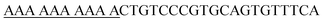Open Access Article
Open Access ArticleZika virus lateral flow assays using reverse transcription-loop-mediated isothermal amplification†
Gna Ahn‡
a,
SeonHyung Lee‡b,
Se Hee Leea,
Yun Hee Baekc,
Min-Suk Songc,
Yang-Hoon Kim*ab and
Ji-Young Ahn *ab
*ab
aDepartment of Microbiology, Chungbuk National University, 1 Chungdae-Ro, Seowon-Gu, Cheongju 28644, South Korea. E-mail: jyahn@chungbuk.ac.kr; kyh@chungbuk.ac.kr; Fax: +82-43-264-9600; Tel: +82-43-261-2301
bDepartment of Biological Sciences and Biotechnology, Chungbuk National University, 1 Chungdae-Ro, Seowon-Gu, Cheongju 28644, South Korea
cCollege of Medicine, Medical Research Institute, Chungbuk National University, 1 Chungdae-Ro, Seowon-Gu, Cheongju 28644, South Korea
First published on 18th May 2021
Abstract
Owing to the global spread of the Zika virus (ZIKV) infection, field-ready diagnostics are urgently warranted. In this study, we sought to detect ZIKV using reverse transcription loop-mediated isothermal amplification (RT-LAMP). Briefly, we performed and optimized ZIKV RT-LAMP for the analysis of biological samples (PBS, urine, and plasma). Based on our findings, this method could detect ZIKV RNA in 40 min at 63 °C without any off-target amplification. After performing specificity tests using BtsI restriction enzyme digestion, the feasibility of ZIKV RT-LAMP was determined via end-point detection with different sample matrices. Thereafter, a lateral flow assay (LFA) was conducted to directly detect the ZIKV RT-LAMP products. Based on the LFA reaction, hybridization occurred between the AuNPs:polyadenylated (polyA10)-ZIKV probe and the LAMP amplicons. Subsequently, we optimized the assay parameters, including the concentration of AuNPs and migration matrices (glass fiber and nitrocellulose membrane). By employing a specific AuNP:polyA10-ZIKV LAMP probe, we could demonstrate the purpose and utility of primary and secondary antibodies. Owing to LFA, the resultant ZIKV RT-LAMP products were rapidly and simply assayed in less than 5 min. Further, no preparation step was required to achieve LAMP-probe hybridization, highlighting the utility of this method for field-ready ZIKV diagnosis. Collectively, our findings suggest that ZIKV RT-LAMP combined with LFA could serve as a rapid, accurate, and independent point-of-care detection method for preventing ZIKV outbreaks.
Introduction
The Zika virus (ZIKV) belongs to the virus family Flaviviridae and genus Flavivirus. Its genome is comprised of approximately 10.7 kb positive-sense single-stranded RNA that encode seven nonstructural proteins and three structural proteins.1,2 Similar to other flaviviruses, such as Dengue (DENV), West Nile (WNV), Japanese Encephalitis viruses (JEV), and yellow fever (YFV), which are also clinically significant, the ZIKV infection is transmitted by mosquitoes.3 ZIKV was first isolated from a rhesus monkey in the Zika forest located in East Africa in 1947. Currently, ZIKV is classified as distinct clades in Africa and Asia.3,4 As ZIKV causes sporadic human infections that exhibit as mild and self-limiting febrile illness, recent outbreaks have been characterized by variable clinical syndromes and rapid spread throughout the South Pacific and Latin America, particularly Brazil. As a result, there is a growing public health concern globally with regard to this virus.3,5 Besides common symptoms such as fever, rash, myalgias/arthralgia, and conjunctivitis, recent ZIKV-infected patients were found to have clinical complications, such as sickle cell disease and post-ZIKV Guillain–Barré syndrome.6,7 Importantly, there are reports of congenital ZIKV infection that causes the development of microcephaly among newborns.8The unavailability of treatment and vaccines for ZIKV infection, has highlighted the need for rapid and sensitive diagnostics to effectively control and prevent the spread of the virus. Unlike other mosquito-borne viruses, ZIKV spreads to new hosts via saliva, semen, urine, blood, and maternal transmission.3 Further, most ZIKV-infected patients remain asymptomatic, which implies the importance of host as veiled reservoirs of ZIKV. The development of a feasible, simple and cheap diagnostic method, especially in a resource-limited-setting, is highly recommended by the World Health Organization.9 Based on the clinical relevance and the common genetic features shared between ZIKV and other viruses in the Flaviviridae family, including DENV, WNV, JEV, and YFV, ZIKV detection methods should not only be accurate, but also distinctly recognize ZIKV alone.10 Standard serologic tests often display inadequate sensitivity and cross-reactivity for patients previously exposed to other viruses in the Flaviviridae family.11 Therefore, nucleic acid amplification tests (NAAT) have been recognized as a more appropriate diagnostic method for ZIKV detection.
Our group and others have shown that loop-mediated isothermal nucleic acid amplification (LAMP) can be used to diagnose infectious agents, including viruses, viroids, and bacteria from various clinical samples.12–14 In the present study, we opted to use reverse transcription LAMP (RT-LAMP), which employs six primers that bind to specific regions on the DNA strands, to detect the presence of ZIKV. Based on our experience, the LAMP method is highly specific and requires only a single temperature for dsDNA product amplification. Recently, several published reports have demonstrated the use of RT-LAMP for diagnosing virus infection; this simple and rapid process is accompanied by a short reaction time. However, one of the following detection techniques must be employed for signal read-out in LAMP: turbidity testing, running the amplification product on an agarose gel, or adding intercalating dyes such as SYBR Green, EvaGreen and pH indicators.15,16 Notably, the specific advantages of each of these techniques are dependent on their operating purposes. In our experiment, we selected a simple and disposable paper-based colorimetric lateral flow assay (LFA). The point-of-care test employed for disease diagnosis must be inexpensive to produce, portable, and easy to use. Further, it must be able to detect and measure small quantities of samples.17–19 Paper-based LFA is the most commercially available diagnostic kit. In fact, it can be used to diagnose and identify various samples, such as viruses, pathogens, proteins, and nucleic acids of small volumes, in a short time in situ. Gold nanoparticles (AuNPs), quantum dots, and fluorescence have been used to confirm the detection of analytes.20,21 AuNPs are used in various applications, because they are easy to produce, are associated with low manufacturing costs, and have excellent sensitivity for application in the field. Binding probes that conjugate to AuNPs can use various substances such as antibodies, nucleic acids, and proteins, depending on the analytes or artificially manufactured parts that can be specifically bonded to a specific part of the sample.
In the present study, we sought to verify the applicability of ZIKV RT-LAMP, which incorporates real-time RT-LAMP, endonuclease digestion, and visualization with a fluorescent indicator, SYBR Green I, for rapid detection of ZIKV. In addition, we report a rapid and specific LFA for detecting ZIKV RT-LAMP products from human specimens, such as urine and serum. Herein, nitrocellulose membrane-based LF strips were found to produce clear and visible signals for positive reactions, enabling immediate detection (within 5 min) of amplified an products with AuNP:polyA10-ZIKV LAMP probe.
Materials and methods
Preparation of viruses and spike samples
RT-LAMP and LFA were carried out to detect the presence of ZIKV (ATCC/VR-1859) and JEV (KBPV-VR-27, Knrrc, South Korea) in biological samples. To determine viral infectious titers, a plaque assay was performed using Vero E6 cells (ATCC, CRL 1586). Briefly, Vero cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM) with 10% fetal bovine serum (FBS). Each virus was individually prepared via 10-fold serial dilution in the infection medium (minimal essential medium). Individual cells were seeded in 6-well plates (6 × 105 cells per well) containing DMEM with 10% FBS and incubated at 37 °C for 24 h in a CO2 incubator. The prepared virus diluent was used to infect cells, which were incubated for 2 h at 37 °C to allow virus inoculation. A mixture of agar overlay was added to each well and plates were incubated for 5 to 7 days. Cells were fixed post-infection with 10% formaldehyde and stained with crystal violet after agar removal. The simulated clinical samples were prepared by mixing ZIKV viruses with normal human serum (Sigma Aldrich, USA), urine (Innovative Research, 46![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430, Peary Court Novi, MI, USA) or PBS. Serial dilutions of viruses were performed to achieve a final concentration of 1 × 10−1 PFU mL−1.
430, Peary Court Novi, MI, USA) or PBS. Serial dilutions of viruses were performed to achieve a final concentration of 1 × 10−1 PFU mL−1.
Viral RNA extraction
Viral RNA was extracted from 200 μL of each virus stock using the RNeasy Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's protocol. After extraction, RNA was stored at 80 °C until use. To prepare the virus RNA standards, the target region was amplified using the TOPscript™ One-step RT-PCR Kit (Enzynomics, Republic of Korea), generating a 240 bp (ZIKV) and a 264 bp (JEV) fragment, respectively (ESI Fig. S3†). RNA transcripts were generated using the MegaScript kit (Ambion, Thermo Fisher, Waltham, MA) according to the manufacturer's instructions. The products were then treated with Turbo DNase for 15 min at 37 °C to remove any template DNA. The purified RNA was diluted 10-fold and frozen at −80 °C after the copy numbers of RNA transcripts were calculated according to the concentrations determined with a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific).Optimization of reverse-transcription LAMP (RT-LAMP) and real-time RT-LAMP for ZIKV detection
We analyzed the sequences of each virus retrieved from the NCBI database using the CLC Main Workbench (Qiagen, United States). Envelop protein-coding genes for ZIKV were selected as target regions for reverse transcription RT-LAMP. Primer sets for efficient RT-LAMP reaction were designed using the Opensource program, Eiken's PrimerExplorerV4 software version 4 (http://primerexplorer.jp/e). These primer sets consisted of two inner primers (Forward, FIP; Backward, BIP), two outer primers (Forward, F3; Backward, B3), and two loop primers (Forward, LF; Backward, LB). The inner primers, FIP (and BIP), were composed of F2 (B2) regions and a complementary sequence for the F1(B1) region in the template. The primer sets are listed in Table 1. The FIP primers were modified by adding biotin to the 5′-end for the LFA. The regions of the ZIKV-specific probes designed for the LFA were selected in the LAMP amplified regions and confirmed via sequence comparison in Clustal Omega (Multiple Sequence Alignment). All primers and probes were synthesized via commercial services offered by Cosmogenetech Co., Ltd.The LAMP reaction mixture consisted of 2.5 μL of 10× LAMP reaction buffer, 2 μL of each 2 mM dNTP, 1.5 μL of 100 mM extra MgSO4, 1 μL of inner primer (FIP and BIP), 1 μL of outer primer (F3 and B3), 1 μL of loop primer (LF and LB), 1 μL of 200U M-MLV reverse transcriptase, 1 μL of 8U Bst polymerase, 1 μL of virus RNA isolates (3 × 105 to 3 × 10−1 copies per μL), and distilled water; the final volume was 25 μL. The RT-LAMP reaction mixture was incubated at 63 °C for 40 min and heated at 85 °C for 1 min for reaction termination. LAMP products (1 μL) were then digested with the restriction enzyme, BtsI (NEB, USA), according to the manufacturer's protocol. The digested LAMP products were analyzed using 1% agarose gel electrophoresis. SYBR Green I (information) was used as a colorimetric RT-LAMP product visualization method. Thereafter, 1 μL of 400× SYBR Green I was mixed with 10 μL of LAMP product; the color change was confirmed by UV-on state in gel-doc and UV-off state by the naked eye.
For the real-time RT-LAMP assay, 1 μL of SYBR Green I (25×) was added to the real-time LAMP reaction mixture for use on the CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) programmed at 63 °C for 60 min. Visualization of the RT-LAMP products was achieved using 1% agarose gel electrophoresis at 100 V cm−1 in 0.5× TAE buffer. The specificity of the RT-LAMP assay was confirmed using JEV. Viral RNA isolates were extracted from the cell culture supernatants and spiked human samples (serum and urine). An end-point detection was also performed to determine the capability of the detection methods using RNA templates extracted from virus-spiked human samples.
Real-time RT-PCR for ZIKV
To evaluate the sensitivity of our multiplex RT-LAMP assay for application to real-time RT-PCR approaches, the extracted RNA samples were serially diluted from 3 × 105 to 3 × 101 copies per μL. Real-time RT-PCR was performed using TOPreal™ One-step RTqPCR Kit (Enzynomics, Republic of Korea) with 10 pmol of outer primers (F3 and B3, see Table 1) and 1 μL of serially diluted RNA. The following real-time RT-PCR conditions were carried out on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA): reverse transcription at 55 °C for 30 min, initial denaturation at 95 °C for 5 min, 40 cycles of three steps: denaturation at 95 °C for 30 s, annealing at 60 °C for 30 s, elongation at 72 °C for 30 s, and final elongation at 72 °C for 5 min. The results were analyzed using CFX manager software.AuNP synthesis
Gold(III) chloride hydrate (HAuCl4), trisodium citrate, hydrochloric acid (HCl), and nitric acid (HNO3) were purchased from Sigma-Aldrich (USA). All glassware were cleaned with freshly prepared aqua regia solution (hydrochloric acid and concentrated nitric acid, HCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HNO3, 3
HNO3, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and rinsed with deionized water prior to use. Citrate-stabilized AuNPs with an average size of 13 nm were synthesized according to citrate reduction.22 Briefly, 100 mL of HAuCl4 (0.01% (w/w)) was added to a rinsed flask and boiled under continuous stirring. An additional 5 mL of 38.8 mM trisodium citrate was added to HAuCl4. The mixed solution was boiled and stirred for 20 min and subsequently cooled to room temperature. Finally, the solution was filtered through a 0.2 μm ultra-filtration membrane (GVS Filter Technology, UK) for aggregate removal. The concentration of AuNPs was calculated using Beer's law. The extinction coefficient of the 13 nm AuNPs was 2.7 × 108 M−1 cm−1. The AuNPs were stored in dark glass bottles at 4 °C until use.
1) and rinsed with deionized water prior to use. Citrate-stabilized AuNPs with an average size of 13 nm were synthesized according to citrate reduction.22 Briefly, 100 mL of HAuCl4 (0.01% (w/w)) was added to a rinsed flask and boiled under continuous stirring. An additional 5 mL of 38.8 mM trisodium citrate was added to HAuCl4. The mixed solution was boiled and stirred for 20 min and subsequently cooled to room temperature. Finally, the solution was filtered through a 0.2 μm ultra-filtration membrane (GVS Filter Technology, UK) for aggregate removal. The concentration of AuNPs was calculated using Beer's law. The extinction coefficient of the 13 nm AuNPs was 2.7 × 108 M−1 cm−1. The AuNPs were stored in dark glass bottles at 4 °C until use.
Poly-adenylated (A10) ZIKV probe
Nonthiolated poly-adenine (polyA10)-linked ZIKV RT-LAMP probes ((5-A10-CTG TCC CGT GCA GTG TTT CA-3) (Cosmogenetech, Korea)) were attached to the surface of gold nanoparticles (AuNP, 13 nm) according to a salt-aging method.12 Briefly, 8.1 nM citrate-stabilized AuNPs were incubated with 5 μM polyadenylated (polyA10) ZIKV LAMP probe overnight at room temperature. The conjugate was then dispersed in 0.1 M NaCl for 30 h at 4 °C, and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C for the removal of the unbound barcodes. Following the removal of the supernatant, the successful conjugates were resuspended in deionized water. The AuNP:polyA10-ZIKV LAMP probe was confirmed using wavelength scanning (DS-11 FX+ Spectrophotometer/Fluorometer, Denovix, USA). The salt-induced aggregation of polyA10-ZIKV LAMP probe and the hybrids were tested by adding 3 mM MgSO4, using TEM analysis.
000 rpm for 20 min at 4 °C for the removal of the unbound barcodes. Following the removal of the supernatant, the successful conjugates were resuspended in deionized water. The AuNP:polyA10-ZIKV LAMP probe was confirmed using wavelength scanning (DS-11 FX+ Spectrophotometer/Fluorometer, Denovix, USA). The salt-induced aggregation of polyA10-ZIKV LAMP probe and the hybrids were tested by adding 3 mM MgSO4, using TEM analysis.
Lateral flow (LF) strip fabrication and LF assay (LFA) test
LF strips were fabricated according to a previous study.23 Streptavidin was purchased from Sigma-Aldrich (USA). The 2× SSC powder was purchased from Takara (Japan). The LF strip was provided by Sejong-Bio (Cheongju, South Korea); this strip consisted of three parts: a cotton sample pad, an absorption pad, and a nitrocellulose or glass fiber pad. The distance between the test zone and control zone was maintained at 0.5 cm. The mixture for the control zone was prepared in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratio with streptavidin
4 ratio with streptavidin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) biotin-ZIKV complementary primer (5′-biotin TGA AAC ACT GCA CGG GAC AG-3′). Briefly, 90 μL of control solution (10 nmol ZIKV complementary probe-biotin and 2.5 nmol streptavidin) was prepared in PBS buffer solution and incubated at room temperature for 1 h with slight rotation. For the test zone, 0.5 μL of streptavidin (1 mg mL−1) was used. The strips were completely dried at 37 °C for 2 h with a desiccant. For RT-LAMP combined with LFA, the AuNP:polyA10-ZIKV LAMP probe was used for wavelength scanning analysis at 520 nm.
biotin-ZIKV complementary primer (5′-biotin TGA AAC ACT GCA CGG GAC AG-3′). Briefly, 90 μL of control solution (10 nmol ZIKV complementary probe-biotin and 2.5 nmol streptavidin) was prepared in PBS buffer solution and incubated at room temperature for 1 h with slight rotation. For the test zone, 0.5 μL of streptavidin (1 mg mL−1) was used. The strips were completely dried at 37 °C for 2 h with a desiccant. For RT-LAMP combined with LFA, the AuNP:polyA10-ZIKV LAMP probe was used for wavelength scanning analysis at 520 nm.
To optimize the concentration of the AuNP:polyA10-ZIKV LAMP probe, 1 μL of purified ZIKV LAMP products (500 ng μL−1) was added to 24 μL of distilled water and 50 μL of 2× SSC buffer. Thereafter, 25 μL of the AuNP:polyA10-ZIKV LAMP probe was added to a microplate well, and the dried strip was dipped into the well to detect signal generation. The UV-vis spectroscopic value of the solutions containing the AuNP:polyA10-ZIKV LAMP probe-ZIKV RT-LAMP hybrids was recorded to determine the migration time (especially within 5 min) on the LF strip.
Results and discussion
Optimization of ZIKV RT-LAMP
To establish a consensus sequence, ZIKV sequences were collected from the NCBI database. After the collected sequences were aligned using the Megalign program, the consensus ZIKV sequence was derived (ESI Fig. S1†). For ZIKV RT-LAMP, we targeted a conserved region in the envelope protein-coding gene and designed highly specific primers (Fig. 1(A) and Table 1). The 1814–2058 bp region of the envelope protein-coding gene containing the BtsI restriction enzyme site was selected as the target region. The following LAMP primers were designed based on the selected target site: 1 pair of inner primer, 1 pair of outer primer, and 1 pair of loop primers using Primer Explorer version 5.0. FIP was labeled with biotin at the 5′-end for the LFA. These primers were used to test several reaction conditions for ZIKV RT-LAMP optimization (ESI Fig. S2†). For ZIKV RT-LAMP, the primer concentration was fixed at 0.4 μM for the outer, inner, and loop primers and the best amplification efficiency was achieved at 63 °C. The results of the ZIKV RT-LAMP assay were confirmed using the SYBR Green I colorimetric method and gel electrophoresis. The colorimetric method is widely used after LAMP, instead of gel electrophoresis, to reduce the assay time.12 SYBR Green I is a representative reagent for confirming nucleic acid amplification.24 SYBR Green I changes from orange to green in the UV off state and green fluorescence is achieved in the UV on state as nucleic acid amplification progresses. Because of the color change that occurs when SYBR Green I was mixed with the LAMP products, immediate observation of the results by the naked eye can be achieved. SYBR Green I showed the most efficient color change when 1 μL of the 400× concentration was mixed with 10 μL of the RT-LAMP products. Regardless of UV irradiation, SYBR Green I was found to be sensitive at low concentrations of the LAMP product. However, the LAMP signal read-out with SYBR Green I has been criticized for lack of specificity. Further, the SYBR Green I fluorescence signal-readouts did not exhibit any remarkable reaction time- or concentration dependent changes.14,25,26 To validate the specific amplification of ZIKV using RT-LAMP, we designed primers to include a unique BtsI restriction enzyme site within the target region. Prior to applying gene-specific enzyme digestion to the LAMP products, we carried out an enzyme digestion of the PCR product. After digestion, a 240 bp PCR band was digested and a new 120 bp band was generated (Fig. 1(B), PCR panel). Sequence analysis showed that the 120 bp dsDNA segments corresponded to the ZIKV-specific products. Subsequently, ZIKV RT-LAMP products was employed for BtsI enzyme digestion. The smear of the banded products was visible in the stained agarose gel band (Fig. 1(B), RT-LAMP panel). As shown in Fig. 1(C) and (D), the target ZIKV genes could be amplified to a remarkable amount in 40 min under the optimized RT-LAMP conditions. After incubation at 63 °C, an s-shaped curve graph was obtained for the reaction mixture containing ZIKV RNAs, suggesting successful amplification of the target viral RNAs. The specificity of the ZIKV RT-LAMP assay was confirmed by testing RNA isolates of JEV, a closely-related flavivirus. However, only RNAs from ZIKV-infected cells (i.e., no RNA from JEV-infected cells) had positive RT-LAMP reactions, indicating specificity of the reaction for ZIKV (ESI Fig. S3†). Based on our findings, specific enzyme digestion can contribute to a decrease in the false-positive rate of the LAMP assay; this is because the BtsI restriction enzyme site does not exist in the envelope protein-coding region of viruses, such as Dengue and JEV. Such findings demonstrate that SYBR Green I-based on/off signals and real-time RT-LAMP enable simple colorimetric detection and real-time monitoring of target amplification without gel electrophoresis analysis.Sensitivity of RT-LAMP for detecting ZIKV
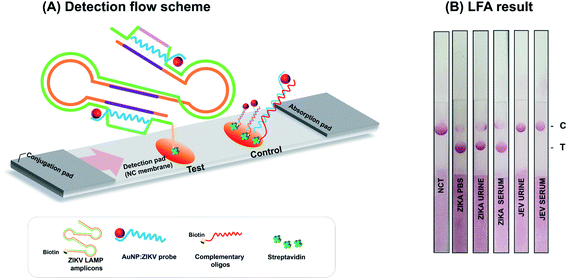
High sensitivity is critical in point-of-care screening tests for ZIKV. In fact, more specific follow-up tests can be performed after this initial screening to confirm infection. The sensitivity of ZIKV RT-LAMP was evaluated by preparing serial ten-fold dilution of ZIKV RNA isolates as templates, from 3 × 105 copies of RNA isolates. As shown in Fig. 2 (at 63 °C), ZIKV could be detected at 3 × 100 copies of RNA; this was identified as the lower detection limit and was achieved within 40 min of incubation (Fig. 2(A)). No off-target amplification occurred in the non-template controls. Amplification using real-time RT-LAMP was also confirmed by agarose gel electrophoresis analysis (Fig. 2(B)). Conventional real-time RT-PCR revealed that the specific primer set (F3 and B3) tended to amplify ZIKV less sensitively than ZIKV RT-LAMP (Fig. 2(C)). For example, 3 × 100 copies of ZIKV RNA could be detected as amplified products after 40 min. These findings indicate the high sensitivity of the optimized RT-LAMP conditions, which can be applied to the rapid and sensitive detection of ZIKV. | ||
| Fig. 2 Sensitivity test of the ZIKV RT-LAMP using viral RNA samples containing 3 × 10−1 to 3 × 105 copies per μL of isolates. (A) With the optimized temperature (63 °C), real-time RT-LAMP could amplify the ZIKV RNAs in 40 min (see black arrow) at an initial concentration of 3 × 100 copies per μL. (B) Amplification was confirmed by gel electrophoresis as described in the Materials and methods section. PC; positive control for ZIKV dsDNA (see ESI Fig. S3(A)†), NC; non-template control. (C) Comparative analysis of RT-LAMP and rtRT-PCR. Calibration curve for rtRT-PCR was shown in ESI Fig. S4.† | ||
End-point detection of ZIKV LAMP
To determine whether ZIKV RT-LAMP can be employed to detect viral RNA in human samples, urine and serum samples were retrieved for testing. Briefly, commercial human serum and urine were used in the ZIKV RT-LAMP experiments. All experiments using ZIKV were performed in a BSL3 laboratory. Viral RNAs were extracted from each sample and subjected to RT-LAMP. Based on gel electrophoresis and SYBR signal read-outs, ZIKV was detected in both urine and serum (ESI Fig. S5†). Such findings indicate that our ZIKV RT-LAMP assay can be used to diagnose ZIKV infection using urine and serum samples.ZIKV RT-LAMP assays were carried out in duplicate to determine the stochastic detection frequencies at the endpoint of the assay. Briefly, ZIKV was spiked into PBS, serum, and urine. As a result, the sensitivity was identified to be much higher in RT-LAMP than real-time (rt) RT-PCR (Table 2, ESI Fig. S4†). At 1 × 102 PFU, real-time RT-PCR displayed a very low detection accuracy. However, RT-LAMP had a detection rate ≥ 80% in all sample types. At 1 × 101 PFU, real-time RT-PCR showed a Ct value that could be deemed undetectable while RT-LAMP had a detection rate greater than 50%. At the 1 × 100 PFU concentration, only a minor detection occurred in both real-time RT-PCR and RT-LAMP. Furthermore, among the sample types tested, the virus-spiked serum was found to have the lowest detection rate in both real-time RT-PCR and RT-LAMP. Therefore, to more accurately detect ZIKV, a sample of urine or purely purified RNA should be employed instead of a serum sample.
| PBS | Serum | Urine | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a N.D: non-detected. | ||||||||||||
| Virus titer (PFU mL−1) | 1 × 103 | 1 × 102 | 1 × 101 | 1 × 100 | 1 × 103 | 1 × 102 | 1 × 101 | 1 × 100 | 1 × 103 | 1 × 102 | 1 × 101 | 1 × 100 |
| rtRT-PCR (Ct value) | 22.5 | 29.5 | 32.5 | N.D | 16.1 | 30.2 | 32.5 | N.D | 25.7 | 29.2 | 32.1 | N.D |
| RT-LAMP (end-point discrimination) | 20/20 | 19/20 | 13/20 | 1/20 | 20/20 | 16/20 | 12/20 | 1/20 | 20/20 | 18/20 | 14/20 | 1/20 |
| Percentage of RT-LAMP (%) | 100 | 95 | 65 | 5 | 100 | 80 | 60 | 5 | 100 | 90 | 70 | 5 |
ZIKV RT-LAMP LFA
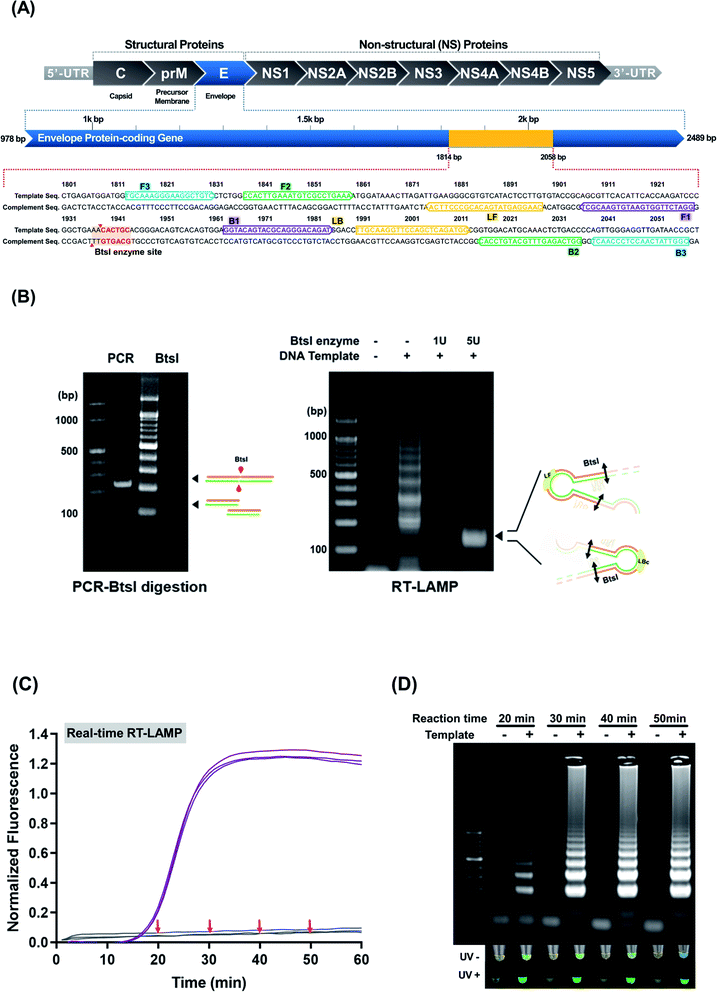
Prior to the LFA, an AuNP:polyadenylated (polyA10) ZIKV probe was prepared for use as a visualizing signal indicator. The colloidal state of the AuNP:polyA10 ZIKV probe displayed the highest peak value at a wavelength of 520 nm (ESI Fig. S6(A)†). Our previous study found that the instantaneous aggregation of polyA probe functionalized AuNPs were gradually decreased after hybridization with target LAMP amplicons.12 Moreover, accumulating evidence has demonstrated that the polyA tail can effectively mediate the anchoring of DNA and the appended recognition probe adopts an upright conformation that favors DNA hybridization.27 Thus, we performed TEM characterization of the ZIKV RT-LAMP hybrids with AuNP:polyadenylated (polyA10) ZIKV probe. An analysis of TEM image revealed that no serious salt-induced aggregation was observed in the presence of the ZIKV RT-LAMP products (500 ng in the total reaction volume, 100 μL), as shown in ESI Fig. S6(B).† This indicates that the AuNP:polyA10 ZIKV probes were high enough to hybridize with ZIKV RT-LAMP target in upright orientation.LF was based on the immobilization of streptavidin for each of the test and control zones (Fig. 3). In the test zone, the immobilized streptavidin can bind to the biotinylated ZIKV RT-LAMP products that are hybridized to the AuNP:polyA10-ZIKV probe on their specific targets. However, in the control zone, the complementary DNA sequences react to the AuNP:ZIKV probe. Because the complementary DNA sequences were biotinylated, they could bind to the streptavidin control zone, generating a red signal that is visible to the naked eye in both the control and test sets (Fig. 3(A)). These analytic sites on the strip were formed by dropping reagent solutions using micro-pipet, presenting a feasible method for detecting ZIKV RT-LAMP amplicons. To establish an appropriate concentration of the AuNP:polyA10 ZIKV probe, 3 different concentrations of the AuNP:polyA10-ZIKV probe (4.1 nM, 5.2 nM, and 10.4 nM) were tested with spotted LF strips. Thereafter, the intensities of the test zone were quantified as a percentage of the intensities in the control zone; the results of three independent experiments were plotted (data not shown). Based on our final optimized condition, we found the concentration of the AuNP:ZIKV probe (5.2 nM) that displayed the strongest signal with a specific U-shaped binding mode on the test zone.
To improve assay performance, two membranes (i.e., nitrocellulose and grass fiber membranes) were evaluated to determine the material that is optimal for the ZIKV RT-LAMP LF strips. The nitrocellulose membrane-based LF strips produced clear and visible signals for positive reactions, thereby enabling immediate detection (within 5 min) of the amplified products with the naked eye (ESI Fig. S7†). To ensure specificity for ZIKV RT-LAMP, JEV was tested at the high concentration of 103 PFU mL−1. As the negative control or JEV sample was not amplified, the specificity for ZIKV was demonstrated. The amplification results were confirmed using 1% agarose gel electrophoresis (ESI Fig. S3†) and LF detection Fig. 3(B).
When compared with previous studies using combined LAMP and LFA for ZIKV detection, this study has advantages in terms of signal detection probe and LFA strip composition (Table 3). The polyA10 tailed ZIKV probes were adsorbed simply and strongly on gold nanoparticles (AuNPs). In order to increase the ZIKV RT-LAMP LF specificity, the sequence-specific probe was designed. Our probe-based ZIKV RT-LAMP LF can reduce the number of false positives.
| Viruses | Method | Detection limit | Sample composition | LFA strip composition | Ref. | ||
|---|---|---|---|---|---|---|---|
| Target | Signal detection | Test | Control | ||||
| DENV | RT-LAMP | 101 copy | FAM and biotin labeled RT-LAMP | Streptavidin coated AuNP | Anti-FAM | Biotin | 28 |
| JEV | RT-LAMP | 5 pg | FITC and biotin labeled RT-LAMP | AuNP labeled anti FITC Ab | Streptavidin | Anti-biotin Ab | 29 |
| ZIKV | RT-LAMP | 100 copy | Digoxigenin labeled RT-LAMP | Streptavidin coated AuNPs | Anti-digoxigenin antibody | Biotin | 30 |
| ZIKV | RT-LAMP | 3 × 100 copy | Biotin labeled RT-LAMP | PolyA10 tailed ZIKV probes plus AuNPs | Streptavidin | Biotinylated oligos (complementary to ZIKV probe) | Our study |
Conclusion
A real-time RT-LAMP assay was developed to detect ZIKV and achieve immediate LF detection of amplified products using an AuNPs:polyA10 ZIKV probe. To confirm that the ZIKV RT-LAMP products correspond to the ZIKV target sequence, the amplified products were digested with BtsI restriction endonuclease and analyzed by agarose gel electrophoresis. Thereafter, ZIKV RT-LAMP assays were carried out to determine the stochastic detection frequencies at the endpoint of the assay. At 1 × 103 PFU, the detection rate of ZIKV RT-LAMP in the human samples was 100%. An LFA was also developed and employed to achieve rapid and reliable detection of ZIKV RT-LAMP. Target-specific RT-LAMP could be observed visually after the addition of an AuNP:polyA10 ZIKV probe. As a result, a high authentication capacity was achieved in the routine workflow. The strategies and methods employed herein can be extended to other mosquito-borne virus targets by simply changing the LAMP primers, ultimately providing an inexpensive diagnostic platform for point-of-care assays.Author contributions
Gna Ahn, SeonHyung Lee, and Se Hee Lee performed the experiments and analyzed the data; Yun Hee Baek and Min-Suk Song contributed materials/analysis tools; Yang-Hoon Kim and Ji-Young Ahn wrote the paper.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MEST) (NRF-2019R1A2C1010860) and a Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2020R1A6A1A06046235).References
- A. R. Plourde and E. M. Bloch, Emerging Infect. Dis., 2016, 22, 1185–1192 CrossRef CAS PubMed.
- D. Margaret and T. C. Pierson, Fields Virology, 2013, 2, 747–794 Search PubMed.
- H. M. Lazear and M. S. Diamond, J. Virol., 2016, 90, 4864–4875 CrossRef CAS PubMed.
- G. W. Dick, S. F. Kitchen and A. J. Haddow, Trans. R. Soc. Trop. Med. Hyg., 1952, 46, 509–520 CrossRef CAS PubMed.
- C. B. Coyne and H. M. Lazear, Nat. Rev. Microbiol., 2016, 14, 707–715 CrossRef CAS PubMed.
- V. M. Cao-Lormeau, A. Blake, S. Mons, S. Lastere, C. Roche, J. Vanhomwegen, T. Dub, L. Baudouin, A. Teissier, P. Larre, A. L. Vial, C. Decam, V. Choumet, S. K. Halstead, H. J. Willison, L. Musset, J. C. Manuguerra, P. Despres, E. Fournier, H. P. Mallet, D. Musso, A. Fontanet, J. Neil and F. Ghawche, Lancet, 2016, 387, 1531–1539 CrossRef.
- E. C. f. D. P. a., Rapid risk assessment, Zika virus disease epidemic: potential association with microcephaly and Guillain-Barré syndrome, second update, 8 February 2016, European Centre for Disease Prevention and Control, Stockholm, 2016 Search PubMed.
- C. V. Ventura, M. Maia, V. Bravo-Filho, A. L. Gois and R. Belfort Jr, Lancet, 2016, 387, 228 CrossRef.
- W. H. Organization, Zika virus, microcephaly and Guillain-Barré syndrome situation report, 2016 Search PubMed.
- S. I. Yun and Y. M. Lee, J. Microbiol., 2017, 55, 204–219 CrossRef CAS PubMed.
- D. G. D Musso, Clin. Microbiol. Rev., 2016, 29, 487–524 CrossRef PubMed.
- S. H. Lee, G. Ahn, M. S. Kim, O. C. Jeong, J. H. Lee, H. G. Kwon, Y. H. Kim and J. Y. Ahn, ACS Comb. Sci., 2018, 20, 472–481 CrossRef CAS PubMed.
- S. H. Lee, Y. H. Baek, Y. H. Kim, Y. K. Choi, M. S. Song and J. Y. Ahn, Front. Microbiol., 2016, 7, 2166 Search PubMed.
- J. H. K. S. Lee, B. K. Han, W. Kim, B. K. Cho, S. M. Woo, Y. H. Kim and J. Y. Ahn, Mol. Cell. Toxicol., 2020, 16, 263–270 CrossRef.
- J. Fischbach, N. C. Xander, M. Frohme and J. F. Glokler, Biotechniques, 2015, 58, 189–194 CrossRef CAS PubMed.
- N. A. Tanner, Y. Zhang and T. C. Evans Jr, Biotechniques, 2015, 58, 59–68 CrossRef CAS PubMed.
- M. Zarei, Biosens. Bioelectron., 2018, 106, 193–203 CrossRef CAS PubMed.
- Z. Li, H. Chen and P. Wang, Analyst, 2019, 144, 3314–3322 RSC.
- S. Lee, J. H. Kim, B. N. Kim, J. Y. Ahn, S. Kim, B. K. Cho, Y. H. Kim and J. Min, J. Toxicol. Environ. Health Sci., 2020, 12, 297–304 CrossRef.
- Y. Zhang, H. Kong, X. Liu, J. Cheng, M. Zhang, Y. Wang, F. Lu, H. Qu and Y. Zhao, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 1685–1693 CAS.
- X. Gong, J. Cai, B. Zhang, Q. Zhao, J. Piao, W. Peng, W. Gao, D. Zhou, M. Zhao and J. Chang, J. Mater. Chem. B, 2017, 5, 5079–5091 RSC.
- C. Chun, J. Joo, D. Kwon, C. S. Kim, H. J. Cha, M. S. Chung and S. Jeon, Chem. Commun., 2011, 47, 11047–11049 RSC.
- J. R. Choi, J. Hu, Y. Gong, S. Feng, W. A. Wan Abas, B. Pingguan-Murphy and F. Xu, Analyst, 2016, 141, 2930–2939 RSC.
- S. H. Lee, S. H. Lee, K. Won, M. S. Kim, H. Ryu, Y. H. Kim and J. Y. Ahn, J. Biomed. Nanotechnol., 2020, 16, 166–178 CrossRef CAS PubMed.
- R. Kumvongpin, P. Jearanaikool, C. Wilailuckana, N. Sae-ung, P. Prasongdee, S. Daduang, M. Wongsena, P. Boonsiri, W. Kiatpathomchai, S. S. Swangvaree, A. Sandee and J. Daduang, J. Virol. Methods, 2016, 234, 90–95 CrossRef CAS PubMed.
- M. Goto, E. Honda, A. Ogura, A. Nomoto and K.-I. Hanaki, BioTechniques, 2009, 46, 167–172 CrossRef CAS PubMed.
- H. Pei, F. Li, Y. Wan, M. Wei, H. Liu, Y. Su, N. Chen, Q. Huang and C. Fan, J. Am. Chem. Soc., 2012, 134, 11876–11879 CrossRef CAS PubMed.
- Y. Xiong, Y. Luo, H. Li, W. Wu, X. Ruan and X. Mu, Int. J. Infect. Dis., 2020, 95, 406–412 CrossRef CAS PubMed.
- J. Deng, J. Pei, H. Gou, Z. Ye, C. Liu and J. Chen, J. Virol. Methods, 2015, 213, 98–105 CrossRef CAS PubMed.
- D. Lee, Y. Shin, S. Chung, K. S. Hwang, D. S. Yoon and J. H. Lee, Anal. Chem., 2016, 88, 12272–12278 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra01227d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2021 |