of EVs isolation
Subsequently, research in the study of extracellular vesicles has grown rapidly in the last few years, and there has been a vast development in the number of techniques for the isolation and characterization of extracellular vesicles, although many of them are poorly standardized. EVs are not homogeneous in size, origin and molecular constituents with a substantial overlap in the size and phenotype between diverse populations of EVs.6,42Prominent methods are shown in Table 1 for EVs isolation from different sources, such as the cell culture medium, serum, plasma, milk, urine and others. Different isolation methods of exosomes to isolate from different types of cancer in clinical application are also mentioned in Table 3.
| Sources of exosomes | Prominent methods of isolation | Ref. |
|---|---|---|
| Blood plasma | (1) Size exclusion chromatography | 88 |
| (2) Ultracentrifugation | ||
| CNS fluid | (1) Differential ultracentrifugation | 60 and 82 |
| (2) Precipitation-based technology | ||
| (3) Size exclusion chromatography | ||
| Saliva | (1) Differential centrifugation | 59 |
| (2) Gel filtration | ||
| Milk (human & bovine) | (1) Density gradient centrifugation | 109 |
| (2) Size exclusion chromatography | ||
| Cell culture filtrate | (1) Ultracentrifugation | 72 |
| (2) Immunomagnetic extraction | ||
| (3) Immunoblotting techniques | ||
| Urine | (1) Ultracentrifugation | 86 |
| (2) Nanomembrane ultrafiltration |
2.1. Exosome isolation by ultracentrifugation
Centrifugation-based techniques are being considered as the best method to isolate the exosomes.13,40 Centrifugation is used for the sedimentation of particulate matter, such as vesicles in solution. Various types of vesicles present in a sample are separated based on the size and mass, so bigger and denser components migrate away from the axis of the centrifuge and minor and less-dense components migrate toward the axis of the centrifuge. Ultracentrifugation is a classical technique that is the most commonly used method for the purification of exosomes, making use of the high centrifugal force for studying the properties of small biological particles. Ultracentrifugation can be characterized based on the principles of separation: (a) differential ultracentrifugation and (b) density gradient ultracentrifugation (Fig. 1).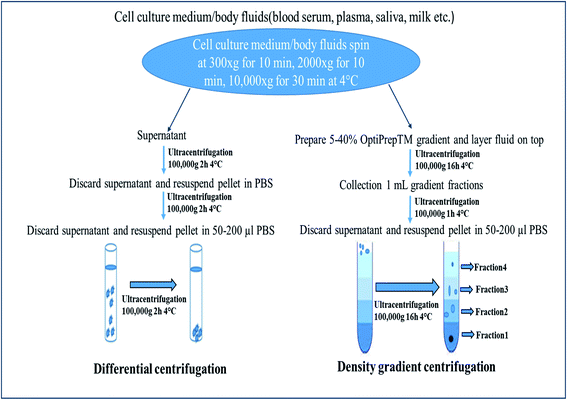 | ||
| Fig. 1 Ultracentrifugation methods: differential centrifugation and density gradient centrifugation. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g to 20
000×g to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g to isolate vesicles larger than 100 nm (microvesicles); then finally (d) at 100
000×g to isolate vesicles larger than 100 nm (microvesicles); then finally (d) at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g to 200
000×g to 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g to isolate vesicles smaller than 100 nm (small EVs: exosomes).58,60
000×g to isolate vesicles smaller than 100 nm (small EVs: exosomes).58,60This is the most commonly used protocol for the isolation of exosomes from cell culture media, but must be altered when the EVs are suspended in viscous fluids, such as plasma or saliva.59 The fluid (such as blood serum, plasma, urine, saliva) is simply collected by the usual means, then stored for up to 5 days at 4 °C in a falcon tube or glass bottle up to succeed with the exosome isolation process. Because of the viscosity of some fluids, it is obligatory to dilute them and to exceed the speed and lengths of centrifugations.40 Small EVs are pelleted at low speed at the bottom of the tube, whereas the top of the tube contains the larger vesicles, with the possible co-precipitation of protein aggregates, apoptotic bodies, and other types of EVs with a high-speed spin. The co-precipitation results in less sample purity and contagion of exosomes with other particles. One potential solution is the use of multiple resuspending and recentrifuging steps of each pellet in a PBS (phosphate-buffered saline) buffer to remove some of these contaminants (Fig. 1).88
2.2. Precipitation based EVs isolation
Membrane particle precipitation-based EV isolation is an interesting method because of the modest procedure and high yield. Presently, there are numerous methods for isolating plasma EVs.78 Several commercial EV Isolation Kits, such as the ExoQuick and Invitrogen Total Exosome isolation reagent, miRCURY™ Exosome Isolation Kit, are based on the precipitation of membrane particles. According to a worldwide survey accomplished in 2015 by the ISEV, precipitation-based methods are used particularly for biological samples with a small volume.79 Generally, the polymeric precipitation method has been focused on the estimation of RNA and protein content.80 The polymeric precipitation methods are established on the creation of a mesh-like network that embeds EVs with a size ranging from 60 to 180 nm. Precipitation based methods are applicable to body fluids and culture media at large scale. In particular, these methods may have an advantage in the exposure of biomarkers in vesicles derived from small biological samples.81 The purity of EVs remains a question in this method, as it may also precipitate the viruses and few proteins in the sample.2.3. Chromatography-based method
2.4. Isolation of exosomes by immunological separation technique
For the specific isolation of exosomes, there is an alternate method based on numerous proteomic studies of the molecular composition of exosomes that can be used as affinity purification with antibodies to CD63, CD81, CD82, CD9, Alix, annexin, EpCAM and Rab5.39,40 Johnstone et al., 1991 reported that during the late stage of erythrocyte differentiation, exosomes were initially originated and predicted as functioning in shedding the transferrin receptor (CD71). Various studies of exosomal constituents revealed the presence of numerous proteins on the exosomal membrane.68 These proteins can be ultimate markers for the extraction of exosomes due to immunoaffinity interactions between the proteins and antibodies, which should be specific for a particular marker present on the exosomal membrane.40 Current advances in the exosome purification field have shown that antibody-coated magnetic beads can be efficiently employed to extract exosomes from antigen-presenting cells [APC]. For this purpose, selecting a proper exosomal membrane marker is one of the most significant steps in these immunoassays. It was revealed that members of the tetraspanin family, such as CD81, CD9, and CD63 that exist on the exosomal membrane, can be used for the efficient immuno-capture of exosomes.69–71This method is applicable for the downstream analysis of nucleic acids and proteins, enabling the identification of EGFR (epidermal growth factor receptor) exon 19 and 21 and KRAS codons 12 and 13 mutations, following the isolation of extracellular vesicles from the blood plasma of non-small-cell lung-cancer (NSCLC) patients.73
2.5. Rapid isolation with a novel synthetic peptide
A new method has been reported, which has the capability to aggregate the exosomes from the medium within 30 minutes.41 Ghosh et al., (2014) synthesized and validated a novel class of peptides named as venceremin (Vn), showing specific affinity for canonical heat shock proteins that cover the surface of exosomes and EVs. The Vn peptides were shown to efficiently capture Hsp-containing EVs from cell culture growth media, plasma and urine by electron microscopy, atomic force microscopy, proteomic profiling, and NTA. This technology is a more efficient isolation method than current methods. The peptide Vn 96 can isolate EVs derived from cells due to their high content of Hsps under stress conditions, such as cancer, infection, cardiovascular, neurodegenerative and metabolic diseases in human or animal body fluids. After the examination of purified EVs, this method follows the recent clinical diagnostic trend for biomarker screening with non-invasive measures envisioned for early disease detection.412.6. Isolation by microfluidic device
Microfluidics technology considered by the engineered manipulation of fluids at the nanoscale level has revealed significant potential for improving diagnostics and biological research. Certain properties of microfluidic technologies, such as fast sample processing and the accurate control of fluids in an assay, have made them attractive applicants to substitute traditional experimental methodologies.74 Microfluidic devices enable the manipulation of minor amounts of samples and components in their channels, and allow for the rapid and low-cost separation and detection of targets. So, microchip devices are a verified technology for cellular handling as they can offer accurate spatial and temporal control in a significantly miniaturized platform. These devices can be simply made by employing standard micro-fabrication tools, which lowers the production cost and makes it commercially viable. Based on these properties, various methods have been implicated to extract and detect the exosomes.All of these techniques have some advantages over other techniques, and also have some limitations with the time of isolation, purity of sample, and handling errors, which are mentioned in Table 2. So, for the high purity and quick isolation of EVs, we have to rectify these limitation with some new methods or have to do more work for better results on those described methods.
| Isolation techniques | Purity | Ease of use | Time | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|---|---|
| Differential centrifugation | Low | Moderate | Long | Considered to be a gold standard and reliable method. Allows for the analysis of large sample volumes and multiple samples at the same time | Time-consuming procedures, high equipment cost, high centrifugation speed, resulting in exosome damage. Protein contamination and low EV yields | 58 |
| Density-gradient centrifugation | High | Difficult | Long | The method separates low-density exosomes from other extracellular vesicles | Very high sensitivity to the centrifugation time | 60 |
| Immunoaffinity-based separation (surface-modified) | High | Moderate | Intermediate | The method isolates exosomes directly from the cell culture supernatant or bodily fluids | The method cannot be applied to large-volume samples. Protein contamination | 69–71 |
| Size exclusion chromatography | High | Easy | Intermediate | SEC results in a good recovery of vesicles with almost complete removal of contaminants, no expensive equipment is needed | Apo lipoprotein leftover | 81 and 82 |
| Thus, isolation of vesicles by SEC is quick, cheap and easy | ||||||
| Microfluidic device (ExoChip, ciliated nanowire-on-microvillar, microfluidic filtration) | High | Moderate | Intermediate | These devices isolate and detect exosomes in parallel with a very short time procedure | Many of these devices suffer from low throughput due to the single-channel and limited lifespan of the microfluidic chips, which are typically shortened due to blocking or clogging | 76, 77 and 47 |
3. Characterization of EVs
The most commonly used conventional methods for the detection and quantification of EVs that use micro- and nano-based techniques to improve these EVs detection have been recently developed. The advantages and disadvantages of the detection methods for these given techniques are highlighted in Table 4.| Biological fluid | Disease | Method of isolation | Ref. |
|---|---|---|---|
| Cell culture medium | Lung cancer | Centrifugation with magnetic bead capture | 58 |
| Human-prostate cancer | Sequential centrifugation and antibody-conjugated magnetic microbeads | 61 | |
| Ovarian cancer | Modified magnetic activated cell sorting (MACS) procedure | 29 | |
| Breast cancer | Immuno-based separation with HER2 antibody-coated paramagnetic beads | 62 | |
| Plasma | Melanoma cancer | Immunoaffinity-based method | 63 and 64 |
| Size exclusion chromatography | |||
| Urine | Prostate cancer | Ultracentrifugation followed by filtration | 65 |
| Prostate cancer | Vn-96-peptide-based EV isolation | 66 | |
| Serum | Non-small cell lung cancer (NSCL) | Commercial Kit (Invitrogen) | 67 |
| Plasma | Brain tumor-glioblastoma multiforme (GBM) | Immunoaffinity-based microfluidic device | 75 |
| Non-small cell lung cancer (NSCL) | Liquid nanoprobe system (LNP) | 72 |
| Name of technique | Detectable size | Advantages | Disadvantages | References |
|---|---|---|---|---|
| ELISA | Exosome surface proteins are detected by antibodies | Sensitive and highly specific for exosomal protein | Monoclonal antibodies are required | 36 |
| Quick and convenient | Possibility of false positive/negative results | |||
| Required reagents are cheap | Nonspecific binding & cross reactivity | |||
| Equipment are widely available & relatively inexpensive | Time-consuming | |||
| Flow cytometry | Size range ≥ 150 nm | Non-specialized laboratory instruments. fluorescently labeled extracellular vesicles | Low detection threshold limit (≥150 nm) | 37 |
| Nanoparticle tracking | Size range: 10 nm to 2 μm | Fluorescent-NTAs are easily available | Vesicle aggregates are difficult to determine | 88 |
| Analysis (NTA) | Absolute particle concentration | |||
| Measures the exact size, shows high resolution | ||||
| qNano (TRPS) | Size range < 40 nm to 10 μm | Provide absolute EV concentration & size distribution | Different pore sizes are required for different biological samples | 36 |
| Can measure the charge on the particle surface | It can detect particles of exosomal size | |||
| Electrochemical detection | Size ranges > 4 nm to 10 μm | Highly sensitive, specific and required low cost of detection | Complicated fabricated steps are involved | 103 |
| Required a small volume of sample | Not easy to handle | |||
| DLS | 3 nm to 7 μm | This technique is relatively fast, convenient and mostly used for the study of a colloidal system | Unable to determine the particle-mixture in large difference in the ratio b/w the diameter of particles | 33 |
| Raman spectroscopy | Vibrating molecules of the sample interact with photons | Raman spectroscopy (SERS) can distinguish ELVs derived from different origins, the detection of individual vesicles from a colloidal suspension and is used for clinical diagnostics | Raman spectroscopy is considered to be a very inefficient process because it requires a high sample concentration, high laser power and long signal integration time | 35 |
3.1. ELISA
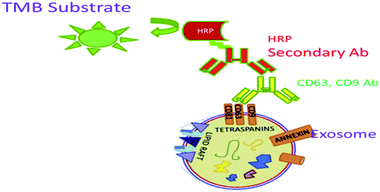
ELISA is an in vitro technique widely used for the detection and quantification of small EVs. Here, exosomes are directly immobilized on micro-well plates and the plates are blocked with blocking agents. An antibody (e.g., anti-CD9) is added to the wells, which recognizes and binds to certain antigens (e.g., CD9) that are available on the external surface of the exosomes. Subsequently, an HRP-linked detection antibody is applied via an enzymatic signal amplification process and specific read out.87 TMB (3,3′,5,5′-tetramethylbenzidine), a colorimetric substrate, is used for the assay read out (Fig. 7).88,89 Logozzi et al. (2009) designed a sandwich ELISA to detect and quantify the exosomes in cell culture media, as well as in plasma samples with tidying proteins CD63, Rab-5b and caveolin-1 (tumor-associated marker).49An ExoView instrument from Nanoview uses a Tetraspanin Kit functionalized with antibodies against CD9, CD63, CD81, CD41a plus IgG negative control. It uses single particle interferometric reflectance imaging sensing (SP-IRIS) to measure the size of the single EVs with high precision by analyzing the enhanced interferometric signal of the bound EVs when attached to a layered substrate. By using this methodology, EVs as small as 50 nm can be imaged and measured. It has a strong peak-to-peak resolving capability and can more accurately resolve distinct vesicles in a heterogeneous sample.96 It is easily able to quantify the exosomes present in cell culture supernatants. The presence of the CD63 protein on the exosome surface makes the detection promising in a dose-dependent manner. However, exosome detection and quantification by Exotest displayed a higher sensitivity for the detection of the CD63 protein with respect to western blotting (WB) analysis. Therefore, exosomes can be monitored more specifically by using commercially available pre-coated plates for exosome capture and their quantification by using ELISA. This assay typically employs a primary antibody like (CD9 or CD63) diluted in a sample buffer (0.5% 273 BSA PBS) for 2 h at 4 °C. After extensive washing, the HRP-conjugated secondary antibody is immobilized on the plate, which recognizes the conjugate to the primary ones. Upon addition of the BM Blue POD substrate, the optical densities are recorded at 450 nm using a microplate reader. Quanta Blu Fluorogenic Peroxidase is another substrate that can be used for the specific read out in a non-stopped mode using excitation/emission maxima of 325/420.90
3.2. Flow cytometry
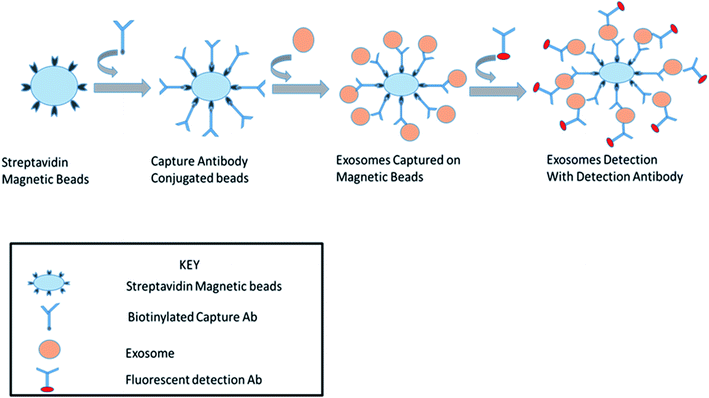
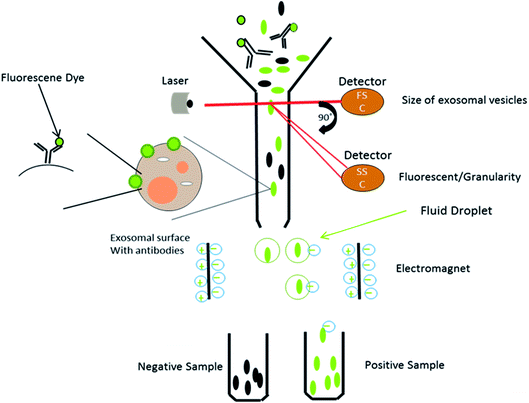
This is a well-established technology for the high-throughput examination and quantification of EVs.91 The flow cytometry approach is primarily based totally at the recording of fluorescence and light scattering through individual small extracellular vesicles, i.e., nanosized particles that are present in the suspension.49 It provides detected scattering power in arbitrary units, inferring a correlation between the detected scattering power and the diameter of the vesicles. To establish this affiliation, it measures the relative power in arbitrary units (a.u.) and in parallel, and considers the absolute power (mW) of light that is scattered by beads of known diameter and refractive index in the pathway of the detectors.58 Attributable to the small size of the exosomes, they could be immediately visualized with electron microscopy and not by light microscopy. It was shown that exosomes released from antigen-presenting cells having MHCII molecules, like HLA DP, DQ, DR and monoclonal antibodies, which recognize that the said molecules are attached to specific magnetic beads. With the help of these magnetic beads, exosomes can be isolated from the cell-free supernatant (Fig. 8). Exosomes are likely to be present on the beads and afterward stained with conjoined antibodies, and analysed by flow cytometry.92 This technique allows for the sorting of exosomal vesicles based on fluorescent labeling. By means of specific antibodies tagged with fluorescent dyes, the target exosomes can be captured and sorted.58,93 For instance, exosomes are too tiny to be detected by the current flow cytometry tools.94 Owing to these detection limitations, the Amnis Cell Stream Flow Cytometer is employed, which contains the Amnis Time Delay Integration (TDI) image capturing system. This detection technology allows the Cell Stream Instrument to combine the advantages of high throughput flow cytometry with high sensitivity to submicron particles. Meanwhile, recent advance techniques like NanoFCM provide a versatile and powerful platform for the quantitative analysis of extracellular vesicles. The Flow NanoAnalyzer platform is distinctively sensitive, and enables the quantitative and multiparameter analysis of single EVs down to 40 nm. The NanoFCM-based approach is used for the early diagnosis of colorectal cancer by using the surface protein, which will provide an advanced technique and method for the EVs research, as well as their clinical applications.953.3. Nanoparticle tracking analysis
Nanoparticle Tracking Analysis (NTA) is a technique for the real-time visualization of nanoparticles present in liquids and for their direct analysis.96 Nanoparticles show Brownian motion, and this technique relates the nanovesicle size with their motion. Using this method, small vesicles are visualized by light scattering and using a light microscope. NTA software tracked the vesicles in motion and a video was taken, and their size and total concentration was calculated.97 A laser ray interacts with the nanovesicles (exosomal particles), and a charge coupled camera is used for capturing the scattered light by the moving particles. The Stokes–Einstein equation is used by a special type of NTA software, which relates the movement to the particle size (Fig. 9).88 Gardiner et al. used the human placental exosomes and efficiency of NTA. A large amount of microvesicles and nanovesicles (exosomes) is released by the placenta in maternal blood during pregnancy. The maternal surface of the placenta was perfused with a buffer, and vesicles were prepared by ultracentrifugation for further analysis. Biological vesicles at 50 nm in size can be detected by this technique. Before NTA analysis, placental vesicles stored in PBS are thawed back to room temperature (18–25 °C).93 An important aspect of NTA is that it can detect a larger amount of cellular vesicles, as compared to several other approaches like electron microscopy. NTA has shown good results in many research programs, but in clinical aspects, NTA has many limitations as well. NTA analysis requires a lengthy procedure for data acquisition. The analysis time was also lengthy, as it took 10 minutes to analyze 1000 nanovesicles, whereas flow cytometry requires less than a second to analyze the same number.88 However, the extracellular vesicles can be analyzed for the size, concentration and some other parameters via ZetaView fluorescence mode. The particles to be examined ought to be adequately well purified. Furthermore, these vesicles can be specified with membrane dyes, primary and secondary fluorescently labeled antibodies, targeting their surface markers. By using a TWIN-laser machine, even phenotyping experiments may be done on extracellular vesicles with a double-stained pattern. For experimentation, two different fluorescent dyes coupled to the different antibodies are used to bind specifically to tetraspanins at the membrane of vesicles (e.g., CD63-Alexa405 & CD81-Alexa488).98 In NTA, bleaching problems of the exosomal fluorescent dyes (green fluorescent proteins) were also noticed due to the long analysis time period.88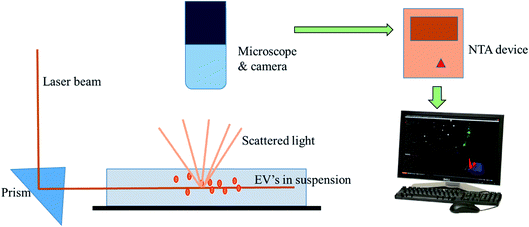 | ||
| Fig. 9 Nanoparticle tracking analysis. It depicts the ability of NTA to track the Brownian motion of the exosomes in the suspension. | ||
3.4. Tunable resistive pulse sensing
Tunable resistive pulse sensing (TRPS) is a technique developed for sensing the nanoparticles to establish the size of nanovesicles.104 This allows the measurement of a single particle from a bimolecular analytes or colloids, which are driven over pores one at a time.99 A polyurethane membrane was used, on which tunable pores were synthesized, and this membrane was also flexible in nature and caused optimization of the pore size. The fabrication technique was used for determining the shape and size of the pore. There was a change in the ionic current transiently inside the pore when a single nanoparticle passed through the pore.88 qNano proved to be a consistent tool for the detection of extracellular vesicles and quantification, providing an accurate analysis of the vesicles having a size from 40 nm to 10 μm. During the traversing of a particle through the pore, a transient current pulse is generated, which is directly proportional to the particle volume, which allows for the most accurate and repetitive measurement of the size. Particles having the size in the sub nm range can be resolved efficiently by using the TRPS under the ideal conditions. It gives an aligned estimation of the true size and concentration distribution of nanoparticles. It provides an extra layer of essential information by providing the concentration of particles within each size band. It concurrently measures both particle size and zeta potential reproducibly on a particle-by-particle basis with excessive precision and accuracy, presenting an amazing and powerful technique for the life sciences. The above technique can be applied for various applications, like investigating and understanding nano-bio interactions, establishing physico-chemical equivalence, and the discrimination of sub-populations within a sample.100 Robert Vogel et al. (2016) has standardized a method to determine the concentration of EVs using TRPS. The method is based on determining the EV concentration as a function of a well-defined size range. For this method, blood plasma EVs are isolated and purified using size exclusion columns (qEV) and sequentially measured with tunable resistive pulse sensing (TRPS). The measured mean liposome (which is not filtered with qEV) and EV (i.e., filtered with qEV) concentrations had coefficients of variance (CV) of 23.9% and 52.5%, respectively. The increased variance of the EV concentration measurements could be ascribed to the usage of qEVs and the poly-disperse nature of the EVs. The results of this study determine the feasibility of this standardized methodology to enable analogous and reproducible EV concentration measurements.1013.5. Electrochemical detection
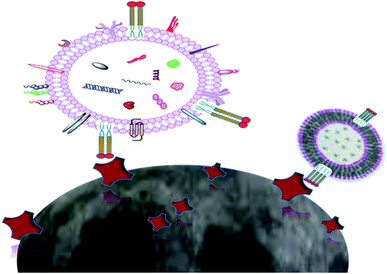
The electrochemistry-based detection of EVs is considered to be a highly favorable tool for the detection and analysis of nanovesicles and is highly sensitive, specific and required low cost for detection.88 A glucometer is an example of an electrochemical readout sensor recently developed, which is used to detect the glucose level in the body, and the detection of other analytes may be approached by this technique.102 However, the exosome analysis by this technique is quite challenging. Here, an antibody is used, which recognize the antigen present on the exosomal surface, and a measurable electro-active signal is obtained by using the signal transducer, which detects and quantifies the exosome. An integrated magneto-electrochemical sensor (iMEX) is an electrochemical system that was recently developed for exosome analysis. Magnetic selection and electrochemical detection are two main processes that are utilized during the iMEX sensor analysis. Exosomes are captured using magnetic beads coated with anti-CD9, CD63, and CD81, which recognize and bind to tetraspanin proteins (CD9, CD63, CD81) acting like antigen proteins on the exosomal surface, and detection was done by using electrochemical sensing. The iMEX assay used only 10 μl of sample, and results are obtained within 1 hour.103 iMEX is a potential electrochemical method used to detect the EV and exosomes from ovarian cancer cells, showing fast and on-spot analysis, and is accelerating toward routine clinical testing.99 The electrochemical sandwich immune sensor is an alternative method for the detection of exosomes, and relies on the immobilization of the gold electrode with tetraspanin (CD63) and magnetic beads attached with α-CD63 antibodies conjugated with HRP, producing electrochemical signals (Fig. 10).57 α-CD9 antibodies can also be pre-functionalized to the gold electrode, after which the sample is spread and analyzed from different organisms. Finally, the HRP-conjugated α mouse IgG antibody is applied. HRP oxidises the TMB, and electrochemical signals are monitored. Another important aspect of the sandwich electrochemical immunoassay is that it discriminates between microvesicles (MV) and exosomes, and can detect up to 200 particles per microliter.88 Recently, a new electrochemical method has been designed to quantify the exosomes using quantum dots (QDs) functionalized with a specific type of antibody, showing a high sensitivity detection limit of 100 exosomes per microliter. The QD method shows several hundred times more sensitivity as compared with other bioaffinity-based assays. As compared to conventional techniques of exosome analysis, the electrochemical technique is rapid, cost effective and requires a small volume of sample. However, some complicated fabricated steps are involved during the electrochemical platform setup, which makes it a little typical. The electrochemical technique together with a microfluidic platform can be used in conjugation, and can turn out to be an efficient tool for disease diagnosis applications by using exosomal biomarkers.88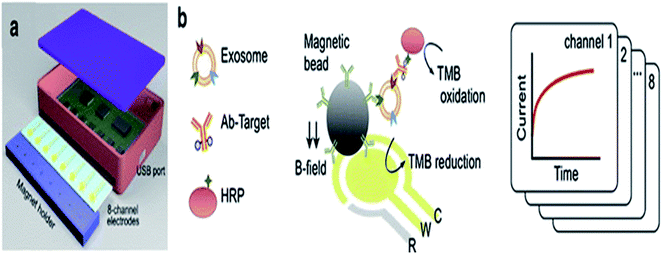 | ||
| Fig. 10 (a) iMEX platform (b) Biosensor. Exosomes were harvested with antibody-conjugated magnetic beads by a magnet on the electrodes. Exosomes were detected by binding the HRP-labeled antibodies with an exosomal surface protein, i.e., (CD9). HRP oxidized the TMB and electrical signal.103 | ||
3.6. Dynamic light scattering
Dynamic Light Scattering (DLS) is frequently referred to as photon correlation spectroscopy. This technique is commonly utilized for determining the particle size in a colloidal suspension. Suspended particles in a solvent undergo Brownian motion. The particles present in the liquid caused a localized change in the refractive index. The autocorrelation functional equation can be applied for the measurement of intensity variation produced by the particles.105The size distribution of EVs is measured by DLS geared up with a strong state He–Ne laser at 633 nm wavelength. The intensity of the scattered light is measured at 173 °C. All measurements were undertaken in triplicates at 25 °C. Data processing and analysis are performed using the Zeta sizer. This technique is primarily based on the dimensions of Brownian motion concerning the hydrodynamic diameter via the Stokes–Einstein equation. However, DLS makes use of Rayleigh scattering for the estimation of the particle diameter.106
3.7. Raman spectroscopy
Raman spectroscopy is a technique recently applied for the detection of EVs to analyze the vibrational modes of the nanovesicles by measuring the non-elastic scattering effect of a monochromatic laser light.107 The scattered photons causing the Raman spectrum contain the information of the molecular composition of the sample. However, the Raman spectroscopy technique is considered very inefficient because of the high sample concentration requirement in combination with the high laser power and the long time required for signal integration.108 This technique is therefore not feasible and is a rare event. The obtained signal intensity is very low and hard to distinguish. To overcome these problems, the Raman spectroscopy signal can be fortunately enhanced by using surface-enhanced Raman spectroscopy (SERS), which enhanced the electrical signal by applying gold nanoparticles as a surface treatment to amplify the Raman signal. SERS analysis has been performed for numerous studies, ranging from molecules to cell lines.3.8. Plasmonic exosome detection (nPLEX)
Hyungsoon Im et al. (2014) designed the nPLEX sensor to attain the label-free detection of small EVs, where the basic sensing unit consists of a periodic matrix of nanoholes patterned in a gold (Au) film. The structure of nPLEX, a periodic array of sub-wavelength gaps in a metal film, produces exceptional surface plasmons whose termination depth is equivalent to the exosome size, making this technique innovative and well-suited for a sensitive, label-free exosome detection. For clinical applications, with a further turn of events and approval, nPLEX could be valuable for investigating exosomes as a malignant growth biomarker, for diagnostics and for assessing tumor reaction to therapy. A quick, multiplexed protein investigation of exosomes could enhance the early ailment detection and treatment monitoring. A typical nPLEX array has 12 channels, and every channel is functionalized with unique antibodies for EpCAM, CD24, CD63 and IgG control. The entire 12 × 3 array is imaged using the portable imager system (Fig. 11). The extracted exosomes can be easily removed from the tool for downstream examinations, for example, genomic profiling. Together, these methodologies will facilitate extensive exosomal examinations by yielding both proteomic and genetic data.47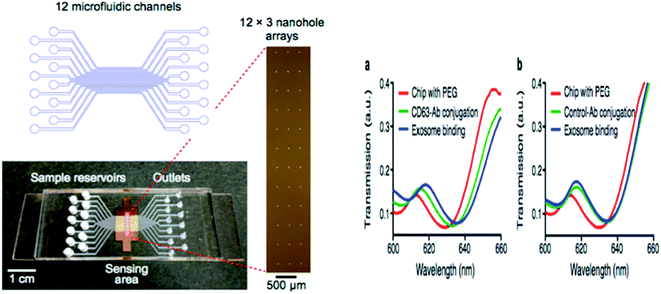 | ||
| Fig. 11 Device configuration. A 12-channel fluidic cell (top) was placed on top of a glass slide containing nanohole arrays (bottom). A total of 36 measurement sites were arranged into a 12 × 3 array format (right) with each fluidic channel encompassing three measurement sites (left side). The nPLEX chips were conjugated with either CD63 (a) or IgG control (b) antibodies. Exosomes from human ovarian cancer cells (CaOV3) were subsequently introduced. Transmission spectral shifts associated with the antibody conjugation and specific exosome binding were measured. Similar spectral shifts were observed for both CD63 and the control antibody conjugation, which indicated a similar extent of antibody grafting onto the sensor surface. Exosome binding, however, was only observed with the CD63-specific chip (a); the control chip (b) displayed negligible binding (right side).47 | ||
4. Conclusion and future perspective
We have precisely summed up the recent advances in the isolation and characterization techniques for extracellular vesicles. Despite progressive advances, it is apparent that none of the current techniques for exosome isolation is a versatile model. The major technical challenge in EVs detection in clinical applications is to exactly detect disease-specific EVs from the pool of EVs that are derived from normal cells. We admit that by conjoining the outstanding components of different exosome quantification techniques and by inventive assay design in a multiplexed system that is adept at the selective isolation of various EVs subtypes in heterogeneous samples, it will be possible to open a new path in the detection of extracellular vesicles and related research. Obviously, researchers have constantly struggled to develop such techniques. The usage of micro- and nano-technologies designed to overcome these technical challenges are ongoing. By aiding the quick sample preparation and molecular evaluation from small sample volumes, these platforms can facilitate and entirely connect the clinical potential of these circulating biomarkers in cancers or different pathological conditions and beyond. We believe that, in the near future, the ongoing endeavor toward the improvement of high-performance exosome detection techniques will result in an idyllic next-generation platform that can be regularly used for EVs examination for both research and clinical purposes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
R. B. acknowledges the UGC, Govt. of India for financial support in the form of MANF.References
- J. R. Sotelo and K. R. Porter, An electron microscope study of the rat ovum, J. Biophys. Biochem. Cytol., 1959, 5, 327, DOI:10.1083/jcb.5.2.327.
- A. B. Novikoff, E. Essner and N. Quintana, Golgi apparatus and lysosomes, Fed. Proc., 1964, 23, 1010–1022 CAS.
- S. J. Gould and G. Raposo, As we wait: coping with an imperfect nomenclature for extracellular vesicles, J. Extracell. Vesicles, 2013, 220389, DOI:10.3402/jev.v2i0.20389.
- G. Van Niel, I. Porto-Carreiro, S. Simoes and G. Raposo, Exosomes: a common pathway for a specialized function, J. Biochem., 2006, 140, 13–21 CrossRef CAS.
- G. Raposo and W. Stoorvogel, Extracellular vesicles: exosomes, microvesicles, and friends, J. Cell Biol., 2013, 200(4), 373–383 CrossRef CAS.
- J. Skog, T. Würdinger, S. van Rijn, D. H. Meijer, L. Gainche, M. Sena-Esteves, W. T. Curry Jr, B. S. Carter, A. M. Krichevsky and X. O. Breakefield, Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers, Nat. Cell Biol., 2008, 10(12), 1470–1476 CrossRef CAS PubMed.
- K. C. Miranda, D. T. Bond, M. McKee, J. Skog, T. G. Păunescu, N. Da Silva, D. Brown and L. M. Russo, Nucleic acids with in urinary exosomes/microvesicles are potential biomarkers for renal disease, Kidney Int., 2010, 78(2), 191–199 CrossRef PubMed.
- M. Tkech and C. Thery, Communication by Extracellular Vesicles: Where We Are and Where We Need to Go, Cell, 2016, 164, 1226–1232 CrossRef PubMed.
- B. T. Pan and R. M. Johnstone, Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor, Cell, 1983, 33, 967 CrossRef CAS.
- B. H. Sung, T. Ketova, D. Hoshino, A. Zijlstra and A. M. Weaver, Directional cell movement through tissues is controlled by exosome secretion, Nat. Commun., 2015, 6, 7164 CrossRef CAS PubMed.
- J. W. Clancy, A. Sedgwick, C. Rosse, V. Muralidharan-Chari, G. Raposo, M. Method, P. Chavrier and C. D'Souza-Schorey, Regulated delivery of molecular cargo to invasive tumour-derived microvesicles, Nat. Commun., 2015, 6, 6919 CrossRef CAS PubMed.
- C. Théry, A. Regnault, J. Garin, J. Wolfers, L. Zitvogel, P. Ricciardi-Castagnoli, G. Raposo and S. Amigorena, Molecular characterization of dendritic cell-derived exosomes: selective accumulation of the heat shock protein hsc73, J. Cell Biol., 1999, 147, 599, DOI:10.1083/jcb.147.3.599.
- C. Théry, M. Boussac, P. Véron, P. Ricciardi-Castagnoli, G. Raposo, J. Garin and S. Amigorena, Proteomic analysis of dendritic cell-derived exosomes: a secreted subcellular compartment distinct from apoptotic vesicles, J. Immunol., 2001, 166, 7309, DOI:10.4049/jimmunol.166.12.7309.
- L. Zitvogel, A. Regnault, A. Lozier, J. Wolfers, C. Flament, D. Tenza, P. Ricciardi-Castagnoli, G. Raposo and S. Amigorena, Eradication of established murine tumors using a novel cell-free vaccine: dendritic cell derived exosomes, Nat. Med., 1998, 4, 594–600 CrossRef CAS PubMed.
- G. Raposo, H. W. Nijman, W. Stoorvogel, R. Liejendekker, C. V. Harding, C. J. Melief and H. J. Geuze, B lymphocytes secrete antigen-presenting vesicles, J. Exp. Med., 1996, 183, 1161, DOI:10.1084/jem.183.3.1161.
- P. J. Peters, H. J. Geuze, H. A. vanderDonk and J. Borst, A new model for lethal hit delivery by cytotoxic T lymphocytes, J. Immunol., 1990, 11, 28, DOI:10.1016/0167-5699(90)90008-W.
- P. J. Peters, J. Borst, V. Oorschot, M. Fukuda, O. Krähenbühl, J. Tschopp, J. W. Slot and H. J. Geuze, Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes, J. Exp. Med., 1991, 173, 1099, DOI:10.1084/jem.173.5.1099.
- N. Blanchard, D. Lankar, F. Faure, A. Regnault, C. Dumont, G. Raposo and C. Hivroz, TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex, J. Immunol., 2002, 168, 3235, DOI:10.4049/jimmunol.168.7.3235.
- G. Raposo, D. Tenza, S. Mecheri, R. Peronet, C. Bonnerot and C. Desaymard, Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation, Mol. Biol. Cell, 1997, 8, 2631, DOI:10.1091/mbc.8.12.2631.
- D. Skokos, S. Le Panse, I. Villa, J. C. Rousselle, R. Peronet, B. David, A. Namane and S. Mécheri, Mast cell-dependent B and T lymphocyte activation is mediated by the secretion of immunologically active exosomes, J. Immunol., 2001, 166, 868, DOI:10.4049/jimmunol.166.2.868.
- G. Van Niel, G. Raposo, C. Candalh, M. Boussac, R. Hershberg, N. Cerf-Bensussan and M. Heyman, Intestinal epithelial cells secrete exosome-like vesicles, Gastroenterology, 2001, 121, 337, DOI:10.1053/gast.2001.26263.
- B. Fevrier, D. Vilette, F. Archer, D. Loew, W. Faigle, M. Vidal, H. Laude and G. Raposo, Cells release prions in association with exosomes, Proc. Natl. Acad. Sci. U. S. A., 2004, 21, 21, DOI:10.1073/pnas.0308413101.
- J. Wolfers, A. Lozier, G. Raposo, A. Regnault, C. Théry, C. Masurier, C. Flament, S. Pouzieux, F. Faure, T. Tursz, E. Angevin, S. Amigorena and L. Zitvogel, Tumor-derived exosomes are a source of shared tumor rejection antigens for CTL cross-priming, Nat. Med., 2001, 7, 297–303 CrossRef CAS PubMed.
- B. Riteau, F. Faure, C. Menier, S. Viel, E. D. Carosella, S. Amigorena and N. Rouas-Freiss, Exosomes bearing HLA-G are released by melanoma cells, Hum. Immunol., 2003, 64, 1064, DOI:10.1016/j.humimm.2003.08.344.
- D. G. Halme and D. A. Kessler, FDA regulation of stem-cell-based therapies, N. Engl. J. Med., 2006, 355, 1730–1735 CrossRef CAS PubMed.
- D. W. G. Harron, Technical requirements for registration of pharmaceuticals for human use: the ICH process, in The Textbook of Pharmaceutical Medicine, ed. J. P. Griffin, Blackwell Publishing Ltd, Oxford, New Jersey, USA, 2007, pp. 552–564 Search PubMed.
- J. Ancans, Cell therapy medicinal product regulatory framework in Europe and its application for MSC-based therapy development, Front. Immunol., 2012, 3, 253, DOI:10.3389/fimmu.2012.00253.
- T. Lener, M. Gimona, L. Aigner, V. Börger, E. Buzas and G. Camussi, et al., Applying extracellular vesicles based therapeutics in clinical trials – an ISEV position paper, J. Extracell. Vesicles, 2015, 4, 30087, DOI:10.3402/jev.v4.30087.
- K. Mizutani, R. Terazawa, K. Kameyama, T. Kato, K. Horie, T. Tsuchiya, K. Seike, H. Ehara, Y. Fujita, K. Kawakami, M. Ito and T. Deguchi, Isolation of Prostate Cancer-related Exosomes, Anticancer Res., 2014, 34, 3419–3424 CAS.
- C. Sotiriou and P. Lajos, Gene-expression signatures in breast cancer, N. Engl. J. Med., 2009, 360(8), 790–800 CrossRef CAS PubMed.
- A. M. Friel, C. Corcoran, J. Crown and L. O'Driscoll, Relevance of circulating tumor cells, extracellular nucleic acids, and exosomes in breast cancer, Breast Cancer Res. Treat., 2010, 123(3), 613–625 CrossRef CAS.
- D. D. Taylor and C. Gercel-Taylor, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecoloncol, 2008, 110(1), 13–21 CrossRef CAS PubMed.
- J. Lötvall, A. F. Hill, F. Hochberg, E. I. Buzás, D. Di Vizio, C. Gardiner, Y. S. Gho, I. V. Kurochkin, S. Mathivanan, P. Quesenberry, S. Sahoo, H. Tahara, M. H. Wauben, K. W. Witwer and C. Théry, Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles, J. Extracell. Vesicles, 2014, 3, 26913, DOI:10.3402/jev.v3.26913.
- R. Szatanek, J. Baran, M. Siedlar and M. Baj-Krzyworzeka, Isolation of extracellular vesicles: determining the correct approach, Int. J. Mol. Med., 2015, 36(1), 11–17 CrossRef CAS PubMed.
- A. Clayton, J. Court, H. Navabi, M. Adams, M. D. Mason, J. A. Hobot and B. Jasani, Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry, J. Immunol., 2001, 247(1–2), 163–174 CAS.
- A. Sabapatha, C. Gercel-Taylor and D. D. Taylor, Specific isolation of placenta-derived exosomes from circulation of pregnant women and their immune regulatory consequences, Am. J. Reprod. Immunol., 2006, 56, 345–355 CrossRef CAS PubMed.
- M. L. Merchant, D. W. Powell, D. W. Wilkey, T. D. Cummins, J. K. Deegens, I. M. Rood and J. B. Klein, Microfiltration isolation of human urinary exosomes for characterization by MS, Proteomics: Clin. Appl., 2010, 4(1), 84–96 CAS.
- P. A. Gonzales, H. Zhou, T. Pisitkun, N. S. Wang, R. A. Star, M. A. Knepper and P. S. Yuen, Isolation and purification of exosomes in urine, Methods Mol. Biol., 2010, 89–99 CrossRef CAS.
- C. E. Yoo, G. Kim, M. Kim, D. Park, H. J. Kang, M. Lee and N. A. Huh, Direct extraction method for microRNAs from exosomes captured by immunoaffinity beads, Anal. Biochem., 2012, 431(2), 96–98 CrossRef CAS PubMed.
- C. Théry, S. Amigorena, G. Raposo and A. Clayton, Isolation and characterization of exosomes from cell culture supernatants and biological fluids, Curr. Protoc. Cell Biol., 2006, 3, 22–30 Search PubMed.
- A. Ghosh, M. Davey, I. C. Chute, S. G. Griffiths, S. Lewis, S. Chacko, D. Barnett, N. Crapoulet, S. Fournier, A. Joy, C. Michelle, M. C. Caissie, A. D. Ferguson, M. Daigle, M. V. Meli, S. M. Lewis and R. J. Ouellette, Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for Heat Shock Proteins, PLoS One, 2014, 9, e110443, DOI:10.1371/journal.pone.0110443.
- E. Van der Pol, A. N. Böing, E. L. Gool and R. Nieuwland, Recent developments in the nomenclature, presence, isolation, detection and clinical impact of extracellular vesicles, J. Thromb. Haemostasis, 2016, 14(1), 48–56 CrossRef CAS PubMed.
- I. Lozano-Ramos, I. Bancu, A. Oliveira-Tercero, M. P. Armengol, A. Menezes-Neto, H. A. DelPortillo, R. Lauzurica-Valdemoros and F. E. Borràs, Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples, J. Extracell. Vesicles, 2015, 4(1), 27369, DOI:10.3402/jev.v4.27369.
- C. Chen, J. Skog, C. H. Hsu, R. T. Lessard, L. Balaj, T. Wurdinger and D. Irimia, Microfluidic isolation and transcriptome analysis of serum microvesicles, Lab Chip, 2010, 10(4), 505–511 RSC.
- R. Cantin, J. Diou, D. Bélanger, A. M. Tremblay and C. Gilbert, Discrimination between exosomes and HIV-1: purification of both vesicles from cell-free supernatants, J. Immunol. Methods, 2008, 338(1–2), 21–30 CrossRef CAS PubMed.
- S. S. Kanwar, C. J. Dunlay, D. M. Simeone and S. Nagrath, Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes, Lab Chip, 2014, 14(11), 1891–1900 RSC.
- H. Im, H. Shao and Y. Park, et al., Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor, Nat. Biotechnol., 2014, 32, 490–495 CrossRef CAS PubMed.
- L. L. Lv, Y. Cao, D. Liu, M. Xu, H. Liu, R. N. Tang, K. L. Ma and B. C. Liu, Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery, Int. J. Biol. Sci., 2013, 9(10), 1021, DOI:10.7150/ijbs.6100.
- M. Logozzi, A. De Milito, L. Lugini, M. Borghi, L. Calabrò, M. Spada, M. Perdicchio, M. L. Marino, C. Federici, E. Iessi, D. Brambilla, G. Venturi, F. Lozupone, M. Santinami, V. Huber, M. Maio, L. Rivoltini and S. Fais, High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients, PLoS One, 2009, 4(4), e5219, DOI:10.1371/journal.pone.0005219.
- R. A. Dragovic, C. Gardiner, A. S. Brooks, D. S. Tannetta, D. J. Ferguson, P. Hole, B. Carr, C. W. Redman, A. L. Harris, P. J. Dobson, P. Harrison and I. L. Sargent, Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis, J. Nanomed. Nanotechnol., 2011, 7(6), 780–788 CrossRef CAS PubMed.
- K. T. Rim and S. J. Kim, Quantitative analysis of exosomes from murine lung cancer cells by flow cytometry, Eur. J. Cancer Prev., 2016, 21(3), 194, DOI:10.15430/JCP.2016.21.3.194.
- M. He, J. Crow, M. Roth, Y. Zeng and A. K. Godwin, Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology, Lab Chip, 2014, 14(19), 3773–3780 RSC.
- S. Jeong, J. Park, D. Pathania, C. M. Castro, R. Weissleder and H. Lee, Integrated magneto–electrochemical sensor for exosome analysis, ACS Nano, 2016, 10(2), 1802–1809 CrossRef CAS PubMed.
- Y. Sharda, K. Boriachek, M. N. Islam, R. Lobb, A. Möller, M. M. Hill, M. S. A. Hossain, N. T. Nguyen and M. J. A. Shiddiky, An Electrochemical Method for the Detection of Disease Specific Exosomes, ChemElectroChem, 2017, 4, 967–971 CrossRef.
- Y. G. Zhou, R. M. Mohamadi, M. Poudineh, L. Kermanshah, S. Ahmed, T. S. Safaei, J. Stojcic, R. K. Nam, E. H. Sargent and S. O. Kelley, Interrogating circulating microsomes and exosomes using metal nanoparticles, Small, 2016, 121(6), 727–732 CrossRef PubMed.
- F. Wei, J. Yang and D. T. W. Wong, Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM), Biosens. Bioelectron., 2013, 44, 115–121 CrossRef CAS PubMed.
- X. Doldán, P. Fagúndez, A. Cayota, J. Laíz and J. P. Tosar, Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification, Anal. Chem., 2016, 88(21), 10466–10473 CrossRef PubMed.
- P. E. Van der, A. N. Böing, P. Harrison, A. Sturk and R. Nieuwland, Classification, functions, and clinical relevance of extracellular vesicles, Pharmacol. Rev., 2012, 64, 676–705 CrossRef PubMed.
- R. J. Berckmans, A. Sturk, L. M. van Tienen, M. C. Schaap and R. Nieuwland, Cell-derived vesicles exposing coagulant tissue factor in saliva, Blood, 2011, 117, 3172–3180 CrossRef CAS PubMed.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes, Methods, 2012, 56, 293–304 CrossRef CAS PubMed.
- K. T. Rim and J. K. Soo, Quantitative analysis of exosomes from murine lung cancer cells by flow cytometry, J. Cancer Prev., 2016, 21, 194, DOI:10.15430/JCP.2016.21.3.194.
- D. D. Taylor and C. Gercel-Taylor, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecol. Oncol., 2008, 110, 13–21 CrossRef CAS.
- K. Koga, K. Matsumoto, T. Akiyoshi, M. Kubo, N. Yamanaka, A. Tasaki, H. Nakashima, M. Nakamura, S. Kuroki, M. Tanaka and M. Katano, Purification, characterization and biological significance of tumor-derived exosomes, Anticancer Res., 2005, 25, 3703–3707 CAS.
- M. A. J. van Eijndhoven, J. M. Zijlstra, N. J. Groenewegen, E. E. Drees, S. van Niele, S. R. Baglio, D. Koppers-Lalic, H. van der Voorn, S. F. Libregts, M. H. Wauben and R. X. de Menezes, Plasma vesicle miRNAs for therapy response monitoring in Hodgkin lymphoma patients, J. Clin. Invest., 2016, 1(19), e89631 Search PubMed.
- S. Priyanka, S. Ludwig, L. Muller, C. S. Hong, J. Kirkwood, S. Ferrone and T. L. Whiteside, Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma, J. Extracell. Vesicles, 2018, 7, 1435138 CrossRef PubMed.
- I. V. Bijnsdorp, O. Maxouri, A. Kardar, T. Schelfhorst, S. R. Piersma, T. V. Pham, A. Vis, R. J. Van Moorselaar and C. R. Jimenez, Feasibility of urinary extracellular vesicle proteome profiling using a robust and simple, clinically applicable isolation method, J. Extracell. Vesicles, 2017, 6, 1313091, DOI:10.1080/20013078.2017.1313091.
- J. Nilsson, J. Skog, A. Nordstrand, V. Baranov, L. Mincheva-Nilsson, X. O. Breakefield and A. Widmark, Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer, Br. J. Cancer, 2009, 100, 1603–1607 CrossRef CAS.
- S. Taverna, M. Giallombardo, I. Gil-Bazo, A. P. Carreca, M. Castiglia, J. Chacártegui, A. Araujo, R. Alessandro, P. Pauwels, M. Peeters and C. Rolfo, Exosomes isolation and characterization in serum is feasible in non-small cell lung cancer patients: critical analysis of evidence and potential role in clinical practice, Oncotarget, 2016, 7, 28748–28760 CrossRef PubMed.
- J. Conde-Vancells, E. Rodriguez-Suarez, N. Embade, D. Gil, R. Matthiesen, M. Valle, F. Elortza, S. C. Lu, J. M. Mato and J. M. Falcon-Perez, Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes, J. Proteome Res., 2008, 7, 5157–5166 CrossRef CAS PubMed.
- K. R. Jakobsen, B. S. Paulsen, R. Baek, K. Varming, B. S. Sorensen and M. M. Jørgensen, Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma, J. Extracell. Vesicles, 2015, 26659 CrossRef PubMed.
- M. P. Oksvold, K. W. Pedersen, B. Kierulf, I. Manger, M. Li, A. Vlassov, N. Roos, A. Kullmann and A. Neurauter, Direct Isolation of Exosomes from Cell Culture: Simplifying Methods for Exosome Enrichment and Analysis, Arch. Med., 2015, 6, 18 Search PubMed.
- M. P. Oksvold, A. Neurauter and K. W. Pedersen, Magnetic bead-based isolation of exosomes, RNA Interference, Humana Press, 2015, pp. 465–481 Search PubMed.
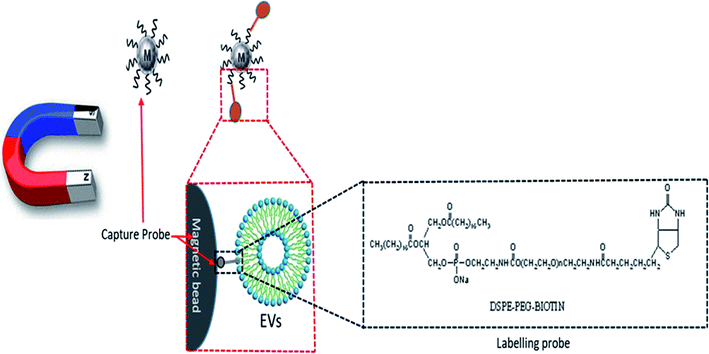
- Y. Wan, G. Cheng, X. Liu, S. J. Hao, M. Nisic, C. D. Zhu, Y. Q. Xia, W. Q. Li, Z. G. Wang, W. L. Zhang, S. J. Rice, A. Sebastian, I. Albert, C. P. Belani and S. Y. Zheng, Rapid magnetic isolation of extracellular vesicles via lipid-based nanoprobes, Nat. Biomed. Eng., 2017, 1, 1–11 CrossRef.
- E. K. Sackmann, A. L. Fulton and D. J. Beebe, The present and future role of microfluidics in biomedical research, Nature, 2014, 507, 181–189 CrossRef CAS PubMed.
- M. E. Piyasena and S. W. Graves, The intersection of flow cytometry with microfluidics and microfabrication, Lab Chip, 2014, 14, 1044–1059 RSC.
- Z. Wang, H. J. Wu, D. Fine, J. Schmulen, Y. Hu, B. Godin and X. Liu, Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles, Lab Chip, 2013, 13, 2879–2882 RSC.
- R. T. Davies, J. Kim, S. C. Jang, E. J. Choi, Y. S. Gho and J. Park, Microfluidic filtration system to isolate extracellular vesicles from blood, Lab Chip, 2012, 12, 5202–5210 RSC.
- J. Karttunen, M. Heiskanen, V. N. Ferrandis, S. D. Gupta, A. Lipponen, N. Puhakka, K. Rilla, A. Koistinen and A. Pitkänen, Precipitation-based extracellular vesicle isolation from rat plasma co-precipitate vesicle-free microRNAs, J. Extracell. Vesicles, 2018, 8, 1555410, DOI:10.1080/20013078.2018.1555410.
- C. Gardiner, D. D. Vizio, S. Sahoo, C. Théry, K. W. Witwer, M. Wauben and A. F. Hill, Techniques used for the isolation and characterization of extracellular vesicles: results of a worldwide survey, J. Extracell. Vesicles, 2016, 5, 32945, DOI:10.3402/jev.v5.32945.
- N. Karimi, A. Cvjetkovic, S. C. Jang, R. Crescitelli, M. A. H. Feizi, R. Nieuwland, J. Lötvall and C. Lässer, Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins, Cell. Mol. Life Sci., 2018, 75, 2873–2886 CrossRef CAS.
- M. C. Deregibus, F. Figliolini, S. Antico, P. M. Manzini, C. Pasquino, M. De Lena and G. Camussi, Charge-based precipitation of extracellular vesicles, Int. J. Mol. Med., 2016, 38, 1359–1366 CrossRef CAS PubMed.
- A. N. Böing, E. Van Der Pol, A. E. Grootemaat, F. A. Coumans, A. Sturk and R. Nieuwland, Single-step isolation of extracellular vesicles by size-exclusion chromatography, J. Extracell. Vesicles, 2014, 23430, DOI:10.3402/jev.v3.23430.
- T. Baranyai, K. Herczeg, Z. Onódi, I. Voszka, K. Módos, N. Marton and Z. Pálinkás, Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods, PLoS One, 2015, 10, e0145686, DOI:10.1371/journal.pone.0145686.
- The qEV Automatic Fraction Collector (AFC), https://izon.com/automatic-fraction-collector Search PubMed.
- B. J. Benedikter, F. G. Bouwman and T. Vajen, et al., Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies, Sci. Rep., 2017, 7, 15297, DOI:10.1038/s41598-017-15717-7.
- E. Oeyen, K. V. Mol, G. Baggerman, H. Willems, K. Boonen, C. Rolfo, P. Pauwels, A. Jacobs, K. Schildermans, W. C. Cho and I. Mertens, Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine, J. Extracell. Vesicles, 2018, 7(1), 1490143 CrossRef PubMed.
- E. Engvall and P. Perlmann, Enzyme-linked immunosorbent assay (ELISA) quantitative assay of immunoglobulin G, Immunochemistry, 1971, 8, 871–874 CrossRef CAS.
- K. Boriachek, M. N. Islam, A. Möller, C. Salomon, N. T. Nguyen, M. S. A. Hossain, Y. Yamauchi and M. J. A. Shiddiky, Biological functions and current advances in isolation and detection strategies for exosome nanovesicles, Small, 2018, 14, 1702153, DOI:10.1002/smll.201702153.
- Complete Characterization of Exosomes and Extracellular Vesicles (EVs), https://www.nanoviewbio.com/ Search PubMed.
- N. Zarovni, A. Corrado, P. Guazzi, D. Zocco, E. Lari, G. Radano, J. Muhhina, C. Fondelli, J. Gavrilova and A. Chiesi, Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immune capture approaches, Methods, 2015, 87, 46–58 CrossRef CAS PubMed.
- U. Erdbrügger and J. Lannigan, Analytical challenges of extracellular vesicle detection: a comparison of different techniques, Cytometry, Part A, 2016, 89, 123–134 CrossRef.
- H. K. Kim, K. S. Song, E. S. Lee, Y. J. Lee, Y. S. Park, K. R. Lee and S. N. Lee, Optimized flow cytometric assay for the measurement of platelet microparticles in plasma: pre-analytic and analytic considerations, Blood Coagulation Fibrinolysis, 2002, 13, 393–397 CrossRef CAS PubMed.
- M. Kesimer, M. Scull, B. Brighton, G. De Maria, K. Burns, W. O'Neal and J. K. Sheehan, Characterization of exosome-like vesicles released from human tracheobronchial ciliated epithelium: a possible role in innate defense, FASEB J., 2009, 23, 1858–1868 CrossRef CAS PubMed.
- M. H. Julius, T. Masuda and L. A. Herzenberg, Demonstration that antigen-binding cells are precursors of antibody-producing cells after purification with a fluorescence-activated cell sorter, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 1934–1938 CrossRef CAS PubMed.
- M. P. Hunter, N. Ismail, X. Zhang, B. D. Aguda, E. J. Lee, L. Yu, T. Xiao, J. Schafer, M. T. Lee, T. D. Schmittgen, S. P. Nana-Sinkam, D. Jarjoura and C. B. Marsh, Detection of microRNAexpression in human peripheral blood microvesicles, PLoS One, 2008, 3, e3694, DOI:10.1371/journal.pone.0003694.
- C. Lässer, M. Eldh and J. Lötvall, Isolation and characterization of RNA-containing exosomes, J. Visualized Exp., 2012, 59, e3037, DOI:10.3791/3037.
- S. Zhu, L. Ma, S. Wang, C. Chen, W. Zhang, L. Yang, W. Hang, J. P. Nolan, L. Wu and X. Yan, Light-scattering detection below the level of single fluorescent molecules for high-resolution characterization of functional nanoparticles, ACS Nano, 2014, 8, 10998–11006 CrossRef CAS PubMed.
- B. Carr, P. Hole, A. Malloy, P. Nelson and J. Smith, Applications of nanoparticle tracking analysis in nanoparticle research: a mini-review, Eur. J. Pharm. Sci., 2009, 14, 45 Search PubMed.
- R. Vogel, G. Willmott, D. Kozak, G. S. Roberts, W. Anderson, L. Groenewegen and M. Trau, Quantitative sizing of nano/microparticles with a tunable elastomeric pore sensor, Anal. Chem., 2011, 83, 3499–3506 CrossRef CAS PubMed.
- Measure the Zeta Potential of Individual Particles with TRPS, https://izon.com/nanoparticlemeasurement Search PubMed.
- R. Vogel, F. A. Coumans, R. G. Maltesen, A. N. Böing, K. E. Bonnington, M. L. Broekman, M. F. Broom, E. I. Buzás, G. Christiansen, N. Hajji and S. R. Kristensen, A standardized method to determine the concentration of extracellular vesicles using tunable resistive pulse sensing, J. Extracell. Vesicles, 2016, 5, 31242 CrossRef PubMed.
- K. J. Odenthal and J. J. Gooding, An introduction to electrochemical DNA biosensors, Analyst, 2007, 132, 603–610, 10.1039/B701816A.
- S. Jeong, J. Park, D. Pathania, C. M. Castro, R. Weissleder and H. Lee, Integrated magneto-electrochemical sensor for exosome analysis, ACS Nano, 2016, 10, 1802 CrossRef CAS PubMed.
- S. L. Maas, M. L. Broekman and J. de Vrij, Tunable resistive pulse sensing for the characterization of extracellular vesicles, Exosomes and Microvesicles, 2017, pp. 21–33 Search PubMed.
- C. Liu, Y. Yang and Y. Wu, Recent advances in exosomal protein detection via liquid biopsy biosensors for cancer screening, diagnosis, and prognosis, AAPS J., 2008, 20, 41, DOI:10.1208/s12248-018-0201-1.
- E. Serrano-Pertierra, M. Oliveira-Rodríguez, M. Rivas, P. Oliva, J. Villafani, A. Navarro, M. C. Blanco-López and E. Cernuda-Morollón, Characterization of plasma-derived extracellular vesicles isolated by different methods: a comparison study, Bioengineering, 2019, 6, 8, DOI:10.3390/bioengineering6010008.
- C. M. Hoo, N. Starostin, P. West and M. L. Mecartney, A comparison of atomic force microscopy (AFM) and dynamic light scattering (DLS) methods to characterize nanoparticle size distributions, J. Nanopart. Res., 2008, 10, 89–96 CrossRef CAS.
- G. Bryant and J. C. Thomas, Improved particle size distribution measurements using multiangle dynamic light scattering, Langmuir, 1995, 11, 2480–2485 CrossRef CAS.
- K. M. Kanninen, N. Bister, J. Koistinaho and T. Malm, Exosomes as new diagnostic tools in CNS diseases, Biochim. Biophys. Acta, Mol. Basis Dis., 2016, 1862, 403–410 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2021 |