Open Access Article
Open Access ArticleElectrochemical sensing of blood proteins for mild traumatic brain injury (mTBI) diagnostics and prognostics: towards a point-of-care application†
Nadezda Pankratova
,
Milica Jović
 and
Marc E. Pfeifer
*
and
Marc E. Pfeifer
*
University of Applied Sciences and Arts Western Switzerland (HES-SO Valais-Wallis), School of Engineering, Institute of Life Technologies, Diagnostic Systems Research Group, Route du Rawil 64, 1950 Sion, Switzerland. E-mail: marc.pfeifer@hevs.ch
First published on 12th May 2021
Abstract
Traumatic Brain Injury (TBI) being one of the principal causes of death and acquired disability in the world imposes a large burden on the global economy. Mild TBI (mTBI) is particularly challenging to assess due to the frequent lack of well-pronounced post-injury symptoms. However, if left untreated mTBI (especially when repetitive) can lead to serious long-term implications such as cognitive and neuropathological disorders. Computer tomography and magnetic resonance imaging commonly used for TBI diagnostics require well-trained personnel, are costly, difficult to adapt for on-site measurements and are not always reliable in identifying small brain lesions. Thus, there is an increasing demand for sensitive point-of-care (POC) testing tools in order to aid mTBI diagnostics and prediction of long-term effects. Biomarker quantification in body fluids is a promising basis for POC measurements, even though establishing a clinically relevant mTBI biomarker panel remains a challenge. Actually, a minimally invasive, rapid and reliable multianalyte detection device would allow the efficient determination of injury biomarker release kinetics and thus support the preclinical evaluation and clinical validation of a proposed biomarker panel for future decentralized in vitro diagnostics. In this respect electrochemical biosensors have recently attracted great attention and the present article provides a critical study on the electrochemical protocols suggested in the literature for detection of mTBI-relevant protein biomarkers. The authors give an overview of the analytical approaches for transduction element functionalization, review recent technological advances and highlight the key challenges remaining in view of an eventual integration of the proposed concepts into POC diagnostic solutions.
Introduction
Traumatic Brain Injury (TBI) is a leading cause of disability and death in both developed and developing countries.1 Nearly seventy million people suffer from TBI worldwide every year.2 In Europe alone 2.5 million people experience TBI each year, 1 million of them are being hospitalized, while 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people die.1 This results in significant public health implications, given the frequency of head impacts particularly among adolescents and in contact sports.3 The considerable financial burden that arises from TBI depends on many factors, mainly duration of rehabilitation and long-term patient care, costs of prescriptions, therapies, need for medical equipment, costs of employment loss, etc. The overall global economic burden of TBI is estimated at about 400 billion dollars, meaning that 1 out of every 200 dollars generated in the global economy is spent to cover the costs and consequences of TBI.1
000 people die.1 This results in significant public health implications, given the frequency of head impacts particularly among adolescents and in contact sports.3 The considerable financial burden that arises from TBI depends on many factors, mainly duration of rehabilitation and long-term patient care, costs of prescriptions, therapies, need for medical equipment, costs of employment loss, etc. The overall global economic burden of TBI is estimated at about 400 billion dollars, meaning that 1 out of every 200 dollars generated in the global economy is spent to cover the costs and consequences of TBI.1
Diagnosis of TBIs is mainly based on patient's medical history, findings on neurological examination, clinical assessment scales and neuroimaging tools, such as computed tomography (CT) and magnetic resonance imaging (MRI), single-photon emission computed tomography (SPECT), or positron emission tomography (PET) of the brain. Based on the severity of the condition TBIs are typically classified as mild (GCS 13–15), moderate (GCS 9–12) and severe (GCS 3–8), using the Glasgow Coma Scale (GCS) which is an assessment of conscious level of the patient motor, eye, and verbal responses. Mild TBI (mTBI) accounts for 80–90% of all cases and is the most prevalent form of brain injury. Patients with mTBI frequently develop non-specific symptoms, including fatigue, headaches, visual or sleep disturbances, depression, or seizures, which can occur immediately following the injury or after several days or weeks. Occurrence of mTBI, especially repetitive, has been associated with an increased risk of long-term cognitive and neuropathological disorders. Moderate and severe TBIs are easily diagnosed, often evident from patient's history and injury signs or abnormalities detected on the neuroimaging screen. However, diagnosis of mTBI is rather challenging. The GCS can be inaccurate in distinguishing between mild and moderate TBIs and provides a poor prediction of patient outcome (not appropriate for patients with prior neurological conditions). mTBI is typically not associated with any structural changes on brain MRI and is difficult to be assessed by standard diagnostic workup. Furthermore, MRI and CT scans are quite costly and difficult to be made available on-site (undeveloped areas, emergency rooms, battlefield, sport facilities, car accident sites), not to mention the harm of exposure to ionizing radiation during a head CT and difficulties with exposure to strong magnetic fields in patients with metal implants (e.g., pacemaker, heart valve, cochlear implant, etc.). Therefore, there is a growing need in additional diagnostic tools for aiding both diagnosis and prognostics in order to enable an accurate, inexpensive and fast triage and decision-making in the treatment of mTBI.4–8
Biomarkers relevant to mTBI
Biomarkers are promising candidates for aiding the identification of mTBI and prognosis. They could be classified as diagnostic (indicating the presence or absence of a specific physiological/pathophysiological state or disease), prognostic (categorizes patients by degree of risk for disease occurrence or progression of a specific aspect of a disease), or predictive (categorizes patients by their likelihood of response to a particular treatment relative to no treatment). A good biomarker should provide good specificity (be uniquely present in the central nervous system and accurately reflect the extent of brain damage), high sensitivity (high abundance in the analyzed fluid and easy/sensitive detection), as well as be of use for estimating the therapeutic efficacy/intervention.9,10 In the case of mTBI the most promising are brain protein biomarkers that can be safely quantified by analyzing biofluids, such as cerebrospinal fluid (CSF), serum and plasma. mTBI biomarkers are mostly studied in reference to specific injured cell types, including markers of glial cell injury (glial fibrillary acidic protein, GFAP; calcium binding protein B, S100β; myelin basic protein, MBP), axonal and neuronal injury (ubiquitin carboxyl-terminal hydrolase-L1, UCH-L1; neuron-specific enolase, NSE; Tau protein and phosphorylated-Tau [p-Tau]), and due immunological (e.g. antibodies) or inflammatory responses (cytokines).11 Table 1 gives the detailed list of protein biomarkers which have been shown to be relevant to mTBI and mTBI-recovery.12 An extensive survey of blood biomarkers relevant for head (brain) injuries can be found in the recent review article by Zoe S. Gan et al.12 Peptides and cleavage products were omitted in the present work, as well as biomarkers related specifically to severe TBI (sTBI) diagnostics and prognostics. Discussion of autoantibodies as potential biomarkers has been put aside here, mainly due to the fact that not sufficient clinical data has been reported yet in order to confirm diagnostic and prognostic values of autoantibodies for mTBI.13 Nevertheless, it is worth mentioning that some of the autoantibodies have been shown to be relevant e.g. for repetitive sub-concussive events (anti-S100β14,15) or severe trauma (anti-GFAP15,16).| Biomarker | Physiological concentrationa | ||
|---|---|---|---|
| Abbreviation | Full name | Normal | Mild TBI |
| a Physiological concentrations are indicated for human serum, unless otherwise specified. Values reported in samples other than blood/serum/plasma (e.g., sweat, urine, muscle-on-tissue etc.) are not considered.b Based on the results for the protein levels in postmortem cortical tissue, studies conducted for sTBI.c Data for uncomplicated mild TBI.d Based on animal model.e The terms pNF-H and NF–H are used interchangeably in the literature, due to the fact that the NF heavy chain is always phosphorylated.86 | |||
| BDNF | Brain-derived neurotrophic factor | RG: 15.8–79.8 ng mL−1 (ref. 25) MN: 32.7 ng mL−1 (ref. 25) | MD: 8.3 ng mL−1 (ref. 26) |
| CRP | C-reactive protein | MN: 2.1 μg mL−1 MD: 1.2 μg mL−1 (HP) (ref. 27) | Elevated (ref. 29 and 30) |
| MN: 1.4 μg mL−1 (ref. 28) | |||
| GFAP | Glial fibrillary acidic protein | RG: 0.002–0.049 pg mL−1 (ref. 31) MD: 0.004 pg mL−1 (ref. 31) | ≥0.033 pg mL−1 (ref. 31) CO: 22 pg mL−1 (ref. 32) |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor | <145 pg mL−1 (ref. 33) | Elevated (ref. 34)b |
| h-FABP | Heart-fatty acidic binding protein | <5.5 ng mL−1 (ref. 35) MN: 3.78 ng mL−1 (ref. 36) | CO: 2.62 ng mL−1 (HS/HP) (ref. 37) |
| IL-6 | Interleukin-6 | <5.9 ng L−1 (ref. 38) | Elevated (ref. 39) |
| IL-8 | Interleukin-8 | 5–18 pg mL−1 (ref. 40) <62 ng L−1 (HP) (ref. 38) | Elevated (ref. 41) |
| IL-10 | Interleukin-10 | RG: 4.8–9.8 pg mL−1 MN: 7.1 pg mL−1 (ref. 42) | Elevated (ref. 43) |
| MMP-2 | Matrix metallo-proteinase-2 | MN: 251.4 ng mL−1 (HS, PL) (ref. 44) | Elevated (ref. 45 and 46)c |
| MT3 | Metallo-thionein | MN: 0.51 ng mL−1 (ref. 47) | MN: 0.13 ng mL−1 (ref. 47) |
| NCAM | Neuron cell adhesion molecule (CD56) | MN: 54.82 μg mL−1 (ref. 48) | Elevated (ref. 49 and 50)d |
| NFL | Neuro-filament light | MD: 14.5 pg mL−1 (ref. 51) 13 pg mL−1 (ref. 52) | RG: 2.6–246.9 pg mL−1 (ref. 53) MN: 32.1 pg mL−1 |
RG: 4.1–23.5 pg mL−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) (ref. 53) (ref. 53) |
MD: 19 pg mL−1 (ref. 53)c | ||
| NGB | Neuroglobin | MN: 10.31 ng mL−1 (ref. 54) 14.54 ng mL−1 (ref. 55) | MN: 17.58 ng mL−1 (mTBI) (ref. 55) 28.76 ng mL−1 (sTBI) (ref. 55) |
| NRGN | Neurogranin | MD: 0.02 ng mL−1 (ref. 56) | Elevated (ref. 47 and 56) |
| NSE | Neuron-specific enolase | 5–15 ng mL−1 (ref. 57) MN: 7 ng mL−1 (ref. 58) | CO: 20 ng mL−1 (ref. 59) MN: 14 ng mL−1 (mTBI) (ref. 58) |
| MN: 3.5 ng mL−1 (ref. 47) | 20 ng mL−1 (mdTBI) (ref. 58) 32 ng mL−1 (sTBI) (ref. 58) | ||
| S100β | S100β calcium-binding protein | MD: 50 pg mL−1 (HP) (ref. 60) <0.11 pg mL−1 (ref. 38) | ≥100 pg mL−1 (sTBI, HP) (ref. 61) >75 pg mL−1 (ref. 62) |
| CO: 0.042 μg L−1 (HS/HP) (ref. 37) | |||
| T-Tau | Total tau (P- + non-phosphor.) | MN: 86 pg mL−1 (ref. 63) RG: 52.2–215 pg mL−1 (T-Tau) (ref. 63) | MN: 188 pg mL−1 (ref. 63) RG: 52.2–850 pg mL−1 (T-Tau) (ref. 63) |
| MN: 289 pg mL−1 (ref. 64) | Elevated(ref. 65) | ||
| UCH-L1 | Ubiquitin C-terminal hydrolase | MD: 0.09 ng mL−1 (ref. 66) RG: 0.03–0.11 ng mL−1 (ref. 66) | ≥1 ng mL−1 (sTBI, HP) (ref. 61) CO: 327 pg mL−1 (ref. 32) |
| VCAM-1 | Vascular cell adhesion protein 1 | RG: 449–1103 ng mL−1 (ref. 67) MD: 631 ng mL−1 (ref. 67) | Lowered (ref. 39 and 68) |
| Biomarkers without reported EC detection approaches (as of December 2020) | |||
| BMX | Bone marrow tyrosine kinase on | MN: 6.08 pg mL−1 (ref. 69) | MN: 7.47 pg mL−1 (ref. 69) |
| Chromosome X | |||
| CKBB | Creatine kinase B type | <3 pg mL−1 (sTBI, HP) (ref. 61), <6 pg mL−1 (ref. 70) | >6 pg mL−1 (ref. 70), ≥3 pg mL−1 (sTBI, HP) (ref. 61) |
| ICAM-1 | Intracellular adhesion molecule-1 | MN: 236.9 ng mL−1 (ref. 71) RG: 210–306 ng mL−1 (ref. 72) | Elevated (ref. 73) |
| MDA-LDL | Malondialdehyde modified low density | MDA-LDL-to-LDL-C-ratio: 1.16 | n/a |
| Lipoprotein | LDL-C: MN 1270 μg mL−1 (ref. 74) | ||
| NFM | Neurofilament medium | MD: 2.29 ng mL−1 (ref. 75) RG: 0.26–8.57 ng mL−1 (ref. 75) | RG: 0.21–202.2 ng mL−1, MD: 7.89 ng mL−1 (mTBI) (ref. 75) |
| RG: 3.48–45.4 ng mL−1, MD: 13.3 ng mL−1 (sTBI) (ref. 75) | |||
| Nogo-A | Neurite outgrowth inhibitor protein | MN: 128 ng mL−1 (ref. 55) | MN: 220.09 ng mL−1 (mTBI), 315.67 ng mL−1 (sTBI) (ref. 55) |
| pNF-H (NF–H) e | (Phosphorylated) neurofilament heavy protein | RG: 189.59–634.12 pg mL−1 (ref. 76) MN: 311.98 pg mL−1 (ref. 76) | Elevated (ref. 77) |
| E-selectin | E-selectin | MD: 39.6 ng mL−1 (ref. 78) RG: 33.2–44 ng mL−1 (ref. 78) | Elevated (ref. 79)d |
| SNTF | Calpain-derived αII-spectrin N-terminal fragment | Absent from healthy neurons (ref. 80) | Elevated (ref. 80 and 81) |
| Ub | Ubiquitin | <100 ng mL−1 (ref. 82) MN: 29.6 (fUb), 4.1 ng mL−1 (mtUb) (ref. 83) | Elevated (ref. 85) |
| MN: 37.2/126 pg mL−1 (fUb) (ref. 84) MN: 3.4/3.86 pg mL−1 (mtUb) (ref. 84) | |||
The first applications of mTBI biomarkers in medical practice dates from 2015, when S100β has been included in an algorithm of the Scandinavian guidelines to triage patients with mTBI to CT after TBI17 (the cost for S100β analysis in Sweden is 21 euro, while the cost of CT scan is 130 euro).18 Furthermore, in February 2018 the first biomarker core lab blood assay proposed by Banyan Biomarkers has been cleared by the FDA (based on the 2018 ALERT-TBI pivotal trial with 1959 mild-to-moderate TBI patients).19 The latter relies on a chemiluminescent-based ELISA for measuring the concentrations of two proteins, GFAP and UCH-L1, and has been shown to be able to predict the TBI-positive CT scan with the sensitivity of 97.5% and negative predictive value of 99.6%.19 Practically the latter means that in more than 33% of the cases the patients being suspected of brain injury can be ruled out prior to CT scan.19 Following the approval of the test via de novo FDA pathway and into the Class II, a new product code has been created to designate the brain assessment tests. Thus, the subsequent (e.g., POC) tests that have the same use will be classified into the same product code and will be reviewed by the 510(k) regulatory pathway. The search for the ‘ideal’ mTBI biomarkers still faces many challenges, such as insufficient specificity, as well as influence of age, gender, injury severity, pre-existing medical conditions and other individual differences.12,20,21 TBI biomarker discovery today is mainly focused on detection at very early stages after injury (hyper acute and acute TBI), which will allow for implementation of patient treatments at an earlier time point. For the chronic stages of mTBI, Tau protein and phosphorylated-Tau are under examination as markers of neurodegeneration for in vivo detection of neurodegenerative disorders which are possible long-term sequelae of mTBI such as Alzheimer's disease (AD) and chronic traumatic encephalopathy (CTE).22 Neurogranin could be mentioned here as one of the prospective candidates that can be measured in whole blood samples – researchers aim at evaluating its potential role for avoiding CT overuse in mTBI diagnostics. Myelin Basic Protein (MBP) was also highlighted in the literature as potential negative predictive biomarker for the absence of TBI.23 The role of the CRP biomarker within mTBI related publications is dual: on one hand, CRP is often employed as a model analyte for method development, on the other hand, despite being an inflammatory non-specific biomarker it has a potential of being part of a future mTBI multi-biomarker panel.
The choice of body fluid for mTBI biomarker detection is one of the key aspects to be considered. CSF is attractive because it is in contact with the neural interstitial fluid and detection of CSF biomarkers should reflect neural tissue injury.24 On the other hand, the disadvantage is the requirement for lumbar puncture, which is an invasive procedure and unlikely combinable with a decentralized POC diagnostic application.23 As most of the in vitro diagnostic (IVD) assays are approved for use with blood samples the scope of this review will be limited to those (whole blood, serum, plasma). However, it must be noted that mTBI biomarker detection and quantification using blood sampling is still challenging. Once the neural tissue has been injured, the mTBI biomarkers need to pass through a biophysical barrier into the bloodstream. Many biomarkers that have excellent specificity for mTBI may not be present in blood in sufficiently high concentrations to be detectable using currently available assays. Detection of biomarkers in the peripheral blood is limited by the clearance from blood by liver or kidney, proteolytic degradation, and their permeability through the blood brain barrier (depending on the molecular size and charge). Due to above mentioned facts the concentrations and kinetic profiles of mTBI biomarkers in blood are quite difficult to determine.23 While concentrations of some biomarkers continue to rise within days or even weeks, many biomarkers peak early and decline within a few hours after the injury, depending on the molecular and cellular origin and the release mechanism.87,88 Moreover, it must be noted that plasma and serum often contain different amount of proteins and the concentration of the proteins is strongly affected by the blood pre-treatment procedure (anti-clotting factors, clotting reagents).89–91 Some studies indicate that serum samples are not recommended for quantifying certain biomarkers (e.g. some small proteins and peptides) and plasma is very much preferred in general cases.89,92 However, for many biomarkers it has been shown that the serum protein concentrations do correlate with plasma concentrations90 and thus both serum and plasma are being used for biomarker detection. The choice of the blood fraction to be analyzed may have important implications and depends on the target analyte. In some cases, it is quite straightforward. For example, in the case of the BDNF protein which is known to be bound by the platelets in blood, total concentration can be given by analyzing the serum sample, while free (circulating) BDNF can be detected by analyzing the plasma sample. Currently, the detection of mTBI-relevant biomarkers in body fluids is mostly performed using: (1) clinical analyzers in core/centralized labs that run high-throughput immunoassays (e.g., 96-well plate based) predominantly with fluorogenic, chemiluminescent and colorimetric readout modalities, and (2) biosensor-based approaches described in research literature that employ either electrochemical (EC) or spectroscopic detection principles. The Abbott i-STAT is a rare example of an EC-based (portable) in vitro POC diagnostic device for protein quantification (e.g. cardiac troponin I in blood and plasma).93 To the best of our knowledge, all other EC-based POC (portable) diagnostic applications target biomarkers other than proteins, such as e.g. the ‘game-changing’ continuous glucose sensing based on enzymatic amperometric detection. EC measurement of large protein biomarkers appears more challenging due to issues such as nonspecific adsorption of biological fluid, very low abundance of most protein biomarkers, requirement of extremely good specificity due to various interferences of other biomolecules present in physiological samples.94
In the last years, there has been a significant number of publications focusing on the EC sensing techniques for protein detection and quantification. The interest in EC techniques for mTBI research field accounts for the following facts: unlike spectroscopic methods, EC measurements are not affected by sample turbidity, colour, quenching, or interference from absorbing and fluorescent compounds commonly found in biological fluids. EC techniques are easily adaptable to relatively cheap mass production and miniaturization to circuit board levels with low power consumption.95 The low fabrication costs, along with potential high sensitivity, fast response time, small sample volume requirements, low cost of operation, possibility of miniaturization and integration for multianalyte detection have made EC biosensors an attractive tool for mTBI biomarker detection, especially from the point-of-view of possible realization of a POC device for concussion diagnostics.
Table 2 provides a proposal for a target product profile (TPP) of an mTBI POC diagnostic device with presumed key product requirements specifications of the future system. In the next sections we will give an overview of the various sensor designs and types of electrochemical biosensors reported in the literature for detection of mTBI relevant proteins and in the Conclusions and future outlook section we will discuss how they respond to the TPP and their potential implementation into the POC concept.
 |
Presumed key product requirements specifications of a future system for POC diagnostics and prognostics of mTBI. |
| The number of biomarkers necessary to achieve sufficient diagnostic specificity is an assumption based on recent and ongoing clinical studies.21,98–100 | |
| Diagnostic sensitivity ≥ 95% | Number of biomarkers detected (multiplex multivariate analysis) ≥3 (∼5–8) |
| Diagnostic specificity ≥ 75% | Capillary whole blood (finger prick) sample volume ≤50 μL |
| Intra-assay %CV precision ≤10% | Linear range (i.e., upper limit of quantification, ULOQ relative to LDL) ×50 above concentration cut-off value for specific biomarker, e.g., ≥ 1.1 ng mL−1 for GFAP |
| Inter-assay %CV precision ≤15% | Time-to-results ≤10 min |
| Reagent shelf life ≥6 months | Hands-on-time ≤5 min |
| Lower Detection Limit (LDL) 1/10 of the cutoff (CO) value to distinguish mTBI from physiological concentration for specific biomarker, e.g., ≤2.2 pg mL−1 for GFAP | |
Design of electrochemical sensors for detection of protein biomarkers relevant to mTBI
This section of the review provides the details on various (bio)sensor designs and components reported in the literature for detection of 19 mTBI-relevant biomarkers, based on the survey that includes 127 publications published until December 2020. The literature search related to NSE, IL-6, IL-8 and CRP biomarkers has been limited to the last three years (from January 2018 up to December 2020), mainly due to a large number of strategies reported in the past decade, as well as already available reviews summarizing EC strategies for the detection of these specific targets (e.g. CRP;101–103 IL-6;104 CRP, IL-6 and IL-8;105 NSE106).EC (bio)sensor configuration typically comprises two main parts: (i) immobilized recognition element providing the selectivity to the target analyte (T) and (ii) the EC transducer serving as a converter of the biorecognition event to an electronic signal.107,108
The most common transducers reported for mTBI-related biosensors include gold electrodes (disk, microelectrodes, films and interdigitated electrodes109,110), carbon-based (glassy carbon, carbon paste, carbon nanotubes (CNTs)111) and graphene-based materials. Other approaches have been reported employing conductive polymer (CP) nanowires (NWs), such as polypyrrole (PPy) NWs112,113 or silica-NWs.114,115 Metal oxide substrates such as hafnium oxide (HfO2),116,117 aluminium oxide (Al2O3),118 indium tin oxide (ITO)119–121 have been incorporated, more rarely platinum122,123 and molybdenum.124
Regarding the transducer surface modification, researchers often rely on gold nanoparticles (AuNPs) deposition on various substrates to benefit from easily addressable gold-sulphur chemistry and the signal amplification due to the accelerated electron transfer between the electrode and a redox moiety/mediator.125 Gold-sulphur chemistry is often explored with self-assembled monolayers (SAMs) based on various thiol derivatives (e.g., MPA, MHDA, MUA) employed to immobilize bioreceptors on gold electrode surface. The key issue limiting SAM practical applications remains however the overall stability of the monolayer film.126,127 CNTs have also found their application for transducer modification in various composites,128,129 such as AuPd-multiwall carbon nanotubes (MWCNTs) for the detection of the NSE biomarker,128 CNTs with Nafion and glutaraldehyde for the detection of S100β,130 MWCNTs incorporated into molecularly imprinted polymer (MIP) layer for the detection of GFAP131 and MWCNTs with reduced graphene oxide (rGRO) and chitosan for detection of tau-441 protein.132 Nanostructured conducting polymers (CPs) such as PPy-NWs functionalized via diazonium coupling reaction113 or (nano)composite layer (e.g. with AuNPs133,134 or graphene135) based on polyaniline (PANI), poly(3,4-ethylenedioxythiophene) (PEDOT) or 2,2:5,2-terthiophene-3-(p-benzoic acid) (pTTBA) (electropolymerized onto AuNPs136) present also an attractive platform for mTBI biosensor fabrication. Furthermore, transducer (bio)functionalization was also achieved via composites based on ionic liquids, e.g. with TiO2 mesocrystal nanoarchitectures,137 MOF architectures138 or ZnO/porous carbon composite.139 Furthermore, EC sensing immunoassay-based strategies relevant for mTBI often employ magnetic beads (MBs) in order to amplify the signal and increase sensitivity, reduce matrix effects and for multiplexing purposes.105,140–142
Recognition elements
The survey of publications relevant to mTBI biomarkers indicates that antibodies are used as recognition elements in most cases (in 86 out of 127 publications). However, there is an increasing trend towards the application of synthetic recognition structures. For example, aptamers have been employed for detection of CRP,112,125,143–146 IL-6,147 NSE,134 Tau proteins,123,148,149 and UCH-L1 biomarkers.123 The number of aptamers available for different target proteins is much lower than that of antibodies and a systematic screening technology is required in order to discover novel molecules for different biosensing applications.150–152 Other major issues with aptamer-based sensing are the time-dependent and poorly predictable alteration of folded aptamer structure in complex media and sensitivity of aptamer molecules towards degradation catalyzed by ribonucleases. To the best of our knowledge, this biosensing technology has not made it to the market yet, though several companies have been reported to work on the solutions for EC aptamer-based diagnostic devices.153,154 ‘Molecular containers’ like cucurbit[n]urils (CB[n]) have been shown to be promising candidates as recognition elements155–157 and have been explored for detection of MMP-2 biomarker.158 MIPs were also employed in the context of mTBI-relevant biomarkers for the detection of GFAP131 and NSE.159–161 Wang et al.159 suggested an ionic liquid with a pyrrole moiety and NSE as a template to fabricate MIP by electrochemical deposition in aqueous phase without any harsh polymerization initiators. Such sensors had a detection limit of 2.6 pg mL−1 (below cutoff value of NSE biomarker of 20 ng mL−1,59 Table 1). Another interesting strategy for detection of NSE biomarker using AuNPs decorated with epitope-mediated hybrid MIPs has been developed by Pirzada et al.161 who reported an ultrasensitive detection of NSE in human serum with LDL of 25 pg mL−1.It should be noted that non-specific binding (NSB), one of the most frequently encountered problems when designing affinity-based (bio)sensors is particularly important in the context of mTBI, as some of the potential biomarkers are present in blood in very low concentrations (e.g., GFAP cutoff 22 pg mL−1,32 or UCH-L1 cutoff 327 pg mL−1,32 Table 1). The overview and the discussion of the advantages and disadvantages of the most applied strategies to prevent the NSB including the immobilization of ‘blocking’ proteins and chemical approaches is given in ESI Section SI-3.†
Types of electrochemical (bio)sensors for detection of protein biomarkers relevant to mTBI
Potentiometric (bio)sensors
Both ion-selective electrodes (ISE) and non-ISE formats have been proposed for mTBI related protein biosensing.162 The ISE formats are commonly based on registering pH changes resulting from the catalytic reaction. For example, Liang et al.163 proposed an electrochemical immunoassay on a handheld pH meter using glucose oxidase-loaded liposomes (GOx-LS) for signal amplification. A sandwich immunocomplex was composed of a microplate coated with capture antibodies, NSE biomarker as antigen, and detection antibodies labelled with GOx-LS, employed to oxidise glucose into gluconic acid and hydrogen peroxide, leading to a pH change recorded with a pH meter. The authors explored the usage of liposomes with strong encapsulation ability for loading of natural GOx enzyme and for enhancing the catalytic efficiency after the antibody–antigen reaction. This strategy showed an improvement compared to the traditional GOx labelling, enabling low-cost detection of NSE biomarker with the detection limit of 8.9 pg mL−1 (cutoff for mTBI 20 ng mL−1)59 and within the dynamic linear range of 0.01–100 ng mL−1. However, in addition to the multi-step reagent-demanding nature of the proposed sandwich assay, the storage stability is reported to be relatively short owing to the usage of easy-broken liposomes.163Potentiometric sensors based on surface molecular imprinting have been suggested for protein biomarker detection, however to our knowledge not to mTBI specific protein analytes.164
Conductometric (bio)sensors
Very few researchers have adopted conductometric strategies for detection of mTBI-related biomarkers (Table 3 and Table SI-1†). Lin et al. have suggested a CRP-biosensor using conductive polypyrrole nanowire mesh architecture uniformly dispersed within a polymeric matrix with molecularly imprinted cavities hosting immobilized aptamers. Serum CRP levels were analyzed through monitoring the conductance change caused by the polymeric network shrinkage upon target binding, providing ultrasensitive detection of CRP in blood samples from melanoma patients (LDL 9 × 10−14 mg mL−1 in a wide dynamic range up to approximately 10−3 mg mL−1).112 Carbonaro et al.122 proposed an interesting multianalyte label-free approach for detection of GM-CSF (and G-CSF) biomarker based on resistive pulse sensing (RPS) and multiple artificial pores integrated on a chip. The sensor was detecting the size change of latex colloids upon specific antigen–antibody binding on the colloid surface. Another approach based on resistive RPS was proposed by Maugi et al.145 who developed a strategy combining RPS with nanocarriers for detection of CRP biomarker. The surface of a nanocarrier was coated with a mixture of peptide aptamer and a non-binding DNA. The target binding to the aptamer creates a corona around the carrier, shielding the phosphate groups of the DNA and resulting in a change in electrophoretic mobility of the nanocarrier. The advantage of this approach lies in its universality as it can be easily adapted to other proteins without the need for chemical modifications. On the other side, the main limitation is the limit of detection (sub-micromolar level).| Technique | Biomarker/publication year | Transducer | Surface modification | Label | Analysis timea | Sample | Lower detection limitb | Rangec |
|---|---|---|---|---|---|---|---|---|
a The total analysis time (unless clearly indicated in the original publication) is an approximate value estimated by the authors of this review article based on the sum of the duration of individual analytical steps reported in the original publications. For more detailed information please consult ESI Table SI-1. The analysis time serves only an indicative purpose and does not take into account the potential for shortening upon further method optimization.b Lowest reported LDL using EC detection methods; '<x’ corresponds to the lowest concentration analyzed within the working range of the sensor (employing standard addition method and/or a reference material/method for validation, with a decent recovery), actual LDL being possibly lower than the indicated value.c Linear vs. target concentration (CT), if not stated otherwise (e.g., vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT). The upper limit of the range indicated often presents the maximum concentration explored but not the upper detection limit. Please consult original papers for details. See Abbreviations. Complete list of all published electrochemical strategies (with details on biosensor configuration) for the detection of blood protein biomarkers relevant to mTBI is available in ESI SI-1. CT). The upper limit of the range indicated often presents the maximum concentration explored but not the upper detection limit. Please consult original papers for details. See Abbreviations. Complete list of all published electrochemical strategies (with details on biosensor configuration) for the detection of blood protein biomarkers relevant to mTBI is available in ESI SI-1. |
||||||||
| Potentiometry | NSE/2019 (ref. 163) | pH electrode | Ab1/T/Ab2-GOx-LS | GOx-LS | ∼45 min | Buffer | 8.9 pg mL−1 | 0.01–100 ng mL−1 |
| Serum | <0.5b ng mL−1 | (pH vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
||||||
| Conductometry | CRP/2018 (ref. 112) | CuPT-PPy nanowiremesh | NIPAAm-AM/Apt/CRP polymer | Label-free | ∼20 min | Buffer | 9.03 × 10−17 g mL−1 | Non-linear up to ca. 10−8 M |
| Serum | <70b ng mL−1 | |||||||
| Conductometry (RPS) | CRP/2019 (ref. 145) | Nanocarriers: SPBs | Peptide-Apt/non-binding DNA/T | Label-free | ∼90 min | Buffer | n/a | ca. 0.5–2.5 μM |
| SWV/EIS | CRP/2019 (ref. 165) | Au arrays | MPA/(EDC + NHS)/Ab/T | Label-free | ∼30 min | Buffer | 2.25 fg mL−1 (SWV) | 5–220 fg mL−1 |
| 3 fg mL−1 (EIS) | 7–215 fg mL−1 | |||||||
| Serum | 4.5 fg mL−1 (SWV) | 12–166 fg mL−1 | ||||||
| EIS | VCAM-1/2017 (ref. 166) | Au microelectrode | DTSP/Ab/T or DTSP/Ab1/T/Ab2 | Label-free | ∼15 min | Buffer | 8 fg mL−1 | 8 fg mL−1- 800 pg mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Urine | <500b μg mL−1 | |||||||
| EIS | IL-6/2018 (ref. 113) | PPy-NWs layer | PPyPAC/(EDC + NHS)/Ab/T | Label-free | ∼30 min | Buffer | 0.36 pg mL−1 | 1–50 pg mL−1 |
| EIS | MMP-2/2015 (ref. 167) | Au | Pept-SH (target-induced cleavage) | Label-free | ∼60 min | Buffer | 0.5 pg mL−1 | 0.1–400 ng mL−1 (non-linear) |
| Capacitive (impedance derived) | CRP/2020 (ref. 169) | Graphene nanoplate SPE | PANI-PA/Ab/T | Label-free | ∼10 min | Buffer | 0.5 μg mL−1 | 0.25–2 μg mL−1 |
| DPV | MMP-2/2016 (ref. 140) | GCE | MBs/(EDC + NHS)/Pept-SH/AuNPs-DNA1 | MeB-DNA2+Exo III | ∼180 min | Buffer | 0.15 pg mL−1 | 0.5 pg mL−1-50 ng mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Serum | <0.1b ng mL−1 | |||||||
| DPV | Tau/2018 (ref. 173) | Au DE | MPA/(EDC + NHS)/Ab/T/AuNPs–SH–Apt | Label-free | ∼45 min | Buffer | 0.42 pM | 0.5–100 pM |
| Serum | <1.5b pM | |||||||
| DPV | IL-8/2020 (ref. 200) | ITO | β-Ag2MoO4/(EDC + NHS)/Ab/T | Label-free | ∼10 min | Buffer | 90 pg mL−1 | 1 fg mL−1-40 ng mL−1 (two linear ranges) |
| CPA | CRP/2018 (ref. 174) | Carbon SPE | AuNPs/L-cysteine/(EDC + NHS)/Ab/T | Label-free | ∼30 min | Buffer | 17 ng mL−1 | 0.05–23.6 μg mL−1 |
| Serum | <0.932b mg L−1 | |||||||
| CPA | BDNF/2018 (ref. 136) | Carbon SPE | AuNPs/pTTBA/(EDC + NHS)/Ab1/T/Ab2/(EDC + NHS)/TBO-pTTBPA/AuNPs/carbon SPE#2 | TBO | ∼20 min | Buffer | 0.015 ng mL−1 | 0.004–0.6 ng mL−1 |
| Serum | <0.1b ng mL−1 | |||||||
| CPA | Tau/2020 (ref. 178) | Dual SPCE | pABA/(EDC + NHSS)/3D-Au-PAMAM/GA/Ab1/T/Ab2-HRP | Ab2/HRP | ∼120 min | Buffer | 2.3 pg mL−1 (∼pg mL−1)b | 8–5000 pg mL−1 |
| Plasma | ||||||||
| SWV | h-FABP/2012 (ref. 179) | GCE | GRONRs/(EDC + NHS)/Ab1/T/Ab2/GA/TiP–Zn2+-probe | TiP–Zn2+-probe | ∼120 min | Buffer | 3 fg mL−1 | 0.05 pg mL−1 –50 ng mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Serum | <1.7b μg mL−1 | |||||||
| SWV | IL-8/2019 (ref. 201) | Carbon SPE | PEI-AuNPs/GA/Ab1/T/PEI-AuNPs-Ab2-Ag+ | PEI-AuNPs-Ab2-Ag+ | ∼80 min | Buffer | 1 fg mL−1 | 0.5–100 pg mL−1 |
| Serum | <2.5b pg mL−1 | 2.5–50 pg mL−1 | ||||||
| ASV | h-FABP/2017 (ref. 180) | GCE | CD-GS/Ab1/T/Ab2-ZnO-MWCNTs/CdS | ZnO-MWCNT/CdS | ∼120 min | Buffer | 0.3 fg mL−1 | 1.3 fg mL−1-130 ng mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Serum | <5b pg mL−1 | |||||||
| FED (OFET) | GFAP/2017 (ref. 186) | Si/SiO2/pentacene/Au | (PS-MA + PEG)/Ab/T | Label-free | ∼30 min | Buffer | 1 ng mL−1 | 0.5–100 ng mL−1 |
| FED (GFET) | Tau/2020 (ref. 189) | Si/SiO2 | APMES/rGRO/PBASE/Ab/T | Label-free | ∼20 min | Buffer | n/a | 100 fg mL−1-10 ng mL−1 (HP, vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Plasma | 1 pg mL−1 (HP) | |||||||
| FED (FEED) | S100β/2018 (ref. 130) | Carbon SPE | SWCNTs-Nafion-GA/Ab1/T/HRP-Ab2 | HRP | ∼100 min | HS | 10 fg mL−1 | 10 fg mL−1-10 ng mL−1 |
| PEC | MMP-2/2020 (ref. 195) | ITO | Fe3O4@SiO2/(EDC + NHS)/Ab1/T/Ab2/TiO2-AgNPs | TiO2–Ag NPs/Ab2 | ∼120 min | Buffer | 0.34 fg mL−1 | 1 fg mL−1–100 pg mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Serum | <350b pg mL−1 | |||||||
| PEC | NFL/2020 (ref. 196) | Pt NWs on FTO (biocathode) | (MUA + MCH)/Ab, photoanode: FTO/BiVO4–FeOOH | Label-free | ∼60 min | Buffer | 38.2 fg mL−1 | 0.1–1000 pg mL−1 (vs. lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT) CT) |
| Plasma | n/a | |||||||
Capacitive and impedimetric (bio)sensors
Most of the impedimetric/capacitive detection strategies reported for mTBI biomarker detection are based on the faradaic model and capture immunoassay format where the electron-transfer resistance is changed due to interaction between immobilized (capture) antibody and the target molecule. For example, Vilian et al.165 have reported on a gold wire array grown on polycarbonate substrate and functionalized with anti-CRP via self-assembly of 3-mercaptopropionic acid (MPA) for the electron-mediated detection of CRP using both EIS-based and voltammetric techniques (see Fig. 1). The authors reported a LDL of 3 fg mL−1 with EIS with the linear dynamic range of 7 to 215 fg mL−1. This strategy exhibited high selectivity against various potentially interfering species (dopamine, lysine, uric acid, glucose, etc.) and was capable of directly probing trace amounts of the target CRP in human serum.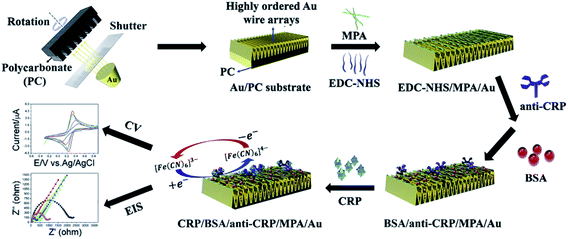 | ||
| Fig. 1 EIS- and CV-based detection of CRP, reprinted from Vilian et al.,165 Copyright (2019), with permission from Elsevier. | ||
A non-faradaic impedimetric strategy has been suggested by Selvam et al.166 for detection of VCAM-1 biomarker. The approach is based on the immobilization of capture antibodies on gold microelectrodes resulting in the formation of a charged electrical double layer (EDL). Binding of the target to the antibody results in highly specific capacitance changes, while the authors have also observed an improvement of the overall signal upon optional addition of a second (detection) antibody to the immobilized immunoassay, resulting in further accumulation or perturbation of charges in the capacitive EDL. The strategy exhibits potential utility for POC applications with an LDL of 8 fg mL−1 and a dynamic range of 8 fg mL−1 to 800 pg mL−1.166 Another non-faradaic approach has been developed by Garcia-Cruz et al.113 who have reported on the fabrication of PPy-NWs using innovative nanocontact printing, allowing for low-cost fabrication of electrodes with highly controllable architecture (see Fig. 2). The impedimetric immunosensor has been designed by immobilizing IL-6 antibodies via diazonium coupling reaction and carbodiimide crosslinker on the PPy-NWs printed using controlled chemical polymerization with an LDL of 0.36 pg mL−1 and a linear range of 1–50 pg mL−1.113 Some other strategies exploit more peculiar transducer substrates for immobilizing an antibody, such as e.g. molybdenum (via cross-linking with EDC and NHS, employed for detection of CRP)124 or HfO2 (via self-assembly, detection of IL-10).116
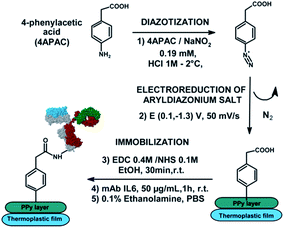 | ||
| Fig. 2 Non-faradaic EIS-based detection of IL-6, reprinted from Garcia-Cruz et al.,113 Copyright (2018), with permission from Elsevier. | ||
Impedimetric detection of mTBI-relevant enzymatic proteins like matrix metalloproteinases (MMPs) is addressed via approaches resting upon a cleavage event of a peptide specific to MMPs after injection of target-containing solution.167
Detection of non-enzymatic proteins (which is the vast majority of target proteins for mTBI) can be as well accomplished via peptide-supported aptasensing (e.g. gold electrode fuctionalization with a ferrocene-tagged peptide, followed by cross-linking with the aptamer143) or by anchoring the recognition molecule (e.g. capture antibody) onto a redox active composite (e.g. graphene oxide and zwitterionic monomer based composite incorporated into a 11-ferrocenyl-undecanethiol monolayer168).
A very recent approach by Baradoke et al.169 for the first time employed a surface-confined redox active polymer (i.e. phytic acid-doped PANI film, see Fig. 3) as a support for reagentless redox capacitive (impedance-derived) sensing of CRP. In this strategy the CRP-sensitive surface has been obtained via glutaraldehyde cross-linking of amine functionalities in the PANI film with the antibody. The construction of the sensory interface by electropolymerization allowed for tuning the surface coverage and capacitive properties of the polymers, which could be used to modulate the assay selectivity, fouling, and sensitivity (LDL 0.5 μg mL−1).
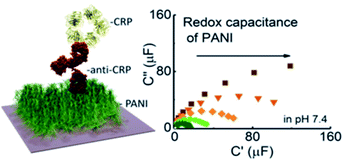 | ||
| Fig. 3 EIS-derived capacitance-based detection of CRP, reprinted with permission from Baradoke et al.169 Copyright (2020) American Chemical Society. | ||
Capacitive and impedimetric measurements represent an attractive platform for practical applications not only because of decreased measurement times and costs due to absence of labelling step, but as well due to enabling continuous real-time sensing which is rarely possible with label-based EC assays.170,171 Their potential for multianalyte mTBI diagnostics was demonstrated by Cardinell et al.172 (detection of GFAP, NSE, S100β and tumor necrosis factor-α), who have characterized mTBI biomarkers in purified solutions (LDL 2–5 pg mL−1) and then verified the detection approach in spiked rat whole blood and plasma solutions (LDL of 14–67 pg mL−1 in 90% whole blood).
Amperometric/voltammetric (bio)sensors
Several approaches have been shown to be applicable for detection of mTBI biomarkers with relatively high clinically relevant concentration ranges. Very few label-free strategies have been reported, e.g. Shui et al.173 have developed an aptamer-antibody sandwich assay by using a tau antibody and an aptamer specific to tau-381 as the recognition element and cysteamine-stabilized gold nanoparticles for signal amplification (see Fig. 4). Detection of tau-381 in buffer and human serum was accomplished using differential pulse voltammetry (DPV) in the presence of [Fe(CN)6]3−/4− with an LDL of 0.42 pM and <1.5 pM (ca. 17 and 60 pg mL−1), respectively, within the linear range from 0.5 to 100 pM. Another label-free approach has been proposed by Thangamuthu et al.174 who employed a simple constant potential amperometry (CPA)-based capture assay for detection of CRP using an antibody-functionalized AuNPs modified carbon screen-printed electrode (SPE). The measurement relies on the decrease of the oxidation current in the presence of redox mediator ([Fe(CN)6]3−/4−) and the authors reported an LDL of 17 ng mL−1 in the range of 0.05–23.6 μg mL−1.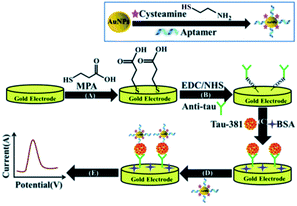 | ||
| Fig. 4 DPV-based detection of tau-381, reprinted from Shui et al.173 Copyright (2018) The Royal Society of Chemistry. | ||
Most amperometric workflows are based on either a labelled competitive assay (e.g. using free and alkaline phosphatase (ALP)-labelled target molecules for detection of GM-CSF biomarker175) or, more commonly, sandwich assay (e.g. horseradish peroxidase (HRP)-labelled microfluidic bead-based enzyme-linked immunosorbent assay for detection of IL-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 176 or Tau proteins177,178). A peculiar dual-probe sandwich-like assay with a single incubation step has been suggested by Akhtar et al.136 for relatively rapid (ca. 20 min) BDNF biomarker detection in the extracellular matrix of neuronal cells (see Fig. 5). In this approach two independently prepared carbon SPE-based probes were placed in front of each other to form a microfluidic channel for the sample solution. The working probe (B) was fabricated by modifying a carbon SPE by covalently attaching capture antibodies to the layer of AuNPs–pTTBA (CP) composite. The bioconjugate probe (A) was prepared from second carbon SPE, modified by drop casting the bioconjugate particles composed of conducting polymer self-assembled onto AuNPs and functionalized with detection antibodies and toluidine blue O (TBO). The method allowed for the detection of BDNF concentrations as low as 100 pg mL−1 (the median serum concentration for mTBI is 8.3 ng mL−1,26 decreased compared to healthy physiological range, see Table 1) spiked in undiluted human serum using CPA.136 The bioconjugate attachment being already available on the bioconjugate probe that can be fabricated in advance, the proposed strategy allows for a single incubation step with the target analyte and thus is more attractive for the realization of POC diagnostic methods, compared to conventional time-consuming sandwich-based approaches.
176 or Tau proteins177,178). A peculiar dual-probe sandwich-like assay with a single incubation step has been suggested by Akhtar et al.136 for relatively rapid (ca. 20 min) BDNF biomarker detection in the extracellular matrix of neuronal cells (see Fig. 5). In this approach two independently prepared carbon SPE-based probes were placed in front of each other to form a microfluidic channel for the sample solution. The working probe (B) was fabricated by modifying a carbon SPE by covalently attaching capture antibodies to the layer of AuNPs–pTTBA (CP) composite. The bioconjugate probe (A) was prepared from second carbon SPE, modified by drop casting the bioconjugate particles composed of conducting polymer self-assembled onto AuNPs and functionalized with detection antibodies and toluidine blue O (TBO). The method allowed for the detection of BDNF concentrations as low as 100 pg mL−1 (the median serum concentration for mTBI is 8.3 ng mL−1,26 decreased compared to healthy physiological range, see Table 1) spiked in undiluted human serum using CPA.136 The bioconjugate attachment being already available on the bioconjugate probe that can be fabricated in advance, the proposed strategy allows for a single incubation step with the target analyte and thus is more attractive for the realization of POC diagnostic methods, compared to conventional time-consuming sandwich-based approaches.
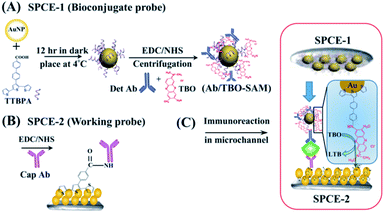 | ||
| Fig. 5 CPA-based detection of BDNF, reprinted from Akhtar et al.,136 Copyright (2018), with permission of Elsevier. | ||
Feng et al.179 have reported on a duplexed sandwich immunoassay for simultaneous detection of h-FABP and troponin I using titanium phosphate nanospheres functionalized with Zn2+ and Cd2+ (respectively) as labels (see Fig. 6). The proposed strategy employs graphene oxide nanoribbons (GRONRs) as a substrate for capture antibody immobilization and enables direct detection of metal ions in the bioconjugates using SWV without acid dissolution and preconcentration (stripping) steps.179 The reported assay allowed for detection of ca. 1.7 μg mL−1 of h-FABP in undiluted serum (cutoff for mTBI 2.6 ng mL−1,37 Table 1), clinically relevant lower target amounts have been quantified in buffer only. Despite of avoiding stripping steps, the protocol still requires more than 100 minutes turnaround time due to incubation steps with both the target and the label-carrying probe.
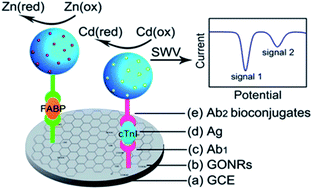 | ||
| Fig. 6 SWV-based detection of h-FABP, reprinted with permission from Feng et al.179 Copyright (2012) American Chemical Society. | ||
Anodic stripping voltammetry (ASV) has been employed for detection of h-FABP,180 IL-8,111,181 MMP-2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 and NSE.183 Cyclic voltammetry (CV) has not been applied in many strategies. Exceptionally, Ramgir et al. have reported on sensitive IL-10 detection using silica-NWs by employing ALP-labelled sandwich assay with the p-nitrophenyl phosphate added as a substrate for detection.114
182 and NSE.183 Cyclic voltammetry (CV) has not been applied in many strategies. Exceptionally, Ramgir et al. have reported on sensitive IL-10 detection using silica-NWs by employing ALP-labelled sandwich assay with the p-nitrophenyl phosphate added as a substrate for detection.114
Field-effect based biosensing devices (Bio-FEDs)
FEDs have a potential in the field of POC diagnostic device development due to their ability to provide instantaneous (possibly real-time) label-free measurements using very small sample volumes, low production cost, high density integration and miniaturization.184,185 Various designs of FET-based sensing devices have been proposed in the literature for detection of mTBI-related biomarkers. Song et al. have developed and organic field effect transistor (OFET)-based biosensor with extended solution gate architecture for label-free detection of GFAP biomarker186 via a strategy for overcoming Debye screening length limitations. The latter has been achieved by mixing the bioreceptor layers with different molecular weight PEGs, which has been previously shown to increase the ‘effective Debye screening length’ for a given ionic strength.186,187 Hao et al. reported on a sensitive and fast (10 min) detection of IL-6 using a graphene-based field-effect transistor (GFET) with the graphene surface covalently functionalized with a negatively charged aptamer undergoing conformational changes upon target binding,188 while Park et al. used reduced graphene oxide FET for detection of T-Tau.189A peculiar strategy based on field-effect enzymatic detection (FEED) reported by Mathew et al. has adopted gating voltage for signal amplification for ultrasensitive detection of S100β.130 In this approach (see Fig. 7), a sandwich assay with an HRP-labelled detection antibody has been realized on a working electrode (WE) using a conventional screen-printed three-electrode cell. An insulated copper wire wound around the WE served as the gating electrode. To achieve voltage-controlled amplification, the WE has been connected to the gating electrode via a DC power supply to apply the gating voltage (VG) yielding an electric field at the WE/solution interface and thus resulting in changes of interfacial charge distribution.130 The proposed technique enabled LDL as low as 10 fg mL−1 in serum which is sufficient for S100β quantification (cutoff for mTBI 42 pg mL−1,37 Table 1).130 Importantly, this is one of very few publications on FED-based architectures reporting its successful application in undiluted serum.
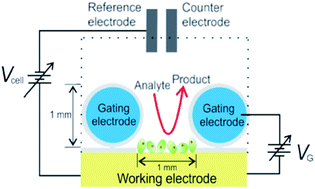 | ||
| Fig. 7 Field effect enzymatic detection of S100β using an insulated copper wire as a gating electrode for signal amplification. Reprinted by permission from Springer Nature, Molecular Diagnosis & Therapy, Mathew et al.,130 copyright (2018). | ||
However, as of today, there are still challenges to be overcome for the FEDs to make their way into medical diagnostics. One of the major issues is obtaining well-defined recognition element structures (reproducible and easily manufacturable solid phase/solution interface)190,191 and overcoming Debye screening in order to provide for direct measurements in undiluted clinical samples with high ionic strength (e.g. blood, serum).190 Passivation in aqueous media is also of great concern (hydrophobicity of passivation materials greatly affects both the stability and overall performance of the sensor191), as well as the problem of noise for nanoscale-devices originating from variations in interfacial charge.191
Photoelectrochemical (PEC) (bio)sensing
The PEC sensor performance relies on photoactive materials that produce photocurrent upon absorbing photons and engage in redox reaction at the WE surface via different transduction mechanisms: formation of electrons/holes, introduction of photoactive species, steric hindrance, in situ induction of light, or resonance energy transfer.146,192,193 PEC-based (bio)sensing, although presenting a promising novel analytical method for biomarker detection, is yet at a very early stage for practical application. Nevertheless, a few examples of PEC sensing strategies have been suggested for detection of mTBI-related biomarkers such as MMP-2,194,195 NFL,196 NSE,197 Tau proteins,149,198 and CRP.146,199 Further discussion of PEC affinity-based detection principles, types of photoactive species and signal transduction mechanisms is outside the scope of this work and can be found e.g. in a recent detailed review by Victorious et al.193Conclusions and future outlook
The aim of this manuscript has been to give a detailed overview of EC approaches developed and used for detection of blood protein biomarkers previously shown to be relevant for diagnostics and prognostics of mTBI with the possible application towards POC testing.12 The relevance of a POC diagnostic mTBI biomarker test cannot be overstated. The optimal approach would likely comprise early biomarker detection (on site of an accident and immediately after injury) with follow-up measurements at various intervals (e.g., in the ambulance, upon arrival at the hospital and pre- or post-diagnostic imaging), allowing for the observation of injury characteristic biomarker increases and decreases. Instead of a single time point measurement, data on the early acute phase trend could be useful both as a diagnostic determinant and as an indicator of injury progression.87As briefly highlighted with an excerpt of a target product profile for a POC diagnostic device for mTBI (see Table 2), the following requirements should be considered when developing a POC biosensor for clinical applications: (i) high sensitivity and specificity to the target analytes; (ii) good reproducibility, reliability and stability of sensor's readings; (iii) short analysis time; (iv) low sample consumption; (v) low cost of production; (vi) portability; (vii) environment-friendly disposable design; (viii) user-friendliness (ease of operation); (ix) preferably requiring no sample pre-treatment, else a pre-treatment as simple as possible; (x) preferably reagent-free, else with small reagent consumption.
The key advantages of (bio)sensors for in vitro POC diagnostics include relatively short analysis time and low reagent consumption (Fig. 8). In respect to the sensor fabrication technology, most of the sensor elements are easily miniaturizable, have low fabrication costs and could be readily integrated into a POC platform. Transducer elements are typically manufactured using microfabrication technology convenient for mass fabrication process, while the electrode functionalization with the recognition elements (e.g., antibodies) is considerably more challenging even before the scale-up of the production process.
Although EC sensors for measurement of non-protein metabolites in whole blood such as glucose, lactate, uric acid, cholesterol, blood gases and electrolytes are commercially available and used routinely for POC diagnostic applications, the development of robust, accurate and highly reproducible strategies remains a challenge for protein biomarkers, especially those relevant to mTBI. The challenge for the POC test development is the significant variation in mTBI biomarker kinetics and the fact that some biomarkers may be more suited to cross the blood brain barrier than others, raising the need for ultrasensitive detection strategies. Detections limits as low as a few fg mL−1 have been reported for the EC detection of certain biomarkers (e.g. 1 pg mL−1 for GFAP;202,203 0.3 fg mL−1 for h-FABP;180 0.32 fg mL−1 for IL-6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 137). As discussed in this article and summarized in Table 3 and SI-1,† various EC sensor designs and analytical approaches have been reported in the literature for detection of 19 mTBI-relevant blood proteins (total 127 publications). However, most of the EC approaches developed so far for detection of mTBI related proteins either suggest serum/blood dilution or have not been tested with clinical samples.204 In the published cases where these performances are claimed to be determined in serum or plasma matrix the question comes up as to what extent these high-sensitivity results are reproducible with multiple biosensor lots manufactured and a statistically relevant number of clinical samples. Moreover, some approaches require a preconcentration step; additional dilution is often required for FET-based biosensing which is limited by high ionic strength of biological fluids due to Debye-screening length, as well as for strategies with short dynamic ranges. In conclusion, the realization of reproducible, sensitive but highly scalable transducer–electrolyte interfaces remains a big challenge. The ongoing studies worldwide are currently aimed at improving the test performance by increasing the sensitivity, specificity, turnaround time and decreasing the costs.
137). As discussed in this article and summarized in Table 3 and SI-1,† various EC sensor designs and analytical approaches have been reported in the literature for detection of 19 mTBI-relevant blood proteins (total 127 publications). However, most of the EC approaches developed so far for detection of mTBI related proteins either suggest serum/blood dilution or have not been tested with clinical samples.204 In the published cases where these performances are claimed to be determined in serum or plasma matrix the question comes up as to what extent these high-sensitivity results are reproducible with multiple biosensor lots manufactured and a statistically relevant number of clinical samples. Moreover, some approaches require a preconcentration step; additional dilution is often required for FET-based biosensing which is limited by high ionic strength of biological fluids due to Debye-screening length, as well as for strategies with short dynamic ranges. In conclusion, the realization of reproducible, sensitive but highly scalable transducer–electrolyte interfaces remains a big challenge. The ongoing studies worldwide are currently aimed at improving the test performance by increasing the sensitivity, specificity, turnaround time and decreasing the costs.
While most designs exploit the antibody–antigen interaction, smaller biorecognition molecules such as DNA/RNA or peptide aptamers, MIPs and other specific synthetic receptors have a promising potential for improving the performance of EC sensors in terms of high specificity and sensitivity, inexpensive and readily scalable cell-free chemical synthesis and low batch-to-batch variability.153,154,205
The lack of stability, enough long shelf-life (often required to exceed 6 months) and deterioration of the analytical performance of the transducer functionalized with biological recognition elements over time could become a serious issue in product development and envisioned commercialization. In fact, POC diagnostic devices for various analytes are known to often underperform in terms of accuracy and precision compared to central laboratory instruments, so a multiplex diagnostic test for mTBI protein biomarkers is likely going to encounter a cumulation of challenges to meet. On the other hand, from the viewpoint of signal processing, multiplexing (i.e., support of multi-biomarker analysis) is more straightforward with EC techniques.
Impedimetric measurements, along with FET-based devices, constitute the main platform for rapid label-free and potentially reagent-free detection of mTBI biomarkers. Nevertheless, the issue of non-specific binding remains challenging for most if not all biosensing strategies and especially for label-free assays since the latter do not discriminate between the signal caused by specific versus non-specific interactions.170,206,207 Despite the inherent advantages offered by label-free techniques, labelled assays continue to be an important direction in the development of sensing strategies providing the benefits of improved selectivity and significantly increased sensitivity using various signal amplification approaches, such as e.g. functionalized nanoparticles. One of the drawbacks for the integration into clinical analysis is the long time required to complete the assay. The latter neither makes them attractive for the development of POC diagnostic applications nor advantageous compared to well-established analytical solutions in laboratory medicine.
Therefore, despite the large and diverse pool of developed EC (bio)sensor designs, for the eventual integration of the proposed concepts into mTBI POC diagnostic device solutions much work still must be accomplished requiring a close collaboration of the researchers in the field of biochemistry with material scientists and nanotechnologists, engineers, and clinicians. Importantly, the development of easy-to-use and affordable tools for detection of specific biomarkers and biomarker panels, could aid not only the diagnostics of well-established disease biomarkers, but as well in the process of evaluation and identification of prognostic value of currently investigated biomarkers and in the establishment of an ‘ideal’ biomarker panel for TBI diagnostics. The interdisciplinary synergy seems to be necessary to overcome the barrier between rapidly progressing academic research and real-life medical diagnostic applications.
A big step toward the first US commercial POC diagnostic test for mTBI was made by Abbott Diagnostics, recently. Following a non-exclusive license agreement with Banyan Biomarkers in 2019, Abbott received FDA 510(k) clearance for the first rapid handheld blood test for concussions in January 2021. The test runs on the i-STAT™ Alinity™ POC device and it measures amperometrically the UCH-L1 and GFAP biomarkers. The UCH-L1 biomarker complements GFAP as each result is produced by a different type of cell and measures distinctive molecular events.208 The results are given 15 min after the plasma sample is inserted in the cartridge. Building on this initial clearance, Abbott is also working on a test that would use whole blood on i-STAT device.209 Further developments, optimization and additional prospective studies are required to assure sufficient diagnostic specificity and sensitivity in evaluating concussions in patients with mTBI (i.e. multiplex panel extension with additional biomarkers or biomarker types like inflammation proteins and brain damage proteins). As other companies, such as NanoDx™210 that uses an ultrasensitive nanowire technology to resistively measure the biomarkers S100β and GFAP, are following on Abbott's heels, electrochemical POC sensing of blood proteins for mTBI may at last experience a much needed push forward.
Author contributions
The manuscript was written through contributions of all authors.Abbreviations
| Ab | Antibody |
| AD | Alzheimer's disease |
| ALP | Alkaline phosphatase |
| AM | Acrylamide |
| Apt | (Oligonucleotide) aptamer for the target (T) |
| APMES | 3-(Ethoxydimethylsilyl)propylamine |
| ASV | Anodic stripping voltammetry |
| AuNPs | Gold nanoparticles |
| BDNF | Brain-derived neurotrophic factor |
| bio-FED(s) | Field-effect based biosensing devices |
| CD-GS | β-Cyclodextrin-graphene sheets |
| CMOS | Complimentary metal-oxide semiconductor |
| CNTs | Carbon nanotubes |
| CO | Cutoff |
| CP | Conductive polymer |
| CPA | Constant potential amperometry |
| CS | Chitosan |
| CSF | Cerebrospinal fluid |
| CT | Computer tomography |
| CV | Cyclic voltammetry |
| DA | Dopamine |
| DE | Disk electrode |
| DPV | Differential pulse voltammetry |
| DTSP | Dithiobis (succinimidyl propionate) |
| EC | Electrochemical |
| EDC | 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide |
| EDL | Electric double layer |
| EIS | Electrochemical impedance spectroscopy |
| Exo III | Exonuclease III |
| Fc | Ferrocene |
| FDA | Food and Drug Administration |
| FED | Field-effect based detection (voltage controlled current amplification) |
| FEED | Field-effect enzymatic detection |
| FET | Field-effect transistor |
| FTO | Fluorine doped tin oxide |
| GA | Glutaraldehyde |
| GCE | Glassy carbon electrode |
| GCS | Glasgow Coma Scale |
| GFAP | Glial fibrillary acidic protein |
| GFET | Graphene-based field-effect transistor |
| GM-CSF | Granulocyte-macrophage colony-stimulating factor |
| GOx | Glucose oxidase |
| GOx-LS | Glucose oxidase-loaded liposomes |
| GR | Graphene |
| GRO | Graphene oxide |
| GRONRs | Graphene oxide nanoribbons |
| h-FABP | Heart-fatty acidic binding protein |
| HP | Human plasma |
| HRP | Horseradish peroxidase |
| HS | Human serum |
| ID | Interdigitated |
| IDE | Interdigitated electrode |
| IL | Ionic liquid |
| IL-6 | Interleukin-6 |
| IL-8 | Interleukin-8 |
| IL-10 | Interleukin 10 |
| ISE(s) | Ion-selective electrode(s) |
| ISFET | Ion-sensitive field-effect transistor |
| ITO | Indium tin oxide |
| IVD | In vitro diagnostics |
| LDL | Lower detection limit |
| LFA | Lateral flow assay |
| MBP | Myelin basic protein |
| MBs | Magnetic beads |
| MD | Median |
| mdTBI | Moderate TBI |
| MeB | Methylene blue |
| MHDA | Mercaptohexadecanoic acid |
| MIP | Molecularly imprinted polymer |
| MMP(s) | Matrix metalloproteinases |
| MN | Mean |
| MOF | Metal–organic framework |
| MPA | 3-Mercaptopropionic acid |
| MRI | Magnetic resonance imaging |
| MT3 | Metallothionein 3 |
| mTBI | mild TBI |
| MUA | 11-Mercaptoundecanoic acid |
| MuxT | Multiple protein biomarker targets detected within the same immunoassay |
| MWCNTs | Multiwalled carbon nanotubes |
| NCAM | Neuron cell adhesion molecule (CD56); |
| NFL | Neuro-filament light |
| NGB | Neuroglobin |
| NHS | N-Hydroxysuccinimide |
| NHSS | Hydroxysulfosuccinimide |
| NRGN | Neurogranin |
| NSB | Non-specific binding |
| NSE | Neuron-specific enolase |
| NWs | Nanowires |
| OFET | Organic field-effect transistor |
| pABA | p-Aminobenzoic acid |
| PAMAM | Poly(amido)amine |
| PANI | Polyaniline |
| PBASE | 1-Pyrenebutyric acid N-hydroxysuccinimide ester |
| PEC | Photoelectrochemical (detection) |
| PEG | Polyethylene glycol |
| PEI | Poly(ethyleneimine) |
| Pept-SH | Thiolated peptide |
| PET | Positron emission tomography |
| POC | Point-of-care |
| PPy | Polypyrrole |
| PPy-NWs | Polypyrrole-nanowires |
| PPyPAC | Polypyrrole electrodes modified by electrodeposition of diazonium salts using 4-aminophenylacetic acid (4APAC) |
| PS–MA | Polystyrene-co-methacrylic acid |
| pTTBA | (2,2:5,2-Terthiophene-3-(p-benzoic acid)) |
| pTTBPA | 4′-([2,2′:5′,2′′-Terthiophen]-3′-yl)-[1,1′-biphenyl]-4-carboxylic acid |
| RG | Range |
| rGRO | Reduced graphene oxide |
| RPS | Resistive pulse sensing |
| RT | Room temperature |
| S100β | Calcium binding protein B |
| SAM(s) | Self-assembled monolayer(s) |
| SH | Thiol group |
| SH-Apt | Thiolated aptamer for the target (T) |
| SI | Supplementary information |
| SL | Single layer |
| SPBs | Superparamagnetic beads |
| SPE | Screen-printed electrode |
| sTBI | Severe TBI |
| Stv | Streptavidin |
| SWCNTs | Single wall carbon nanotubes |
| SWV | Square wave voltammetry |
| T | Target |
| TBI | Traumatic brain injury |
| TBO | Toluidine blue O |
| TPP | Target product profile |
| T-Tau | Total Tau (phosphorylated and non-phosphorylated) |
| UCH-L1 | Ubiquitin carboxyl-terminal hydrolase-L1 |
| undil. | Undiluted |
| VCAM-1 | Vascular cell adhesion protein 1 |
| WE | Working electrode |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by BRIDGE (joint programme conducted by the Swiss National Science Foundation (SNSF) and Innosuisse - the Swiss Innovation Agency), grant no. 40B2-0_181013. The authors thank Igor Boev for his precious help with the TOC graphic.References
- Traumatic Brain Injury. Fact sheets and Policy brief., https://www.center-tbi.eu/, (accessed September 24, 2020) Search PubMed.
- M. C. Dewan, A. Rattani, S. Gupta, R. E. Baticulon, Y.-C. Hung, M. Punchak, A. Agrawal, A. O. Adeleye, M. G. Shrime, A. M. Rubiano, J. V. Rosenfeld and K. B. Park, J. Neurosurg., 2018, 130, 1080–1097 Search PubMed.
- R. Graham, F. P. Rivara, M. A. Ford and C. M. Spicer, Sports-related concussions in youth: improving the science, changing the culture, National Academies Press, 2014 Search PubMed.
- S. Yokobori, K. Hosein, S. Burks, I. Sharma, S. Gajavelli and R. Bullock, CNS Neurosci. Ther., 2013, 19, 556–565 CrossRef CAS PubMed.
- G. S. Tomar, G. P. Singh, D. Lahkar, K. Sengar, R. Nigam, M. Mohan and R. Anindya, Clin. Chim. Acta, 2018, 487, 325–329 CrossRef CAS PubMed.
- R. Sharma, A. Rosenberg, E. R. Bennett, D. T. Laskowitz and S. K. Acheson, PLoS One, 2017, 12, e0173798 CrossRef PubMed.
- K. Blennow, D. L. Brody, P. M. Kochanek, H. Levin, A. McKee, G. M. Ribbers, K. Yaffe and H. Zetterberg, Nat. Rev. Dis. Primers, 2016, 2, 16084 CrossRef PubMed.
- B. I. Martinez, B. I. Martinez and S. E. Stabenfeldt, J. Biol. Eng., 2019, 16, 1–12 Search PubMed.
- S. Yokobori, K. Hosein, S. Burks, I. Sharma, S. Gajavelli and R. Bullock, CNS Neurosci. Ther., 2013, 19, 556–565 CrossRef CAS PubMed.
- K. Blennow, D. L. Brody, P. M. Kochanek, H. Levin, A. McKee, G. M. Ribbers, K. Yaffe and H. Zetterberg, Nat. Rev. Dis. Primers, 2016, 2, 16084 CrossRef PubMed.
- K. K. Wang, Z. Yang, T. Zhu, Y. Shi, R. Rubenstein, J. A. Tyndall and G. T. Manley, Expert Rev. Mol. Diagn., 2018, 18, 165–180 CrossRef CAS PubMed.
- Z. S. Gan, S. C. Stein, R. Swanson, S. Guan, L. Garcia, D. Mehta and D. H. Smith, Front. Neurol, 2019, 10, 446 1–14 Search PubMed.
- M. Raad, E. Nohra, N. Chams, M. Itani, F. Talih, S. Mondello and F. Kobeissy, Neuroscience, 2014, 281, 16–23 CrossRef CAS PubMed.
- N. Marchi, J. J. Bazarian, V. Puvenna, M. Janigro, C. Ghosh, J. Zhong, T. Zhu, E. Blackman, D. Stewart, J. Ellis, R. Butler and D. Janigro, PLoS One, 2013, 8, e56805 CrossRef CAS PubMed.
- F. Kobeissy and R. A. Moshourab, in Brain neurotrauma: molecular, neuropsychological and rehabilitation aspects, CRC Press/Taylor & Francis, 2015, pp. 397–416 Search PubMed.
- Z. Zhang, J. S. Zoltewicz, S. Mondello, K. J. Newsom, Z. Yang, B. Yang, F. Kobeissy, J. Guingab, O. Glushakova, S. Robicsek, S. Heaton, A. Buki, J. Hannay, M. S. Gold, R. Rubenstein, X. M. Lu, J. R. Dave, K. Schmid, F. Tortella, C. S. Robertson and K. K. W. Wang, PLoS One, 2014, 9, e92698 CrossRef PubMed.
- L. Undén, O. Calcagnile, J. Undén, P. Reinstrup and J. Bazarian, BMC Med., 2015, 13, 292–301 CrossRef PubMed.
- O. Calcagnile, A. Anell and J. Undén, BMC Neurol., 2016, 16, 200 CrossRef PubMed.
- J. J. Bazarian, P. Biberthaler, R. D. Welch, L. M. Lewis, P. Barzo, V. Bogner-Flatz, P. G. Brolinson, A. Büki, J. Y. Chen, R. H. Christenson, D. Hack, J. S. Huff, S. Johar, J. D. Jordan, B. A. Leidel, T. Lindner, E. Ludington, D. O. Okonkwo, J. Ornato, W. F. Peacock, K. Schmidt, J. A. Tyndall, A. Vossough and A. S. Jagoda, Lancet Neurol., 2018, 17, 782–789 CrossRef CAS PubMed.
- H. F. Lingsma, J. K. Yue, A. I. Maas, E. W. Steyerberg, G. T. Manley, T.-T. I. including, S. R. Cooper, K. Dams-O'Connor, W. A. Gordon, D. K. Menon, P. Mukherjee, D. O. Okonkwo, A. M. Puccio, D. M. Schnyer, A. B. Valadka, M. J. Vassar and E. L. Yuh, J. Neurotrauma, 2015, 32, 83–94 CrossRef PubMed.
- L. Lagerstedt, L. Azurmendi, O. Tenovuo, A. J. Katila, R. S. K. Takala, K. Blennow, V. F. J. Newcombe, H.-R. Maanpää, J. Tallus, I. Hossain, M. van Gils, D. K. Menon, P. J. Hutchinson, H. Zetterberg, J. P. Posti and J.-C. Sanchez, Front. Neurol, 2020, 11(376), 1–9 Search PubMed.
- A. I. R. Maas, D. K. Menon, P. D. Adelson, N. Andelic, M. J. Bell, A. Belli, P. Bragge, A. Brazinova, A. Büki, R. M. Chesnut, G. Citerio, M. Coburn, D. J. Cooper, A. T. Crowder, E. Czeiter, M. Czosnyka, R. Diaz-Arrastia, J. P. Dreier, A.-C. Duhaime, A. Ercole, T. A. van Essen, V. L. Feigin, G. Gao, J. Giacino, L. E. Gonzalez-Lara, R. L. Gruen, D. Gupta, J. A. Hartings, S. Hill, J.-Y. Jiang, N. Ketharanathan, E. J. O. Kompanje, L. Lanyon, S. Laureys, F. Lecky, H. Levin, H. F. Lingsma, M. Maegele, M. Majdan, G. Manley, J. Marsteller, L. Mascia, C. McFadyen, S. Mondello, V. Newcombe, A. Palotie, P. M. Parizel, W. Peul, J. Piercy, S. Polinder, L. Puybasset, T. E. Rasmussen, R. Rossaint, P. Smielewski, J. Söderberg, S. J. Stanworth, M. B. Stein, N. von Steinbüchel, W. Stewart, E. W. Steyerberg, N. Stocchetti, A. Synnot, B. Te Ao, O. Tenovuo, A. Theadom, D. Tibboel, W. Videtta, K. K. W. Wang, W. H. Williams, L. Wilson and K. Yaffe, InTBIR Participants and Investigators, Lancet Neurol., 2017, 16, 987–1048 CrossRef PubMed.
- A. Wu and W. F. Peacock, Biomarkers for Traumatic Brain Injury, Elsevier Science, 2020 Search PubMed.
- H. Zetterberg and K. Blennow, Nat. Rev. Neurol., 2016, 12, 563 CrossRef CAS PubMed.
- Y. Naegelin, H. Dingsdale, K. Säuberli, S. Schädelin, L. Kappos and Y.-A. Barde, eNeuro, 2018, 5, 1–9 CrossRef PubMed , e0419-17.2018.
- F. K. Korley, R. Diaz-Arrastia, A. H. B. Wu, J. K. Yue, G. T. Manley, H. I. Sair, J. Van Eyk, A. D. Everett, D. O. Okonkwo, A. B. Valadka, W. A. Gordon, A. I. R. Maas, P. Mukherjee, E. L. Yuh, H. F. Lingsma, A. M. Puccio, D. M. Schnyer and D. M. Schnyer, J. Neurotrauma, 2016, 33, 215–225 CrossRef PubMed.
- S. Verma, C.-H. Wang, E. Lonn, F. Charbonneau, J. Buithieu, L. M. Title, M. Fung, S. Edworthy, A. C. Robertson and T. J. Anderson, Eur. Heart J., 2004, 25, 1754–1760 CrossRef PubMed.
- S. Lee, J.-W. Choe, H.-K. Kim and J. Sung, J. Epidemiol., 2011, 21, 161–168 CrossRef PubMed.
- G. A. Quinones-Ossa, H. Padilla-Zambrano, R. Pal, A. Ghosh, L. R. Moscote-Salazar, V. K. Kumar and A. Agrawal, J. Acute Dis., 2019, 8, 1–6 CrossRef.
- R. P. Anada, K. T. Wong, J. J. Jayapalan, O. H. Hashim and D. Ganesan, Electrophoresis, 2018, 39, 2308–2315 CrossRef CAS PubMed.
- U. Missler, M. Wiesmann, G. Wittmann, O. Magerkurth and H. Hagenström, Clin. Chem., 1999, 45, 138–141 CrossRef CAS.
- J. J. Bazarian, P. Biberthaler, R. D. Welch, L. M. Lewis, P. Barzo, V. Bogner-Flatz, P. G. Brolinson, A. Büki, J. Y. Chen, R. H. Christenson, D. Hack, J. S. Huff, S. Johar, J. D. Jordan, B. A. Leidel, T. Lindner, E. Ludington, D. O. Okonkwo, J. Ornato, W. F. Peacock, K. Schmidt, J. A. Tyndall, A. Vossough and A. S. Jagoda, Lancet Neurol., 2018, 17, 782–789 CrossRef CAS PubMed.
- F. Omori, S. Okamura, K. Shimoda, T. Otsuka, M. Harada and Y. Niho, Biotherapy, 1992, 4, 147–153 CrossRef CAS PubMed.
- T. Frugier, M. C. Morganti-Kossmann, D. O'Reilly and C. A. McLean, J. Neurotrauma, 2010, 27, 497–507 CrossRef PubMed.
- U. Hoffmann, F. Espeter, C. Weiss, P. Ahmad-Nejad, S. Lang, M. Brueckmann, I. Akin, M. Neumaier, M. Borggrefe and M. Behnes, BMC Cardiovasc. Disord., 2015, 15, 50 CrossRef PubMed.
- J. Abir, S. Sondes, E. Rania, K. Latifa, B. D. Mokhles, B. Nedia, B. Hadj, M. Manel, K. Souhir, G. Hejer, F. Salima and M. Abdelhedi, Int. J. Pharma Sci. Res., 2017, 8, 1441–1448 CAS.
- L. Lagerstedt, J. J. Egea-Guerrero, A. Bustamante, J. Montaner, A. Rodriguez-Rodriguez, A. El Rahal, N. Turck, M. Quintana, R. Garcia-Armengol, C. M. Prica, E. Andereggen, L. Rinaldi, A. Sarrafzadeh, K. Schaller and J.-C. Sanchez, PLoS One, 2017, 12, e0175572 CrossRef PubMed.
- H. Adrian, K. Marten, N. Salla and V. Lasse, eNeuro, 2016, 3, 1–13 CrossRef PubMed , e0294-16.2016.
- R. P. Berger, S. Ta'Asan, A. Rand, A. Lokshin and P. Kochanek, Pediatr. Res., 2009, 65, 97–102 CrossRef CAS PubMed.
- T. Woodcock and C. Morganti-Kossmann, Front. Neurol, 2013, 4, 18 CAS.
- A. P. Di Battista, S. G. Rhind, M. G. Hutchison, S. Hassan, M. Y. Shiu, K. Inaba, J. Topolovec-Vranic, A. C. Neto, S. B. Rizoli and A. J. Baker, Neuroinflammation, 2016, 13, 40 CrossRef PubMed.
- A. H. Sarris, K.-O. Kliche, P. Pethambaram, A. Preti, S. Tucker, C. Jackow, O. Messina, W. Pugh, F. B. Hagemeister, P. McLaughlin, M.-A. Rodriguez, J. Romaguera, H. Fritsche, T. Witzig, M. Duvic, M. Andreeff and F. Cabanillas, Ann. Oncol., 1999, 10, 433–440 CrossRef CAS PubMed.
- L. Lagerstedt, J. J. Egea-Guerrero, A. Rodríguez-Rodríguez, A. Bustamante, J. Montaner, A. El Rahal, E. Andereggen, L. Rinaldi, A. Sarrafzadeh, K. Schaller and J.-C. Sanchez, PLoS One, 2018, 13, e0193278 CrossRef PubMed.
- K. M. Thrailkill, C. S. Moreau, G. Cockrell, P. Simpson, R. Goel, P. North, J. L. Fowlkes and R. C. Bunn, Clin. Chem. Lab. Med., 2005, 43, 1392–1399 CAS.
- W.-Z. Shi, J.-Y. Ju, H.-J. Xiao, F. Xue, J. Wu, M.-M. Pan and W.-F. Ni, Mol. Med. Rep., 2017, 15, 2129–2135 CrossRef CAS PubMed.
- L. Lorente, Arch Trauma Res., 2015, 4, e30165 Search PubMed.
- W. F. Peacock, T. E. Van Meter, N. Mirshahi, K. Ferber, R. Gerwien, V. Rao, H. I. Sair, R. Diaz-Arrastia and F. K. Korley, Front. Neurol, 2017, 8, 641 CrossRef PubMed.
- H. An, L. Zhou, Y. Yu, H. Fan, F. Fan, S. Tan, Z. Wang, B. Zehre, J. Shi, F. Yang, X. Zhang, Y. Tan and X.-F. Huang, Schizophr. Res., 2018, 192, 457–458 CrossRef PubMed.
- W. Zheng, Q. ZhuGe, M. Zhong, G. Chen, B. Shao, H. Wang, X. Mao, L. Xie and K. Jin, J. Neurotrauma, 2013, 30, 1872–1880 CrossRef PubMed.
- D. L. Emery, R. Raghupathi, K. E. Saatman, I. Fischer, M. S. Grady and T. K. McIntosh, J. Comp. Neurol., 2000, 424, 521–531 CrossRef CAS PubMed.
- A. G. B. Thompson, C. Luk, A. J. Heslegrave, H. Zetterberg, S. H. Mead, J. Collinge and G. S. Jackson, J. Neurol., Neurosurg. Psychiatry, 2018, 89, 955–961 CrossRef PubMed.
- P. Shahim, M. Gren, V. Liman, U. Andreasson, N. Norgren, Y. Tegner, N. Mattsson, N. Andreasen, M. Öst, H. Zetterberg, B. Nellgård and K. Blennow, Sci. Rep., 2016, 6, 36791 CrossRef CAS PubMed.
- G. L. Iverson, P. J. Reddi, J. P. Posti, A.-K. Kotilainen, O. Tenovuo, J. Öhman, H. Zetterberg, K. Blennow and T. M. Luoto, J. Neurotrauma, 2019, 36, 2400–2406 CrossRef PubMed.
- L. Xue, H. Chen, K. Lu, J. Huang, H. Duan and Y. Zhao, J. Neurol. Sci., 2017, 375, 52–57 CrossRef CAS PubMed.
- H. Chen, H.-L. Cao, S.-W. Chen, Y. Guo, W.-W. Gao, H.-L. Tian and L.-X. Xue, Biomarkers, 2015, 20, 495–501 Search PubMed.
- J. Yang, F. K. Korley, M. Dai and A. D. Everett, Clin. Biochem., 2015, 48, 843–848 CrossRef CAS PubMed.
- P. J. Marangos and D. E. Schmechel, Annu. Rev. Neurosci., 1987, 10, 269–295 CrossRef CAS PubMed.
- H. Saidi, A. Dashti, M. A. Aashari, S. G. Gharab, M. Rezai and M. Nasirizadeh, 2019, 7, 15–20.
- F. Cheng, Q. Yuan, J. Yang, W. Wang and H. Liu, PLoS One, 2014, 9, e106680 CrossRef PubMed.
- M. Wiesmann, U. Missler, D. Gottmann and S. Gehring, Clin. Chem., 1998, 44, 1056–1058 CrossRef CAS.
- E. Gordillo-Escobar, J. J. Egea-Guerrero, A. Rodríguez-Rodríguez and F. Murillo-Cabezas, Med. Intensiva Engl. Ed., 2016, 40, 105–112 CrossRef CAS PubMed.
- L. M. Lewis, D. T. Schloemann, M. Lindburg, L. Papa, R. P. Fucetola, J. Bazarian and R. D. Welch, Acad. Emerg. Med., 2017, 24, 710–720 CrossRef PubMed.
- M. Bulut, O. Koksal, S. Dogan, N. Bolca, H. Ozguc, E. Korfali, Y. O. Ilcol and M. Parlak, Adv. Ther., 2006, 23, 12–22 CrossRef CAS PubMed.
- T. T. V. Nu, N. H. T. Tran, E. Nam, T. T. Nguyen, W. J. Yoon, S. Cho, J. Kim, K.-A. Chang and H. Ju, RSC Adv., 2018, 8, 7855–7862 RSC.
- P. Shahim, K. Blennow, H. Zetterberg and Y. Tegner, Br. J. Sports Med., 2017, 51, A6–A7 CrossRef.
- S. Mondello, F. Kobeissy, A. Vestri, R. L. Hayes, P. M. Kochanek and R. P. Berger, Sci. Rep., 2016, 6, 28203 CrossRef CAS PubMed.
- J. W. Ho, R. T. Poon, C. S. Tong and S. T. Fan, World J. Gastroenterol., 2004, 10, 2014–2018 CrossRef CAS PubMed.
- University of Pittsburgh of the Commonwealth System of Higher Education, PCT/US Pat., WO 2017/197028 A1, Axela, Inc, 2017, p. 77.
- Y.-J. Wang, Y.-W. Hsu, C.-M. Chang, C.-C. Wu, J.-C. Ou, Y.-R. Tsai, W.-T. Chiu, W.-C. Chang, Y.-H. Chiang and K.-Y. Chen, BioMed Res. Int., 2014, 2014, 293687 Search PubMed.
- I. M. Skogseid, H. K. Nordby, P. Urdal, E. Paus and F. Lilleaas, Acta Neurochir., 1992, 115, 106–111 CrossRef CAS PubMed.
- Y.-Z. Liu, B. Chen and X.-D. She, World J. Gastroenterol., 1998, 4, 225–227 CrossRef PubMed.
- K. Giannoulis, C. Angouridaki, G. Fountzilas, C. Papapolychroniadis, E. Giannoulis and O. Gamvros, Tech. Coloproctol., 2004, 8, s65–s67 CrossRef PubMed.
- E. G. McKeating, P. J. D. Andrews and L. Mascia, Acta Neurochir., 1998, 71, 200–202 CAS.
- M. Hiki, K. Shimada, H. Ohmura, T. Kiyanagi, A. Kume, K. Sumiyoshi, K. Fukao, N. Inoue, H. Mokuno, T. Miyazaki and H. Daida, J. Cardiol., 2009, 53, 108–116 CrossRef PubMed.
- E. Martínez-Morillo, C. Childs, B. P. García, F. V. Á. Menéndez, A. D. Romaschin, G. Cervellin, G. Lippi and E. P. Diamandis, Clin. Chem. Lab. Med., 2015, 53, 1575–1584 Search PubMed.
- X. Qiao, S. Zhang, W. Zhao, H. Ye, Y. Yang, Z. Zhang, Q. Miao, R. Hu, Y. Li and B. Lu, Medicine, 2015, 94, e1908 CrossRef CAS PubMed.
- K. Shibahashi, T. Doi, S. Tanaka, H. Hoda, H. Chikuda, Y. Sawada, Y. Takasu, K. Chiba, T. Nozaki, Y. Hamabe and T. Ogata, J. Neurotrauma, 2016, 33, 1826–1833 CrossRef PubMed.
- D. Alexiou, A. J. Karayiannakis, K. N. Syrigos, A. Zbar, E. Sekara, P. Michail, T. Rosenberg and T. Diamantis, Am. J. Gastroenterol., 2003, 98, 478–485 CrossRef CAS PubMed.
- J. Wang, E. Su, H. Wang, C. Guo, D. A. Lawrence and D. T. Eitzman, Sci. Rep., 2018, 8, 5639 CrossRef PubMed.
- R. Siman, D. H. Smith, P. Shahim, K. Blennow, H. Zetterberg, Y. Tegner and H. Zetterberg, J. Neurotrauma, 2015, 32, 1294–1300 CrossRef PubMed.
- R. Siman, N. Giovannone, G. Hanten, E. A. Wilde, S. R. McCauley, J. V. Hunter, X. Li, H. S. Levin and D. H. Smith, Front. Neurol, 2013, 4, 190 Search PubMed.
- F. Job, F. Settele, S. Lorey, C. Rundfeldt, L. Baumann, A. G. Beck-Sickinger, U. Haupts, H. Lilie and E. Bosse-Doenecke, FEBS Open Bio, 2015, 5, 579–593 CrossRef CAS PubMed.
- M. Takagi, M. Yamauchi, G. Toda, K. Takada, T. Hirakawa and K. Ohkawa, Alcohol.: Clin. Exp. Res., 1999, 23, 76S–80S CrossRef CAS PubMed.
- K. Takada, H. Nasu, N. Hibi, Y. Tsukada, T. Shibasaki, K. Fujise, M. Fujimuro, H. Sawada, H. Yokosawa and K. Ohkawa, Clin. Chem., 1997, 43, 1188–1195 CrossRef CAS.
- E. N. Anderson, L. Gochenaur, A. Singh, R. Grant, K. Patel, S. Watkins, J. Y. Wu and U. B. Pandey, Hum. Mol. Genet., 2018, 27, 1366–1381 CrossRef CAS PubMed.
- Z. Xu, R. D. Henderson, M. David and P. A. McCombe, PLoS One, 2016, 11, e0164625 CrossRef PubMed.
- H. Adrian, K. Marten, N. Salla and V. Lasse, eNeuro, 2016, 3, 1–13 CrossRef PubMed , e0294-16.2016.
- D. Slavoaca, D. Muresanu, C. Birle, O. V. Rosu, I. Chirila, I. Dobra, N. Jemna, S. Strilciuc and P. Vos, Neurol. Sci., 2020, 41, 2033–2044 CrossRef PubMed.
- H. Tammen, I. Schulte, R. Hess, C. Menzel, M. Kellmann, T. Mohring and P. Schulz-Knappe, Proteomics, 2005, 5, 3414–3422 CrossRef CAS PubMed.
- P. G. Rezaii, G. A. Grant, M. M. Zeineh, K. J. Richardson, M. L. Coburn, A. M. Bet, A. Weber, B. Jiang, Y. Li, K. Ubungen, G. Routh, A. M. Wheatcroft, A. D. Paulino, R. L. Hayes, G. K. Steinberg and M. Wintermark, J. Neurotrauma, 2019, 36, 2407–2416 CrossRef PubMed.
- S.-Y. Hsieh, R.-K. Chen, Y.-H. Pan and H.-L. Lee, Proteomics, 2006, 6, 3189–3198 CrossRef CAS PubMed.
- F.-M. S. Kong, L. Zhao, L. Wang, Y. Chen, J. Hu, X. Fu, C. Bai, L. Wang, T. S. Lawrence, M. S. Anscher, A. Dicker and P. Okunieff, Transl. Lung Cancer Res., 2017, 6, 625–634 CrossRef CAS PubMed.
- cTnI Test Cartridge, https://www.pointofcare.abbott/us/en/offerings/istat/istat-test-cartridges/cTnI, (accessed December 28, 2020) Search PubMed.
- M. Labib, E. H. Sargent and S. O. Kelley, Chem. Rev., 2016, 116, 9001–9090 CrossRef CAS PubMed.
- H. Ju, G. Lai and F. Yan, Immunosensing for Detection of Protein Biomarkers, Elsevier, 2017 Search PubMed.
- WHO, Target product profiles, http://www.who.int/research-observatory/analyses/tpp/en/, (accessed March 3, 2021) Search PubMed.
- P. Cocco, A. Ayaz-Shah, M. P. Messenger, R. M. West and B. Shinkins, BMC Med., 2020, 18, 119 CrossRef PubMed.
- D. Bouvier, C. Oris, M. Brailova, J. Durif and V. Sapin, Clin. Biochem., 2020, 85, 5–11 CrossRef CAS PubMed.
- F. K. Korley, J. K. Yue, D. H. Wilson, K. Hrusovsky, R. Diaz-Arrastia, A. R. Ferguson, E. L. Yuh, P. Mukherjee, K. K. W. Wang, A. B. Valadka, A. M. Puccio, D. O. Okonkwo and G. T. Manley, J. Neurotrauma, 2019, 36, 182–187 CrossRef PubMed.
- J. P. Posti, R. S. K. Takala, L. Lagerstedt, A. M. Dickens, I. Hossain, M. Mohammadian, H. Ala-Seppälä, J. Frantzén, M. van Gils, P. J. Hutchinson, A. J. Katila, H.-R. Maanpää, D. K. Menon, V. F. Newcombe, J. Tallus, K. Hrusovsky, D. H. Wilson, J. Gill, J.-C. Sanchez, O. Tenovuo, H. Zetterberg and K. Blennow, J. Neurotrauma, 2019, 36, 2178–2189 CrossRef PubMed.
- N. K. Bakirhan, G. Ozcelikay and S. A. Ozkan, J. Pharm. Biomed. Anal., 2018, 159, 406–424 CrossRef CAS PubMed.
- H. Sohrabi, H. kholafazad Kordasht, P. Pashazadeh-Panahi, P. Nezhad-Mokhtari, M. Hashemzaei, M. R. Majidi, J. Mosafer, F. Oroojalian, A. Mokhtarzadeh and M. de la Guardia, Microchem. J., 2020, 158, 105287 CrossRef CAS.
- K. Dhara and D. R. Mahapatra, Microchem. J., 2020, 156, 104857 CrossRef CAS.
- M. A. Khan and M. Mujahid, Sensors, 2020, 20, 646 CrossRef PubMed.
- X. Chen, T. Dong, X. Wei, Z. Yang, N. M. Matos Pires, J. Ren and Z. Jiang, Biosens. Bioelectron., 2019, 142, 111453 CrossRef CAS PubMed.
- M. Freitas, H. P. A. Nouws and C. Delerue-Matos, Electroanalysis, 2018, 30, 1584–1603 CrossRef.
- C. Andrade, M. D. Oliveira, T. Faulin, V. Hering and D. S. P. Abdalla, in Biosensors for Health, Environment and Biosecurity, IntechOpen, 2011, pp. 215–240 Search PubMed.
- D. R. Thevenot, K. Toth, R. A. Durst and G. S. Wilson, Pure Appl. Chem., 1999, 71, 2333–2348 CAS.
- Y.-C. Kuo, C.-K. Lee and C.-T. Lin, Biosens. Bioelectron., 2018, 103, 130–137 CrossRef CAS PubMed.
- Y.-C. Kuo, C.-K. Lee and C.-T. Lin, Data Brief, 2018, 17, 1288–1294 CrossRef PubMed.
- E. Cantù, S. Tonello, G. Abate, D. Uberti, E. Sardini and M. Serpelloni, Sensors, 2018, 18, 3719 1–14 CrossRef.
- Z.-T. Lin, Y. Li, J. Gu, H. Wang, Z. Zhu, X. Hong, Z. Zhang, Q. Lu, J. Qiu and X. Wang, Adv. Funct. Mater., 2018, 28, 1802482 CrossRef.
- A. Garcia-Cruz, F. Nessark, M. Lee, N. Zine, M. Sigaud, R. Pruna, M. Lopez, P. Marote, J. Bausells, N. Jaffrezic-Renault and A. Errachid, Sens. Actuators, B, 2018, 255, 2520–2530 CrossRef CAS.
- N. S. Ramgir, P. K. Sekhar, A. Zajac, L. Lee, T. Zhukov and S. Bhansali, Sens. Lett., 2007, 5, 608–611 CrossRef CAS.
- N. S. Ramgir, A. Zajac, P. K. Sekhar, L. Lee, T. A. Zhukov and S. Bhansali, J. Phys. Chem. C, 2007, 111, 13981–13987 CrossRef CAS.
- M. Lee, N. Zine, A. Baraket, M. Zabala, F. Campabadal, R. Caruso, M. G. Trivella, N. Jaffrezic-Renault and A. Errachid, Sens. Actuators, B, 2012, 175, 201–207 CrossRef CAS.
- D. S. Juang, C.-H. Lin, Y.-R. Huo, C.-Y. Tang, C.-R. Cheng, H.-S. Wu, S.-F. Huang, A. Kalnitsky and C.-C. Lin, Biosens. Bioelectron., 2018, 117, 175–182 CrossRef CAS PubMed.
- M. Lee, A. Baraket, N. Zine, M. Zabala, F. Campabadal, R. Caruso, M. G. Trivella, N. Jaffrezic-Renault and A. Errachid, Methods Mol. Biol., 2015, 1172, 49–64 CrossRef CAS PubMed.
- M. Aydın, E. B. Aydın and M. K. Sezgintürk, Biosens. Bioelectron., 2018, 117, 720–728 CrossRef PubMed.
- E. B. Aydın and M. K. Sezgintürk, Anal. Biochem., 2018, 554, 44–52 CrossRef.
- M. Aydın, E. B. Aydın and M. K. Sezgintürk, Macromol. Biosci., 2019, 19, 1900109 CrossRef PubMed.
- A. Carbonaro and L. L. Sohn, Lab Chip, 2005, 5, 1155–1160 RSC.
- University of Pittsburgh of the Commonwealth System of Higher Education, PCT/US Pat., WO 2018/107143 A1, Axela Inc, 2018, p. 25.
- V. Kamakoti, N. R. Shanmugam, A. S. Tanak, B. Jagannath and S. Prasad, Appl. Surf. Sci., 2018, 436, 441–450 CrossRef CAS.
- Z. Wang, P. Dong, Z. Sun, C. Sun, H. Bu, J. Han, S. Chen and G. Xie, J. Mater. Chem. B, 2018, 6, 2426–2431 RSC.
- C. S. Park, R. Colorado, A. C. Jamison and T. R. Lee, in Encyclopedia of Materials: Science and Technology, Elsevier, 2016, pp. 9332–9344 Search PubMed.
- L. Srisombat, A. C. Jamison and T. R. Lee, Colloids Surf., A, 2011, 390, 1–19 CrossRef CAS.
- S. Yin, L. Zhao and Z. Ma, Anal. Bioanal. Chem., 2018, 410, 1279–1286 CrossRef CAS PubMed.
- W. Putzbach and N. Ronkainen, Sensors, 2013, 13, 4811–4840 CrossRef CAS.
- A. S. Mathew, X. Shi and S.-T. Yau, Mol. Diagn. Ther., 2018, 22, 729–735 CrossRef CAS PubMed.
- T. Wang, Y. Fang and Z. He, Int. J. Electrochem. Sci., 2017, 12, 7341–7350 CrossRef CAS.
- X. Li, M. Jiang, J. Cheng, M. Ye, W. Zhang, N. Jaffrezic-Renault and Z. Guo, Microchim. Acta, 2020, 187, 302–309 CrossRef CAS PubMed.
- H. Wang, Z. Ma and H. Han, Bioelectrochemistry, 2019, 130, 107324 CrossRef CAS PubMed.
- Y. Wang, J. Luo, J. Liu, S. Sun, Y. Xiong, Y. Ma, S. Yan, Y. Yang, H. Yin and X. Cai, Biosens. Bioelectron., 2019, 136, 84–90 CrossRef CAS PubMed.
- Q. Zhang, X. Li, C. Qian, L. Dou, F. Cui and X. Chen, Anal. Biochem., 2018, 540–541, 1–8 CAS.
- M. H. Akhtar, K. K. Hussain, N. G. Gurudatt, P. Chandra and Y.-B. Shim, Biosens. Bioelectron., 2018, 116, 108–115 CrossRef CAS.
- N. Liu, H. Yi, Y. Lin, H. Zheng, X. Zheng, D. Lin and H. Dai, Microchim. Acta, 2018, 185, 277 CrossRef.
- S. Dong, H. Cui, D. Zhang and M. Tong, J. Electrochem. Soc., 2019, 166, B193–B199 CrossRef CAS.
- S. Dong, D. Zhang, H. Cui and T. Huang, Sens. Actuators, B, 2019, 284, 354–361 CrossRef CAS.
- D. Wang, Y. Yuan, Y. Zheng, Y. Chai and R. Yuan, Chem. Commun., 2016, 52, 5943–5945 RSC.
- M. Tertis, G. Melinte, B. Ciui, I. Simon, R. Stiufiuc, R. Săndulescu and C. Cristea, Electroanalysis, 2019, 31, 282–292 CrossRef CAS.
- C. Kokkinos, A. Economou and M. I. Prodromidis, TrAC, Trends Anal. Chem., 2016, 79, 88–105 CrossRef CAS.
- J. Piccoli, R. Hein, A. H. El-Sagheer, T. Brown, E. M. Cilli, P. R. Bueno and J. J. Davis, Anal. Chem., 2018, 90, 3005–3008 CrossRef CAS PubMed.
- A. Sinha, T.-Y. Tai, K.-H. Li, P. Gopinathan, Y.-D. Chung, I. Sarangadharan, H.-P. Ma, P.-C. Huang, S.-C. Shiesh, Y.-L. Wang and G.-B. Lee, Biosens. Bioelectron., 2019, 129, 155–163 CrossRef CAS PubMed.
- M. Platt and R. Maugi, Med. Devices Sens., 2020, 3, e10059 Search PubMed.
- X. Zhang, K.-N. Chi, D.-L. Li, Y. Deng, Y.-C. Ma, Q.-Q. Xu, R. Hu and Y.-H. Yang, Biosens. Bioelectron., 2019, 129, 64–71 CrossRef CAS PubMed.
- M. Tertiş, P. I. Leva, D. Bogdan, M. Suciu, F. Graur and C. Cristea, Biosens. Bioelectron., 2019, 137, 123–132 CrossRef PubMed.
- D. Tao, B. Shui, Y. Gu, J. Cheng, W. Zhang, N. Jaffrezic-Renault, S. Song and Z. Guo, Biosensors, 2019, 9, 84 CrossRef CAS PubMed.
- X. Hun and X. Kong, J. Pharm. Biomed. Anal., 2021, 113666 CrossRef CAS PubMed.
- M. Jarczewska, Ł. Górski and E. Malinowska, Anal. Methods, 2016, 8, 3861–3877 RSC.
- A.-E. Radi, Int. J. Electrochem., 2011, 2011, 1–17 CrossRef.
- G. T. Rozenblum, I. G. Pollitzer and M. Radrizzani, Chemosensors, 2019, 7, 57 CrossRef CAS.
- H. Kaur, J. G. Bruno, A. Kumar and T. K. Sharma, Theranostics, 2018, 8, 4016–4032 CrossRef CAS PubMed.
- A. Villalonga, A. M. Pérez-Calabuig and R. Villalonga, Anal. Bioanal. Chem., 2020, 412, 55–72 CrossRef CAS PubMed.
- S. Scarano, S. Lisi, C. Ravelet, E. Peyrin and M. Minunni, Anal. Chim. Acta, 2016, 940, 21–37 CrossRef CAS.
- L. Isaacs, Acc. Chem. Res., 2014, 47, 2052–2062 CrossRef CAS PubMed.
- S. Walker, R. Oun, F. J. McInnes and N. J. Wheate, Isr. J. Chem., 2011, 51, 616–624 CrossRef CAS.
- B.-B. Kou, Y.-Q. Chai, Y.-L. Yuan and R. Yuan, Anal. Chem., 2017, 89, 9383–9387 CrossRef CAS PubMed.
- X. Wang, Y. Wang, X. Ye, T. Wu, H. Deng, P. Wu and C. Li, Biosens. Bioelectron., 2018, 99, 34–39 CrossRef CAS PubMed.
- R. Tchinda, A. Tutsch, B. Schmid, R. D. Süssmuth and Z. Altintas, Biosens. Bioelectron., 2019, 123, 260–268 CrossRef CAS PubMed.
- M. Pirzada, E. Sehit and Z. Altintas, Biosens. Bioelectron., 2020, 166, 112464 CrossRef CAS PubMed.
- X. Luo and J. J. Davis, Chem. Soc. Rev., 2013, 42, 5944–5962 RSC.
- J. Liang, J. Wang, L. Zhang, S. Wang, C. Yao and Z. Zhang, New J. Chem., 2019, 43, 1372–1379 RSC.
- Y. Wang, Z. Zhang, V. Jain, J. Yi, S. Mueller, J. Sokolov, Z. Liu, K. Levon, B. Rigas and M. H. Rafailovich, Sens. Actuators, B, 2010, 146, 381–387 CrossRef CAS.
- A. T. E. Vilian, W. Kim, B. Park, S. Y. Oh, T. Kim, Y. S. Huh, C. K. Hwangbo and Y.-K. Han, Biosens. Bioelectron., 2019, 142, 111549 CrossRef CAS PubMed.
- A. P. Selvam, A. Wangzhou, M. Jacobs, T. Wu, C. Mohan and S. . Prasad, Future Sci. OA, 2017, 3, FSO224 CrossRef CAS PubMed.
- S. Y. Hwang, I. J. Seo, S. Y. Lee and Y. Ahn, J. Electroanal. Chem., 2015, 756, 118–123 CrossRef CAS.
- F. C. B. Fernandes, J. R. Andrade and P. R. Bueno, Sens. Actuators, B, 2019, 291, 493–501 CrossRef CAS.
- A. Baradoke, R. Hein, X. Li and J. J. Davis, Anal. Chem., 2020, 92, 3508–3511 CrossRef CAS PubMed.
- J. S. Daniels and N. Pourmand, Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal., 2007, 19, 1239–1257 CAS.
- P. Skládal, Electroanalysis, 1997, 9, 737–745 CrossRef.
- B. A. Cardinell, C. P. Addington, S. E. Stabenfeldt and J. T. La Belle, Crit. Rev. Biomed. Eng., 2019, 47, 193–206 CrossRef PubMed.
- B. Shui, D. Tao, J. Cheng, Y. Mei, N. Jaffrezic-Renault and Z. Guo, Analyst, 2018, 143, 3549–3554 RSC.
- M. Thangamuthu, C. Santschi and O. J. F. Martin, Biosensors, 2018, 8, 34 CrossRef PubMed.
- E. Crowley, C. O'Sullivan and G. G. Guilbault, Anal. Chim. Acta, 1999, 389, 171–178 CrossRef CAS.
- D. Wu, D. Rios-Aguirre, M. Chounlakone, S. Camacho-Leon and J. Voldman, Biosens. Bioelectron., 2018, 117, 522–529 CrossRef CAS PubMed.
- C. A. Razzino, V. Serafín, M. Gamella, M. Pedrero, A. Montero-Calle, R. Barderas, M. Calero, A. O. Lobo, P. Yáñez-Sedeño, S. Campuzano and J. M. Pingarrón, Biosens. Bioelectron., 2020, 163, 112238 CrossRef CAS.
- V. Serafin, C. A. Razzino, M. Gamella, M. Pedrero, E. Povedano, A. Montero-Calle, R. Barderas, M. Calero, A. O. Lobo, P. Yanez-Sedeno, S. Campuzano and J. M. Pingarron, Anal. Bioanal. Chem., 2021, 413(3), 799–811 CrossRef CAS PubMed.
- L.-N. Feng, Z.-P. Bian, J. Peng, F. Jiang, G.-H. Yang, Y.-D. Zhu, D. Yang, L.-P. Jiang and J.-J. Zhu, Anal. Chem., 2012, 84, 7810–7815 CrossRef CAS PubMed.
- X. Qin, A. Xu, L. Liu, Y. Sui, Y. Li, Y. Tan, C. Chen and Q. Xie, Biosens. Bioelectron., 2017, 91, 321–327 CrossRef CAS PubMed.
- J. Xu, X. Yu, L. Xie and M. Shao, Anal. Bioanal. Chem., 2020, 412, 2599–2606 CrossRef CAS PubMed.
- T. Zheng, R. Zhang, Q. Zhang, T. Tan, K. Zhang, J.-J. Zhu and H. Wang, Chem. Commun., 2013, 49, 7881–7883 RSC.
- Y. Fang, Y. Li, M. Zhang, B. Cui, Q. Hu and L. Wang, Analyst, 2019, 144, 2186–2194 RSC.
- M. J. Schöning and A. Poghossian, Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal., 2006, 18, 1893–1900 Search PubMed.
- S. Chen, Doctoral thesis, Uppsala Universitet, 2013.
- J. Song, J. Dailey, H. Li, H.-J. Jang, P. Zhang, J. T.-H. Wang, A. D. Everett and H. E. Katz, Adv. Funct. Mater., 2017, 27, 1606506 CrossRef PubMed.
- K. Arnold, A. Herrmann, L. Pratsch and K. Gawrisch, Biochim. Biophys. Acta, Biomembr., 1985, 815, 515–518 CrossRef CAS.
- Z. Hao, Y. Pan, C. Huang, Z. Wang and X. Zhao, Biomed. Microdevices, 2019, 21, 65 CrossRef PubMed.
- D. Park, J. H. Kim, H. J. Kim, D. Lee, D. S. Lee, D. S. Yoon and K. S. Hwang, Biosens. Bioelectron., 2020, 167, 112505 CrossRef CAS PubMed.
- A. Zhang, G. Zheng and C. M. Lieber, in Nanowires, Springer, 2016, pp. 255–275 Search PubMed.
- I. M. Bhattacharyya, S. Cohen, A. Shalabny, M. Bashouti, B. Akavayov and G. Shalev, Biosens. Bioelectron., 2019, 132, 143–161 CrossRef CAS PubMed.
- W.-W. Zhao, J.-J. Xu and H.-Y. Chen, Chem. Rev., 2014, 114, 7421–7441 CrossRef CAS PubMed.
- A. Victorious, S. Saha, R. Pandey, T. Didar and L. Soleymani, Front. Chem., 2019, 7, 617 CrossRef CAS PubMed.
- G.-C. Fan, L. Han, H. Zhu, J.-R. Zhang and J.-J. Zhu, Anal. Chem., 2014, 86, 12398–12405 CrossRef CAS PubMed.
- J. Li, Y. Li, L. Xu, X. Fang, H. Yin, Q. Xu, H. Fang, H. Li and W. Wang, Sens. Actuators, B, 2020, 320, 128597 CrossRef CAS.
- K. Kim, G. R. Lee, M. Kim, H. Lim, Y. S. Jung and C. B. Park, ACS Nano, 2020, 14, 10376–10384 CrossRef CAS PubMed.
- R. A. Soomro, N. H. Kalwar, A. Avci, E. Pehlivan, K. R. Hallam and M. Willander, Biosens. Bioelectron., 2019, 141, 111331 CrossRef CAS PubMed.
- K. Kim and C. B. Park, Biosens. Bioelectron., 2020, 154, 112075 CrossRef CAS PubMed.
- M.-J. Li, H.-J. Wang, R. Yuan and Y.-Q. Chai, Chem. Commun., 2019, 55, 10772–10775 RSC.
- N. Pachauri, G. B. V. S. Lakshmi, S. Sri, P. K. Gupta and P. R. Solanki, Mater. Sci. Eng., C, 2020, 110911 CrossRef CAS PubMed.
- T. Putnin, A. Ngamaroonchote, N. Wiriyakun, K. Ounnunkad and R. Laocharoensuk, Microchim. Acta, 2019, 186, 305 CrossRef PubMed.
- S. Khetani, V. Ozhukil Kollath, V. Kundra, M. D. Nguyen, C. Debert, A. Sen, K. Karan and A. Sanati-Nezhad, ACS Sens., 2018, 3, 844–851 CrossRef CAS PubMed.
- S. K. Arya, T. S. Pui, C. C. Wong, S. Kumar and A. R. A. Rahman, Langmuir, 2013, 29, 6770–6777 CrossRef CAS.
- M. Buff, E. Drab and K. Sugihara, Biointerphases, 2019, 14, 011004 CrossRef CAS PubMed.
- A. D. Keefe, S. Pai and A. Ellington, Nat. Rev. Drug Discovery, 2010, 9, 537–550 CrossRef CAS.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Sensors, 2008, 8, 1400–1458 CrossRef CAS PubMed.
- C. Berggren, B. Bjarnason and G. Johansson, Electroanal. Int. J. Devoted Fundam. Pract. Asp. Electroanal., 2001, 13, 173–180 CAS.
- R. Diaz-Arrastia, K. K. W. Wang, L. Papa, M. D. Sorani, J. K. Yue, A. M. Puccio, P. J. McMahon, T. Inoue, E. L. Yuh, H. F. Lingsma, A. I. R. Maas, A. B. Valadka, D. O. Okonkwo, G. T. Manley and TRACK-TBI Investigators, J. Neurotrauma, 2014, 31, 19–25 CrossRef PubMed.
- Abbott Receives FDA 510(k) Clearance for the First Rapid Handheld Blood Test for Concussions, https://abbott.mediaroom.com/2021-01-11-Abbott-Receives-FDA-510-k-Clearance-for-the-First-Rapid-Handheld-Blood-Test-for-Concussions, (accessed March 24, 2021) Search PubMed.
- biodirection_zmuop8, Nanosensor Technology | Our Solution, https://nanodiagnostics.com/our-solution/nanosensor-technology/, (accessed March 29, 2021) Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: SI-1: full list of published electrochemical strategies for the detection of blood protein biomarkers relevant to mTBI; SI-2: summary of key observations and outstanding challenges; SI-3: antifouling approaches in EC sensing. See DOI: 10.1039/d1ra00589h |
| This journal is © The Royal Society of Chemistry 2021 |