Open Access Article
Open Access ArticleEscherichia coli templated iron oxide biomineralization under oscillation†
Panpan Hea,
Junhui Guo*a,
Liwen Leib,
Jiafeng Jianga,
Qichang Lia,
Zhiyi Hub,
Baolian Su bc,
Zhengyi Fu
bc,
Zhengyi Fu *b and
Hao Xie
*b and
Hao Xie *a
*a
aSchool of Chemistry, Chemical Engineering and Life Sciences, Wuhan University of Technology, Wuhan, 430070, China. E-mail: guojunhui@whut.edu.cn; h.xie@whut.edu.cn
bState Key Laboratory of Advanced Technology for Materials Synthesis and Processing, Wuhan, 430070, China. E-mail: zyfu@whut.edu.cn
cLaboratory of Inorganic Materials Chemistry, University of Namur, B-5000 Namur, Belgium
First published on 21st April 2021
Abstract
Motility is significant in organisms. Studying the influence of motility on biological processes provides a new angle in understanding the essence of life. Biomineralization is a representative process for organisms in forming functional materials. In the present study, we investigated the biomineralization of iron oxides templated by Escherichia coli (E. coli) cells under oscillation. The formation of iron oxide minerals with acicular and banded morphology was observed. The surface charge of E. coli cells contributed to the biomineralization process. The surface components of E. coli cells including lipids, carbohydrates and proteins also have roles in regulating the formation and morphology of iron oxide minerals. As-prepared mineralized iron oxide nanomaterials showed activity in photocatalytic degradation of methylene blue as well as in electrocatalytic hydrogen evolution reaction. This study is helpful not only in understanding motility in biological processes, but also in developing techniques for fabricating functional nanomaterials.
Introduction
Motility is one of the most significant characteristics in organisms, especially animals or planktonic microbes. There are numerous studies and reports concerning mechanisms of various types of motility in organisms. However, few reports have been on the influence of motility on biomolecular status and biological processes.Biomineralization is one of the most abundant and essential biological processes in nature for organisms in forming functional materials. More than 60 biominerals have been found including calcium carbonate, calcium oxalate, silica, titanium dioxide, alumina, and iron oxide systems. Biomineralization relies on organisms1–3 and occurs in two ways,4 that is, bio-induced mineralization and bio-controlled mineralization. Bio-induced mineralization (BIM) is a response to changes in mineral saturation in fluids caused by cellular metabolic activity. Bio-impact mineralization or organic matrix-mediated mineralization have import roles in BIM since cells and related organic debris act as templates in regulating the formation of bio-induced minerals with distinct morphologies and structures. Applications of BIM are mostly in ecological restoration and cement-based material restoration.5,6 Bio-controlled mineralization (BCM) usually occurs in specific structures of cells, with strictly controlled composition and morphology of biominerals.4 Typical examples of BCM are the formation of magnetosomes in magnetotactic bacteria as well as structural formation of bones and teeth.4–7 BCM has found wide applications in electrode materials, sewage treatments, ecological restoration, cultural relics restoration, nano-drug carriers and cancer treatments.8–13
Due to the importance of biomineralization in both basic theories and practical applications, biomimetic mineralization attracted attentions of scientists from a broad range of disciplines including biology, chemistry, and materials science. These researches are mainly focused on roles of cell components or compositions in templating or regulating biomimetic mineralization14 and fall into two categories of mineralization systems, that is, in vitro mineralization under functions of specific biomolecules or in vivo mineralization promoting by intact cells or organisms. Biomolecules precisely template or regulate mineralization with their molecular structures or surface groups, which is advantageous in deliberately producing minerals with specific structures, morphologies, or functions. For example, silk fibroin has been used for regulating the superstructure of hemispherical CaCO3 crystals that has potential applications in preparing inorganic materials with new morphologies and special textures.15 Formation of cuprous carbonate on immune complexes was promoted by urease and could be used for detecting colorimetric signals.16 Intact cells or organisms are more efficient in producing biominerals since essential supplies of ions and energy during mineralization are accomplished by the organism that is also a self-regulating system for mineralization. For example, formation of iron ore was observed in iron bacteria and adsorption and transfer of rare earth elements were observed in Saccharomyces cerevisiae cells.17,18 Bacterial cell surface interacts with environmental ions and has multiple roles in bacterial-mediated mineralization. Chemical groups such as hydroxyl, carboxyl, amine and halide on bacterial surface are involved in adsorption and deposition of heavy metal ions as well as morphogenesis of biominerals.19–23 It inspired researchers in using bacterial-mediated mineralization for synthesizing materials with applications in ecological restoration, sewage treatment, drug carrier, etc.24–26
Factors such as temperature, pH, ion strength, biological templates and regulators have been extensively explored in biomineralization. However, there is few reports on effects of motility on biomineralization. Since motility occurs to animals or planktonic microbes all the time, it calls for the need of exploring motility effects on biomineralization.
The present study aims to investigate effects of motility on biomineralization by studying Escherichia coli templated biomineralization of iron oxides under oscillation. E. coli is a model microbe that is broadly used in studying biochemical issues. Natural mineralization was not observed on E. coli surface although it tolerates and adsorbs heavy metal ions.27 In the present study, deposition and mineralization of iron ions was induced on E. coli cell surface and compared between static or oscillatory conditions. Formation of needle-like or band-like nanomaterials of ferric oxide and ferric oxide was observed and characterized on E. coli surface under oscillation. Roles of chemical compositions of E. coli cell surface were explored on morphogenesis of iron minerals during oscillatory mineralization. Both photocatalytic and electro-catalytic performances were evaluated of as-prepared mineralized iron oxide nanomaterials. The present study calls attention to biochemical changes especially biomineralization under motility conditions.
Experimental
Bacteria cultivation
Two bacterial strains were investigated including E. coli DH5a and Bacillus subtilis B168. Bacteria were cultured as previously described.20,21 Briefly, single bacterial colony was inoculated in 3–5 ml of Luria-Bertani (LB) medium, followed by shaking overnight at 37 °C at 220 rpm. Cell suspension was inoculated in LB medium at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, followed by shaking at 220 rpm at 37 °C for 12–18 hours. Cells were harvested by centrifugation at 6000g for 5 minutes at 4 °C. Cells were then washed three times with Tris-buffered saline (TBS) buffer, pH 7.0, before later use.
50, followed by shaking at 220 rpm at 37 °C for 12–18 hours. Cells were harvested by centrifugation at 6000g for 5 minutes at 4 °C. Cells were then washed three times with Tris-buffered saline (TBS) buffer, pH 7.0, before later use.
Surface treatment of E. coli cells
To explore influence of specific chemical compositions (proteins, peptidoglycan, lipids) from bacterial cell surface on biomineralization, E. coli cells were subjected to treatment in different fashions: (1) in 1% trypsin, 50 mM Tris–HCl, pH 7.0, and incubated at 37 °C for 5 hours; or (2) in 2 mg ml−1 lysozyme, 10 mM Tris–HCl, pH 8.0, 20% sucrose, with or without 10 mM EDTA and incubated at 37 °C for 1 hour; or (3) in 95% ethanol solution and incubated at room temperature for 5 minutes; or (4) in 0.5 M Tris–HCl, pH 8.0, 0.1% SDS, 50% (v/v) chloroform, and incubated at room temperature for 20 minutes. Cells were then sedimented by centrifugation at 6000g for 3 minutes and washed three times with deionized water before later use. In the case of fashion (3) treatment, 20% sucrose was included in the wash.Bacteria templated biomineralization of iron oxides under oscillation
Fe3+ solution was prepared by dissolving ferric sulfate or ferric chloride in deionized water at a final concentration of 1 mg ml−1 with the pH adjusting to 2.40. Biomineralization of iron oxide was initiated by supplying with bacterial cells. Typically, 20 ml Fe3+ solution was supplied with 100 mg bacterial cells (wet weight). Mineralization mixtures were immediately subjected to oscillation (110 rpm or 220 rpm) at 37 °C. Mineralization was terminated by removing Fe3+ ions from mineralization mixtures via centrifugation at 5000g, 3 minutes. Mineralized bacterial cell precipitation was washed three times with deionized water and vacuum freeze-dried for 12 hours before later characterization.Liposomes templated biomineralization under oscillation
Soybean lecithin/cholesterol (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w) were dissolved in chloroform/ethanol (7
1 w/w) were dissolved in chloroform/ethanol (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) and evaporated at 37 °C for 3 hours on a rotary evaporator. The lipid was re-suspended in 30 ml deionized water and hydrated by occasionally sonication at 55 °C for 4 hours. Suspension of soybean lecithin/cholesterol (20 mg ml−1) was extruded twice through 0.22 micron filter to generate unilamellar vesicles and stored at 4 °C. Biomineralization was initiated by mixing liposome solution (20 mg ml−1) with Fe3+ solution (1 mg ml−1) at a volume ration of 1
1 v/v) and evaporated at 37 °C for 3 hours on a rotary evaporator. The lipid was re-suspended in 30 ml deionized water and hydrated by occasionally sonication at 55 °C for 4 hours. Suspension of soybean lecithin/cholesterol (20 mg ml−1) was extruded twice through 0.22 micron filter to generate unilamellar vesicles and stored at 4 °C. Biomineralization was initiated by mixing liposome solution (20 mg ml−1) with Fe3+ solution (1 mg ml−1) at a volume ration of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and subjected to oscillation (110 rpm or 220 rpm) at 37 °C. Mineralization was terminated by removing Fe3+ from mineralization mixtures via concentration at 10
4 and subjected to oscillation (110 rpm or 220 rpm) at 37 °C. Mineralization was terminated by removing Fe3+ from mineralization mixtures via concentration at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 minutes. Mineralized liposome precipitation was washed three times with deionized water and vacuum freeze-dried for 12 hours before characterization.
000g, 15 minutes. Mineralized liposome precipitation was washed three times with deionized water and vacuum freeze-dried for 12 hours before characterization.
Characterization of iron biominerals
Biominerals were characterized by scanning electron microscope (SEM), transmission electron microscopy (TEM), X-ray photoelectron spectroscopy (XPS), vibrating sample magnetometry (VSM), thermogravimetric analysis (TGA). Structures and morphologies of biominerals were characterized by the field-emission scanning electron microscopy (FESEM, Hitachi S-4800), most of which were performed using an accelerating voltage of 5 kV and a working distance of 8 mm. Transmission electron microscopy (TEM, JEM-2010HT) and high-resolution transmission electron microscope (HRTEM, JEM-2010FEF) were used to identify the crystalline structure. Chemical states and species of elements on biominerals surface were measured by XPS on an X-ray photoelectron spectrometer (VG Multilab 2000). TGA was performed in a LABSYS evo TGA (Setaram, France) at a heating rate of 5 °C min−1 from 40 °C to 900 °C. Magnetic properties were analyzed by using a vibrating sample magnetometer (Lake Shore 7400 Series VSM).Catalytic measurements of iron biominerals
Photocatalytic degradation of methylene blue was based on ref. 28. Typically, 0.5 g biominerals was added into 50 ml of 0.02 mM methylene blue, pH 3.5, and stirred in dark for 20 minutes, followed by the addition of 0.1 ml hydrogen peroxide. Degradation of methylene blue were initiated by exposing to natural light and assayed by measuring absorbance at 665 nm after centrifugation at a time interval of 5 minutes. Degradation rate (R) was determined by the formula R = (A0 − At)/A0 × 100%, where A0 and At are the absorbance at 625 nm initially and after time t.Electrocatalytic hydrogen evolution was based on ref. 29. A platinum wire was used as a counter electrode and a reversible hydrogen electrode was used as a reference electrode. The working electrode was a glassy-carbon Rotating Disk Electrode (RDE, diameter: 5 mm, area: 0.196 cm2). The Pt loading of all samples on glassy-carbon was 2.0 μg cm−2. Polarization curves were collected in 1 M KOH solutions at a rotation rate of 1600 rpm with a sweep rate of 5 mV s−1.
Results and discussion
Iron biomineralization on E. coli cell surface under oscillation
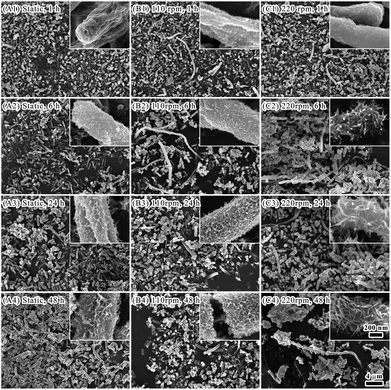
Previous work showed that E. coli surface interacts with various metals ions30 which may facilitate subsequent mineralization. The pH was adjusted to 2.54 (ref. 31) to promote iron mineralization on E. coli surface. Bacterial metabolism and biological processes rely on substance exchanges between bacterial cells and environment, which are greatly improved by oscillation. Bacterial growth in liquid media is benefited by oscillation that is a classic culturing technique in microbiology. In the present study, oscillation was attempted to promote iron biomineralization on E. coli cell surface.Morphology changes of E. coli cells during biomineralization were mainly in two aspects, that is, extreme long fibrous cell body and cell surface (Fig. 1). There was shrinkage of bacterial cells after one hour of biomineralization (Fig. 1). For cells with oscillation, it was observed deposits on cell surface as well as long fibrous cell bodies. After six hours of biomineralization, deposits were also found on surface of static cells. Flocculent protuberance was found on surface of cells from low speed oscillation at 110 rpm. Acicular minerals deposition was observed on surface of cells from high speed oscillation at 220 rpm. Lots of long fibrous cell bodies were observed in samples with oscillations. When extending biomineralization time up to 24 hours and 48 hours, there were flocculent or acicular minerals forming network on surface of cells from both static and oscillating conditions. However, there was less occurrence of long fibrous cell bodies.
Minerals depositing on bacterial surface were characterized by HR-TEM, XPS, and HAADF-STEM. Needle-like substance on E. coli surface was observed by HR-TEM (Fig. 2, Panel A). The root of the needle-like substance was on the cell surface, and grew into a slender needle (Fig. 2, Panel B). FFT algorithm analysis revealed that the crystal plane (004, 311, 111) corresponded to that of Fe3O4 (Fig. 2, Panel C). XRD spectrometry also verified there was crystal plane (311) in the minerals (Fig. S1, see ESI†). It implied that Fe3O4 was contained in minerals depositing on E. coli surface.
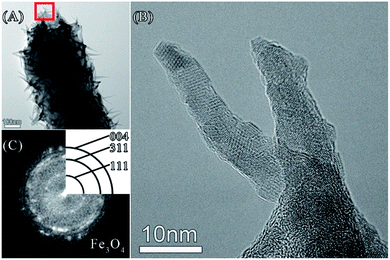 | ||
| Fig. 2 TEM image (Panel A) and HR-TEM image (Panel B, the area indicated by the red box in Panel A). Panel C shows FFT pattern of Panel B. | ||
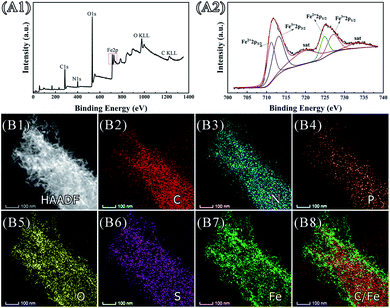
Elements including iron, oxygen, carbon and nitrogen were detected in minerals by using XPS (Fig. 3, Panel A1). Chemical states of iron ions were further analysed (Fig. 3, Panel A2). In comparing with the reference spectra and literature,32,33 it showed 727.05 eV and 712.96 eV corresponding to Fe3+ 2p1/2, Fe3+ 2p3/2, and 724.81 eV, 711.12 eV corresponding to Fe2+ 2p1/2, Fe2+ 2p3/2, respectively. The corresponding satellite peaks are typical spectra of the mixture of trivalent iron and divalent iron. These observations suggested that iron ions exist in two chemical states. Distributions of elements on cell bodies after biomineralization were visualized by using HAADF-STEM and EDX elemental mapping (Fig. 3, Panels B1–B8). Distributions of C, N, P were seen mainly within bacterial cell body due to that these elements are major organic compositions of bacterial cells. Distributions of O, S, Fe were detected on cell surface, which confirmed depositions of iron-containing minerals.
Since there could be formation of Fe3O4, magnetic properties were analysed by means of VSM (Fig. S2, see ESI†). The hysteresis loop was almost a straight line, and the magnetism did not reach saturation under the maximum magnetic field of 25 kOe under the experimental conditions, which indicated the materials were paramagnetic, superparamagnetic or antiferromagnetic. Both the coercivity and residual magnetization have the value of zero, indicating that the material was not ferromagnetic. In comparing with previous studies,34 it was suspected that a superparamagnetic or antiferromagnetic substance containing iron element might have been formed on the surface of bacterial cells.
Factors affecting iron biomineralization on E. coli surface under oscillation
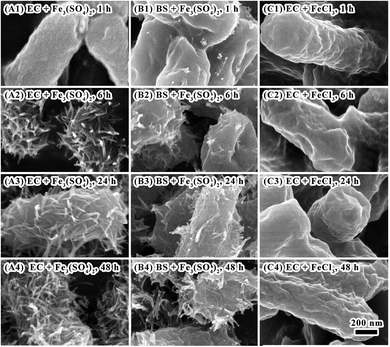
Factors affecting iron biomineralization on E. coli surface under oscillation were evaluated including bacterial strains, iron sources, and surface components of E. coli cells.Bacteria can be classified as either Gram negative or positive based on Gram stain and bacterial cell wall.35 The significant difference between the two groups is that a much larger peptidoglycan (cell wall) present in Gram positives than that in negatives which have more lipoglycans on the surface. The surface of Gram negative bacteria (with pI 4–5) are positively charged in comparing with that of Gram positive ones (with pI 2–3) in mineralization system (with pH 2.4). In the present study, iron biomineralization under oscillation was compared between E. coli (as the representative of Gram negative bacteria) and Bacillus subtilis (as the representative of Gram positive bacteria). Although at the beginning of mineralization (1 hour), there was no much difference of the morphology between the two bacteria (Fig. 4, Panels A1 and B1). After 6 hours of mineralization, there was dense layer of minerals with flocculent or acicular morphology covering on E. coli surface (Fig. 4, Panels A2–A4). While less minerals presented on B. subtilis surface (Fig. 4, Panels B2–B4; Fig. S3, see ESI†). Therefore, the charge of cell surface made significant contributions to iron oxide mineralization. Since element S was observed on E. coli surface during iron mineralization using ferric sulfate as the ferric source (Fig. 3, Panel B6), sulfate might have roles in iron biomineralization. When using ferric chloride as alternative ferric source for iron biomineralization, no mineralization occurred on E. coli cell surface (Fig. 4, Panels C1–C4). This might be due to that the size of sulfate anions is larger than that of chloride anions. It facilitates interactions between sulfate anions and E. coli cell surface and subsequent mineralization and deposition of iron minerals.
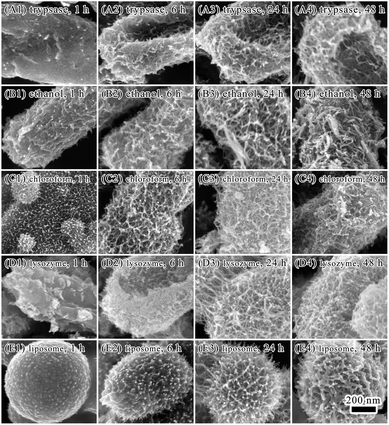
The E. coli cell surface was further explored to understand its influence on biomineralization. Major components on E. coli surface are proteins, carbohydrates, lipids.35 Cells were treated with trypsase or ethanol to eliminate the influences of proteins on iron biomineralization on cell surface. By treating cells with trypsase to degrade proteins on cell surface, most of the cell bodies were sunken (Fig. 5, Panels A1–A4). Minerals forming on cell surface were similar to that on untreated cells, where minerals grew and crosslinked gradually to form a network covering on bacterial surface.
By treating cells with 95% ethanol to dehydrate and denature proteins on cell surface, formation of minerals on cell surface occurred in 1 hour of biomineralization (Fig. 5, Panels B1–B4). Formation of a dense layer with flocculent minerals on cell surface was observed in 6 hours. After 48 hours of mineralization, spindle-like mineral particles were observed on cell surface. It is possible that the formation and size development of acicular or baggy iron oxide nanoparticles occurred in the first 24 hours of biomineralization under the regulation of proteins. Formation of the shape of spindle were after 48 hours of mineralization.
By using chloroform–SDS to dissolve lipids and proteins on cell surface and increase permeability of cell walls, deposition of minerals on cell surface were observed in 1 hour (Fig. 5, Panels C1–C4). The E. coli cell body was encapsulated by a layer of deposited minerals with hairy minerals growing on it. It indicated that treating with chloroform–SDS facilitated absorbance of ferric ions on bacterial surface and improved minerals growth. Although morphology of minerals on chloroform–SDS treated cell surface were similar to that of untreated cells, minerals layer were thicker on the chloroform–SDS treated bacteria after 24 and 48 hours treatment.
Lysozyme can destroy bacterial cell walls by breaking glycoside bonds. EDTA can bind to lipopolysaccharides on cell surface and destabilize cell wall. The combination of lysozyme and EDTA is efficient in removing bacterial cell wall and exposing bacterial cell membrane. After lysozyme–EDTA treatment (Fig. 5, Panels D1–D4), bacterial cells were seriously damaged and no longer able to keep the rod-shaped body. A dense layer of minerals formed on cell surface with a few hairy spherical minerals sporadically grew on the minerals layer. Minerals grew faster on the lysozyme–EDTA treated bacterial cells than untreated ones. If bacterial cells were merely treated by lysozyme, only sporadic small minerals presented on cell surface (Fig. S4, see ESI†).
Lipid and proteins are two major components of cell membrane. To explore the roles of lipid on mineralization under oscillation, reconstitute liposomes of soybean lecithin/cholesterol were subjected to mineralization (Fig. 5, Panels D1–D4, S5 and S6, see ESI†). The size of liposome spheres is between 500 nm and 1 mm. After mineralization of 1 hour, there were granular minerals depositing on surface of liposomes. When extending mineralization time, mineral layer was getting thicker with the deposited minerals changing from granular to inverted spiny strips. The liposomes also became burrs with many spines. The morphology of minerals on the liposome surface was similar to that on E. coli cells upon ethanol or chloroform treatments (Fig. 5, Panels B1–B4). It suggested that lipids may have critical roles on minerals growing on E. coli cell surface. However, lipopolysaccharides and proteins may also be involved in mineralization process. Iron minerals on liposome surface were confirmed by EDAX elemental analysis, which showed that the iron content was 4.08%. The morphology of minerals on cell surface with different treatments was significantly different after 1 hour or 6 hours of mineralization. However, the morphology of minerals on cell surface was similar after 24 and 48 hours of mineralization, despite of treating methods. This could be due to that electrostatic attraction of bacterial surface groups made major contributions to the adsorption of iron ions on bacterial surfaces. Binding of iron ions relied on exposure of surface groups inducing by different treating methods. Therefore, the structure of biomolecules on cell surface can greatly affected mineralization and lead to variability of morphology of minerals in the initial stage of mineralization. Once there was formation of mineral layer on cell surface, it could be template for further mineralization and lead to similar morphology of minerals in the later stage of mineralization.
Bacterial templated iron biominerals with fibrous shape
It was observed the formation of iron biominerals with irregular fibrous shape under oscillation (Fig. 1). These minerals appeared in 1 hour of mineralization (Fig. 1, Panels B1 and C1). After 6 hours of mineralization, there were more fibrous minerals in the oscillation system (Fig. 1, Panels B2 and C2). However, the amount of fibrous minerals significantly reduced after 24 and 48 hours of mineralization (Fig. 1, Panels B3, B4, and C3, C4).The surface of the fibrous minerals was analysed by TEM and HR-TEM (Fig. 6). HR-TEM and FFT algorithm analysis (Fig. 6, Panels A1–A3) showed that these minerals are nano scaled ferric oxide needles that are similar to that deposited on the surface of single E. coli cells. These fibrous minerals were further examined with TEM and EDX mapping. It was showed that the mineral constituted by iron, carbon, nitrogen, phosphorus, sulfur and oxygen elements. Iron element mainly distributed on the surface with carbon element inside. Therefore, it is possible that aggregated E. coli cells formed the main body of the fibrous with iron mineralization on the surface. At the early stage of mineralization, iron oxide deposited on E. coli surface may facilitate the ordered aggregation of cells under oscillation and lead to the formation of fibrous minerals. Along with the biomineralization process, more iron minerals deposited on the cell surface and increased the fragility of fibrous minerals. It made it easy for the fibrous minerals to break under oscillation. Therefore, less fibrous minerals were observed after 24 and 48 hours of mineralization.
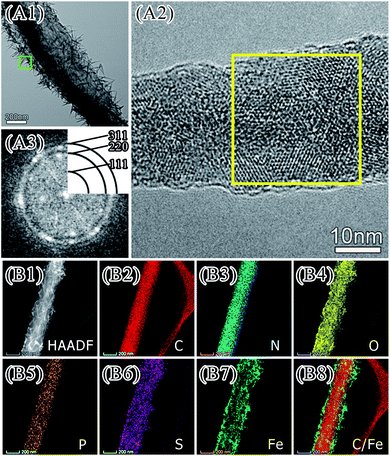 | ||
| Fig. 6 TEM image (Panel A1) and HR-TEM image (Panel A2, the area indicated by the green box in Panel A1). Panel A3 shows FFT pattern of yellow zone of Panel A2. | ||
Catalytic performance of iron biominerals
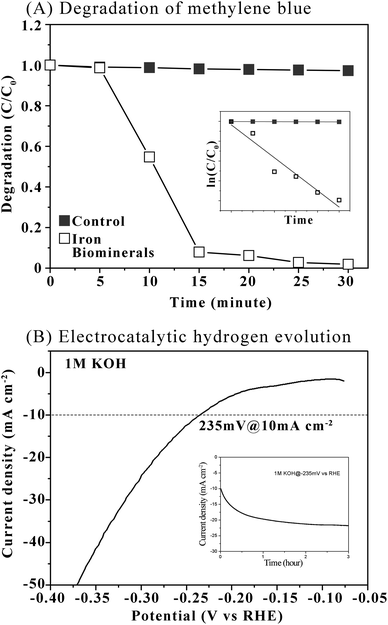
Photocatalytic performance of iron biominerals was evaluated by measuring degradation of methylene blue under visible irradiation (Fig. 7, Panel A). Percentage of degradation (C/C0) = (1 − Ct/C0) × 100, where C0 and Ct in Fig. 7 (Panel A) represent the initial concentration after the adsorption–desorption equilibrium for t minute and the real-time concentration of methylene blue at time t minute, respectively. The degradation of methylene blue reached 92.11% in 15 minutes and 98.11% in 30 minutes. These results demonstrated the reasonable photocatalytic activity of as prepared iron biominerals. The linear relationship between ln(C/C0) and irradiation time (Fig. 7, Panel A, inset) indicated that photocatalytic degradation of methylene blue followed the pseudo first-order law. In the present study, the apparent rate constant was calculated as 0.17 ± 0.05 min−1.The hydrogen evolution reaction (HER) is promising in producing clean and renewable hydrogen resources. In the present study, the HER performance of iron minerals as an efficient, durable, and inexpensive hydrogen evolution electrode was investigated. Polarization curves were collected in 1 M KOH solutions at a rotation rate of 1600 rpm with a sweep rate of 5 mV s−1 at room temperature. Before subjecting to electrocatalytic measurement, iron biominerals were calcinated at 700 °C for 4 hours (Fig. S7, see ESI†). The iron oxide mineral electrode achieved the best activity with the overpotential of 235 mV at the current of 10 mA cm−2 (Fig. 7, Panel B). The catalytic stability of iron oxide minerals was also measured at 10 mA cm−2 in the same electrolyte, which showed that the stable current with a small deviation after one hour at overpotential of −20 mV was presented (Fig. 7, Panel B, inset).
In future, more investigation concerning catalytic performances of acquired biominerals will be carried out to provide detailed information and data including the UV-vis spectra, ROS generation, CV curves.
Conclusions
This article reported effects of motility on biomineralization of iron oxide on E. coli surface. Under oscillation, the bacterial E. coli templated the formation of iron oxide minerals with acicular and banded morphology. The surface charge of E. coli cells contributed to the biomineralization process. The surface components of E. coli cells, that is, lipids, carbohydrates and proteins, regulated the formation and morphology of iron oxide minerals. The morphology of iron oxide minerals formed on liposome surface was similar to that on E. coli cell surface, implying that lipids on E. coli cell surface played a critical role in morphogenesis of iron oxide minerals. As-prepared mineralized iron oxide nanomaterials showed activity in photocatalytic degradation of methylene blue as well as in electrocatalytic hydrogen evolution reaction, implying potential uses of this material in environment and energy.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Supported by National Natural Science Foundation of China (31771032, 51911530153, 51832003).Notes and references
- I. Aksay, M. Traus, S. Manne, I. Honma, N. Yao, L. Zhou, P. Fenter, P. Eisenberger and S. Gruner, Science, 1996, 273, 892 CrossRef CAS PubMed.
- S. Stupp and P. Braun, Science, 1997, 277, 1242 CrossRef CAS.
- X. Zhao, J. Yang, L. McCormick and J. Fendler, J. Phys. Chem., 1992, 96, 9933 CrossRef CAS.
- D. Bazylinski, Encyclopedia of Materials: Science & Technology, 2001, p. 441 Search PubMed.
- W. Muynck, D. Belie and W. Verstraete, Ecol. Eng., 2010, 36, 118 CrossRef.
- A. Amiri and Z. Basaran, Construct. Build. Mater., 2018, 165, 655 CrossRef CAS.
- S. Yao, B. Jin, Z. Liu, C. Shao, R. Zhao, X. Wang and R. Tang, Adv. Mater., 2017, 29, 1605903 CrossRef.
- S. Shankar, A. Ahmad, R. Pasricha and M. Sastry, J. Mater. Chem., 2003, 13, 1822 RSC.
- L. Li, X. Yang, A. Li, T. Zhang and Y. Liu, Appl. Mech. Mater., 2011, 71–78, 2831–2835 CAS.
- D. Tamayo-Figueroa, E. Castillo and P. Brandão, World J. Microbiol. Biotechnol., 2019, 35, 58 CrossRef PubMed.
- T. Li, Y. Hu and B. Zhang, Front. Microbiol., 2018, 9, 1884 CrossRef.
- S. Xie, G. Yin, X. Pu, S. Xie, G. Yin, X. Pu, Y. Hu, Z. Huang, X. Liao, Y. Yao and X. Chen, Curr. Drug Delivery, 2017, 13, 999 Search PubMed.
- S. Kim, L. Palanikumar, H. Choi, M. Jeena, C. Kim and J. Ryu, Chem. Sci., 2018, 9, 2474 RSC.
- I. Weiss and F. Marin, Biomineralization: From Nature to Application, 2008, vol. 4, p. 71 Search PubMed.
- T. Chen, P. Shi, Y. Li, T. Duan, Y. Yu, X. Li and W. Zhu, CrystEngComm, 2018, 20, 2366 RSC.
- B. Li, G. Lai, B. Lin, A. Yu and N. Yang, Sens. Actuators, B, 2018, 262, 789 CrossRef CAS.
- J. Miot, K. Benzerara, G. Morin, A. Kappler, S. Bernard, M. Obst, C. Férard, F. Skouri-Panet, J. Guigner, N. Posth, M. Galvez, G. Brown Jr and F. Guyot, Geochim. Cosmochim. Acta, 2009, 73, 696 CrossRef CAS.
- M. Jiang, T. Ohnuki and S. Utsunomiya, Geomicrobiol. J., 2018, 35, 375 CrossRef CAS.
- K. Saranya, A. Sundaramanickam, S. Shekhar, M. Meena, R. Sathishkumar and T. Balasubramanian, J. Environ. Manage., 2018, 222, 396 CrossRef CAS PubMed.
- P. Dhanwal, A. Kumar, S. Dudeja, H. Badgujar, R. Chauhan, A. Kumar, P. Dhull, V. Chhokar and V. Beniwal, Water Environ. Res., 2018, 90, 424 CrossRef CAS.
- M. Sahmoune, Microchem. J., 2018, 141, 87 CrossRef CAS.
- A. Choińska-Pulit, J. Sobolczyk-Bednarek and W. Łaba, Ecotoxicol. Environ. Saf., 2018, 149, 275 CrossRef PubMed.
- R. Rani and M. Saharay, RSC Adv., 2019, 9, 1653 RSC.
- F. Li, W. Wang, C. Li, R. Zhu, F. Ge, Y. Zheng and Y. Tang, J. Hazard. Mater., 2018, 358, 178 CrossRef CAS PubMed.
- M. Li, X. Cheng and H. Guo, Int. Biodeterior. Biodegrad., 2013, 76, 81 CrossRef CAS.
- M. Ozaki, S. Sakashita, Y. Hamada and K. Usui, Protein Pept. Lett., 2018, 25, 15 CrossRef CAS PubMed.
- T. Beveridge and S. Koval, Appl. Environ. Microbiol., 1981, 42, 325 CrossRef CAS PubMed.
- A. Lassoued, M. Lassoued, B. Dkhil, S. Ammar and A. Gadri, J. Mater. Sci.: Mater. Electron., 2018, 29, 8142 CrossRef CAS.
- J. Ying, G. Jiang, Z. Cano, L. Han, X. Yang and Z. Chen, Nano Energy, 2017, 40, 88 CrossRef CAS.
- T. Beveridge and S. Fyfe, Can. J. Earth Sci., 1985, 22, 1893 CrossRef CAS.
- S. Langley and T. Beveridge, Appl. Environ. Microbiol., 1999, 65, 489 CrossRef CAS PubMed.
- T. Yamashita and P. Hayes, Appl. Surf. Sci., 2008, 254, 2441 CrossRef CAS.
- G. Bhargava, I. Gouzman, C. Chun, T. Ramanarayanan and S. Bernasek, Appl. Surf. Sci., 2007, 253, 4322 CrossRef CAS.
- A. Darbeheshti, M. Ghazi and M. Izadifard, Mater. Res. Express, 2017, 4, 106110 CrossRef.
- G. Tortora, B. Funke, C. Case, D. Weber and W. Bair III, Microbiology: An Introduction, Pearson, 13th edn, 2018 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00847a |
| This journal is © The Royal Society of Chemistry 2021 |