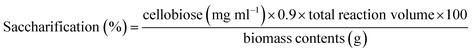Open Access Article
Open Access ArticleEfficient biomass saccharification using a novel cellobiohydrolase from Clostridium clariflavum for utilization in biofuel industry†
Asma Zafar‡
 a,
Muhammad Nauman Aftab‡*b,
Anam Asifb,
Ahmet Karadagc,
Liangcai Pengd,
Hassan Ufak Celebioglue,
Muhammad Sohail Afzalf,
Attia Hamidb and
Irfana Iqbalg
a,
Muhammad Nauman Aftab‡*b,
Anam Asifb,
Ahmet Karadagc,
Liangcai Pengd,
Hassan Ufak Celebioglue,
Muhammad Sohail Afzalf,
Attia Hamidb and
Irfana Iqbalg
aFaculty of Life Sciences, University of Central Punjab, Lahore, Pakistan
bInstitute of Industrial Biotechnology, GC University, Lahore, Pakistan. E-mail: nauman535@yahoo.com; Tel: +92-3444704190
cDepartment of Chemistry, Faculty of Arts and Sciences, Yozgat Bozok University, 66200, Yozgat, Turkey
dBiomass and Bioenergy Research Center, Huazhong Agriculture University, Wuhan, China
eDepartment of Biotechnology, Kutlubey Campus, 74100, Bartin, Turkey
fDepartment of Life Sciences, School of Science, University of Management and Technology (UMT), Lahore, Pakistan
gDepartment of Zoology, Lahore College for Women University, Lahore, Pakistan
First published on 1st March 2021
Abstract
The present study describes the cloning of the cellobiohydrolase gene from a thermophilic bacterium Clostridium clariflavum and its expression in Escherichia coli BL21(DE3) utilizing the expression vector pET-21a(+). The optimization of various parameters (pH, temperature, isopropyl β-D-1-thiogalactopyranoside (IPTG) concentration, time of induction) was carried out to obtain the maximum enzyme activity (2.78 ± 0.145 U ml−1) of recombinant enzyme. The maximum expression of recombinant cellobiohydrolase was obtained at pH 6.0 and 70 °C respectively. Enzyme purification was performed by heat treatment and immobilized metal anionic chromatography. The specific activity of the purified enzyme was 57.4 U mg−1 with 35.17% recovery and 3.90 purification fold. Sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) showed that the molecular weight of cellobiohydrolase was 78 kDa. Among metal ions, Ca2+ showed a positive impact on the cellobiohydrolase enzyme with increased activity by 115%. Recombinant purified cellobiohydrolase enzyme remained stable and exhibited 77% and 63% residual activity in comparison to control in the presence of n-butanol and after incubation at 80 °C for 1 h, respectively. Our results indicate that our purified recombinant cellobiohydrolase can be used in the biofuel industry.
Introduction
Due to the depletion of fossil fuels and increased prices of crude oils, access to renewable energy sources has become crucial to maintain the equilibrium of modern economies.1 Lignocellulose being the most abundant renewable energy reservoir on earth, has the great potential to be utilized as a substrate for biofuel production in the biofuel industry. But the main limitation occurs in its conversion to fermentable sugars due to its rigid and compact structure.2,3 The enzymatic hydrolysis of lignocellulosic biomass into simple sugars that can be fermented into biofuel, requires a complete set of enzymes that includes cellulase, hemicellulases and ligninases.4 To depolymerize the lignocellulosic plant biomass, these enzymes act synergistically and specifically and convert it into simple fermentable sugars for biofuel production at industrial scale.5 Based on their mode of action, the cellulases are divided into three groups: β-glucosidases, cellobiohydrolase and endoglucanase.6 Endoglucanases randomly cleaves amorphous cellulose regions and generates oligosaccharides of various sizes.7,8 Cellobiohydrolases act on the terminals of the cellulose chains and produces either glucose or cellobiose. Lastly, β-glucosidases act on the cellobioses and convert them into glucose.9Cellobiohydrolases are important component of cellulases system and play important role in the production of biofuel from plant biomass. Cellobiohydrolases are also called exo-1-4-p-D-glucanases or cellobiohydrolases due to the generation of cellobioses. These enzymes include both 1,4-p-D-glucan glucohydrolases (EC 3.2.1.74) as well as 1,4-p-D-glucan cellobiohydrolase (EC 3.2.1.91).10 Cellobiohydrolases generally cleave the cellulose strand by attaching to the ends of microfibrils and results in the production of cellobiose. Mostly these enzymes belong to glycoside hydrolase family six, seven and nine, however, some of these enzymes also found in GH-48 and GH-5.11 Cellobiohydrolases typically acts in a directional preference which usually comes from the structural arrangements of the active site of enzyme as well as initial recognition of specific cellulose chain and preferential binding of carbohydrate binding module of the cellulose.12
Cellobiohydrolases are mostly produced by fungal species but some bacteria also contain the genes for the production of exoglucanases. Among bacteria, Clostridia are attracting considerable attention due to their well-known cellulosomes systems.13,14 Cellulosomes are complexes of enzymes which are capable of degrading hemicellulose, cellulose and pectin as well as lignins. Cellulosomes are produced by various anaerobic bacteria such as Acetivibrio cellulolyticus, C. acetobutylicum, Bacteroides cellulosolvens, C. cellulolyticum, C. clariflavum, C. cellulovorans, C. josui, C. thermocellum, C. papyrosolvens, Ruminococcus flavefaciens and R. albus.15
Apart from cellulases, lytic polysaccharide monooxygenases (LPMOs) are key enzymes involved in the degradation of biomass.16 LPMO are copper dependent enzymes that can cleave cellulose to produce nanofibrils. LPMOs are most abundantly found in saprotrophic fungi with around 30 LPMO-encoding genes.17 They are secreted when fungi are grown in the presence of cellulose or lignocellulosic material. Recently, LMPOs from fungal source are cloned, engineered, purified and characterized.18–20
C. clariflavum is a Gram-positive, cellulolytic, thermophilic bacterium, which was first isolated from an anaerobic sewage sludge.21,22 The genome of C. clariflavum contains an abundance of cellulose degrading enzymes. Due to its thermophilic nature and its production of cellulosomal proteins, this bacterium is attracting attention from researchers for biomass degradation.23 Genome analysis showed the high similarity among C. clariflavum and C. thermocellum, which exhibits the model machinery for cellulosome production.24
To enhance the yield, quality and functionality of industrially important enzymes, genetic engineering has played an important role since the last century. For the efficient production of recombinant proteins, the selection of the right expression vector and its host is very important. E. coli is often utilized for expression of exoglucanases gene from different microbes. The optimization of different conditions is another strategy for the maximum production of such a recombinant protein.25,26 Biochemical characterization of enzymes such as pH and temperature stability of enzyme over a wide range, resistance to various ionic and non-ionic detergents and resistance against various organic solvents contributes to its applications towards industries.27,28 Cellobiohydrolases have various industrial applications such as in detergent, food, animal feed, textile and bioenergy industries.29,30
The present study is concerned with the cloning and expression of a cellobiohydrolase gene of C. clariflavum into the E. coli BL21(DE3) strain. We also describe the purification and characterization of recombinant cellobiohydrolase enzyme for its use in industrial processes.
Methodology
Bacterial strain selection
Genomic DNA from C. clariflavum DSM 19732 was obtained from the German Collection of Microorganisms and Cell Culture, DSMZ, Germany (https://www.dsmz.de/). The genomic DNA was used for amplification of the cellobiohydrolase gene, GenBank ID AEV70589.1 (Clocl_4158) (accession number G8LUE1). The pET 21a(+) expression vector was utilized for the expression of cellobiohydrolase gene in E. coli BL21(DE3).Biomass collection
Three biomass including Saccharum munja (soft and hard portions), sugarcane bagasse and sawdust were obtained from different areas of Pakistan. However, hazelnut cob, hazelnut shell and rhododendron plant biomass were provided by our collaborators from Turkey. The biomass was dried and ground in a grinder into very fine particles.31PCR amplification
Primers were designed using Vector NTI software. The sequence of cellobiohydrolase gene, AEV70589.1, (Clocl_4158) and its protein from C. clariflavum is mentioned in ESI file 1.† The restriction sites NdeI and HindIII were added in the forward and reverse primers. Sequences of the forward and reverse primers are given below:Forward primer: 5′ GCCATATGATGACAATAACCTATGAC 3′
Reverse primer: 5′ CGAAGCTTGAATTAGTTCCGCTATAAG 3′
A PCR mixture was prepared that includes genomic DNA (30 ng μl−1), 2 μl forward primer (100 pmol μl−1), 2 μl reverse primer (100 pmol μl−1), 25 μl master mixture and 19 μl distilled water. The PCR was performed for 5 minutes at 95 °C in 35 amplification cycles where each cycle was incubated at 95 °C for 45 seconds, 57 °C for 45 seconds and 72 °C for 90 seconds.32 Agarose gel was used for analysis of amplified PCR product, which was purified using a DNA purification kit (QIAquick).
Cloning of cellobiohydrolase gene in pET-21a(+)
The amplified cellobiohydrolase gene was restricted with HindIII and NdeI enzymes. After digestion, the cellobiohydrolase gene was purified and ligated into pET-21a(+). E. coli BL21(DE3) competent cells were prepared by a standard method and used for transformation of recombinant vector pET-21a(+)/exoglucanase gene.33 The screening of positive clones was performed by digesting isolated plasmid pET 21a(+) using and NdeI and HindIII restriction enzymes.Recombinant cellobiohydrolase expression in E. coli BL21
In order to determine production of recombinant cellobiohydrolase enzyme, E. coli BL21(DE3) cells (transformed) were cultured in Luria–Bertani (LB) medium supplemented with 100 μg ml−1 ampicillin. The medium was induced by 0.5 mM IPTG and incubated at 37 °C for 4 h. The medium was then centrifuged at 6000 rpm, at 4 °C for 10 minutes. The supernatant was discarded and the resulting pellet was suspended into Tris–HCl buffer (pH 7, centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and sonicated for 10 min at 4 °C).34 Expression of the cellobiohydrolase enzyme was determined in extra and intracellular fractions utilizing 12% SDS-PAGE and total proteins were estimated by Bradford method.35
000 rpm and sonicated for 10 min at 4 °C).34 Expression of the cellobiohydrolase enzyme was determined in extra and intracellular fractions utilizing 12% SDS-PAGE and total proteins were estimated by Bradford method.35
Activity assay
Dinitrosalicylic acid (DNS) method was used to determine enzyme activity36 by estimating reducing sugars. A 500 μl aliquot of the sample was incubated with 1.5 ml of 100 mM phosphate buffer, pH 6.5, containing 1.0% CMC (Sigma-Aldrich) at 50 °C for 30 min. Then 500 μl DNS reagent (1% DNS; 0.4 M NaOH; 1 M sodium and potassium tartrate) was added, and the resulting mixture was boiled for 5 min. After the addition of 2.5 ml of distilled water the absorbance was measured in a spectrophotometer at 550 nm. Before measuring the absorbance, spectrophotometer was calibrated using a mixture prepared by 2.0 ml of 100 mM phosphate buffer, pH 6.5, with 1.0% carboxymethylcellulose (CMC). Enzyme activity was defined as “one enzyme unit that is required to liberate 1 μmol of reducing sugar per minute at standard conditions of enzyme assay”.Total protein estimation
The estimation of total protein was carried out using the Bradford method.35 Bradford reagent (5 ml), 100 μl of cellobiohydrolase enzyme and phosphate buffer (900 μl) were added in a test tube. A control was run parallel in which 5 ml Bradford reagent and 1 ml of phosphate buffer was added. Absorbance was observed in a spectrophotometer at 595 nm. For protein estimation, bovine serum albumin was used as the standard.Optimization of cultural conditions
Different parameters were analyzed to determine the optimum conditions for cellobiohydrolase production. The effect of the temperature on enzyme production was optimized by culturing the C. clariflavum strain at various temperatures ranging between 16–42 °C. The effect of the initial pH of the culture medium on enzyme production was evaluated by adjusting pH of the medium with sulphuric acid and sodium hydroxide to obtain pH values ranging between 4.0 to 9.0. The effect of temperature of the culture medium on the enzyme production was observed by culturing the bacteria at various temperatures (16 °C, 23 °C, 30 °C, 37 °C and 42 °C). The effect of time of incubation was tested by incubating the bacterial cultures at 37 °C, at 150 rpm for different time durations (1, 2, 3, 4, 5 and 6 h). The impact of the IPTG addition to the culture medium on the cellobiohydrolase enzyme at various concentration of 0.1, 0.2, 0.3, 0.4, 0.5 and 0.6 during entire culturing of bacteria was also tested.Enzyme purification
Characterization
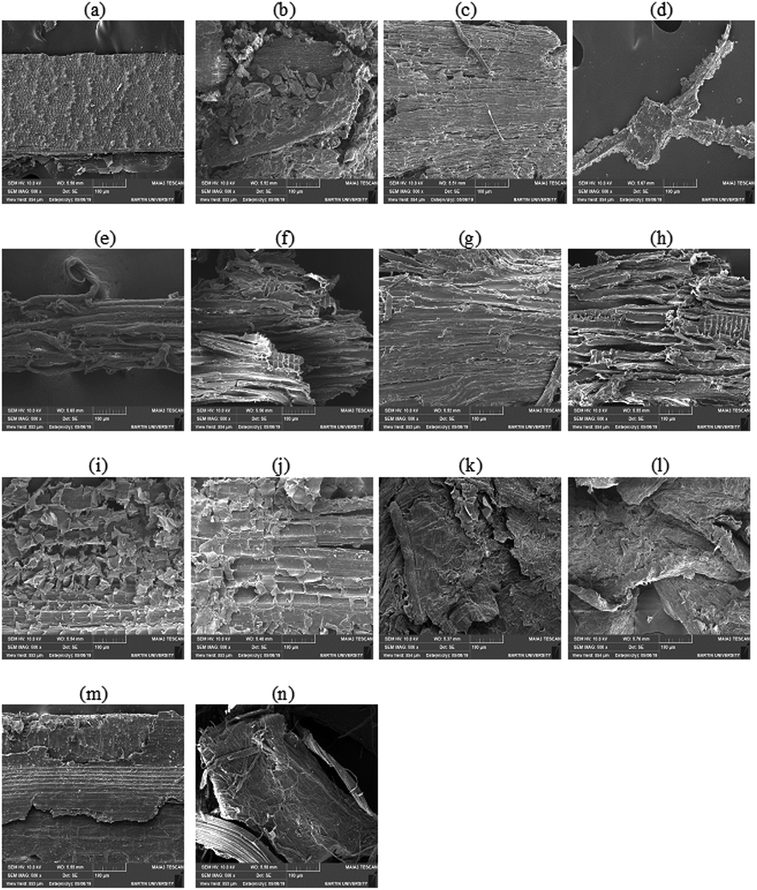
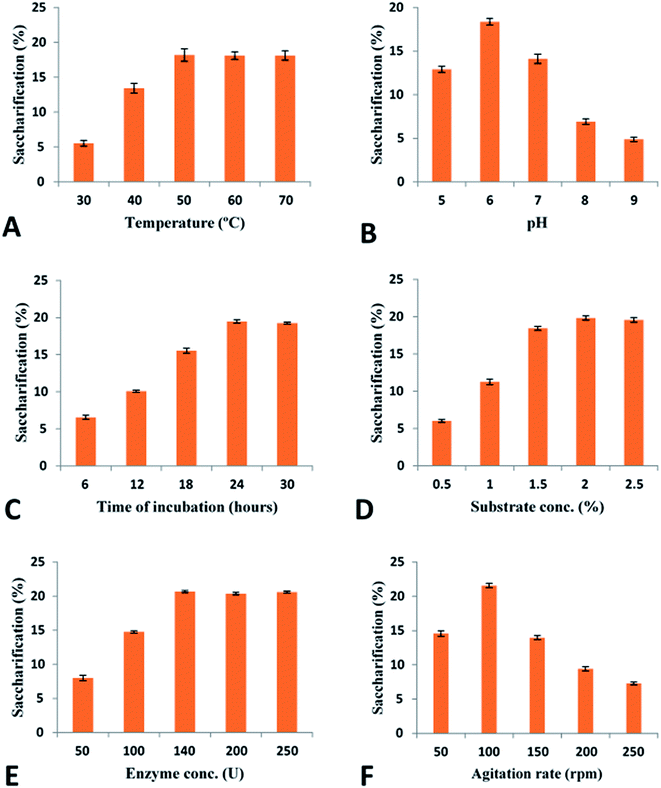
Pretreatment analysis was performed by scanning electron microscopy. For saccharification experiment, 1 g of pretreated biomass was incubated with 50 ml (140 U) (28 mg) enzyme at 50 °C for 24 h in a shaking incubator at 100 rpm. To prevent the contamination by different microorganisms, tetracycline (10 μg ml−1) was added along with cellobiohydrolase enzyme. Various conditions (incubation temperature, pH, incubation period, enzyme units, substrate concentration) were optimized to attain the maximum saccharification of plant biomass. A range of temperature (30–70 °C), pH (5–9), incubation period (6–30 h), substrate concentration (0.5–2.5%), enzyme units (50–250) and agitation speed (50–250 rpm) was used obtain the optimized conditions of plant biomass saccharification. The hydrolysis of plant biomass was checked by measuring the liberated reducing sugars using the protocol given by Miller.36 The percentage saccharification was calculated from the following formula:
Results
Cloning and expression of cellobiohydrolase gene into pET-21a(+)
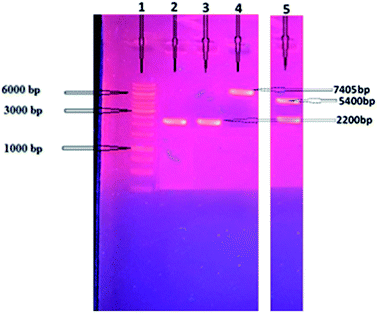
The cellobiohydrolase gene, GenBank ID AEV70589.1, (Clocl_4158) of C. clariflavum was amplified (2200 bp) and then restricted with specific enzymes as mentioned in methodology. The restricted gene was ligated with pET-21a(+) expression vector and then transferred into competent cells of E. coli BL21(DE3). Isolated recombinant plasmids were restricted (7643 bp) to confirm the cloning process (Fig. 1). Double restriction utilizing HindIII and NdeI showed two bands, 5400 bp and 2200 bp, depicting the successful ligation into expression vector. Expression of the cellobiohydrolase was investigated utilizing SDS-PAGE and its activity in extracellular fractions was calculated to be 2.14 U mg−1 by using DNS method.36Purification of recombinant protein
The heat treated denatured host proteins were separated from crude cell lysate by centrifugation and subjected to Protino® Ni-TED columns for further purification. Six His-tagged residues were added at the amino end of the cellobiohydrolase enzyme due to presence of 6 histidine residues located in the pET-21a(+) plasmid at position 140–157. After SDS-PAGE analysis, the purification fold and specific activity of the cellobiohydrolase enzyme was determined to be 3.90 fold and 57.4 U mg−1 respectively (Table 1).| Purification steps | Exoglucanase unit (U) | Total protein (mg) | Specific activity (U mg−1) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|
| Crude exoglucanase | 2780 | 189 | 14.7 | 1 | 100 |
| Heat-treated | 1590 | 55 | 28.90 | 1.96 | 57.19 |
| IMAC | 978 | 17 | 57.4 | 3.90 | 35.17 |
Determination of molecular weight
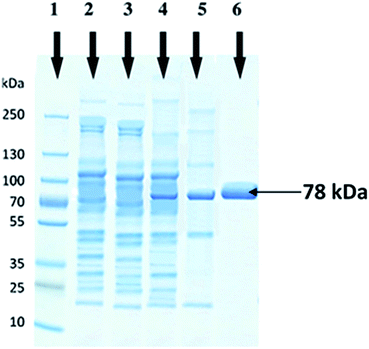
The molecular weight of recombinant cellobiohydrolase enzyme was determined by SDS-PAGE and observed to be 78 kDa. Cell lysates of wild E. coli BL21, non-induced recombinant wild E. coli BL21, induced recombinant wild E. coli BL21 and heat treated partially purified cellobiohydrolase enzyme fraction were also run in parallel on SDS-PAGE (Fig. 2).Optimization of recombinant cellobiohydrolase gene expression
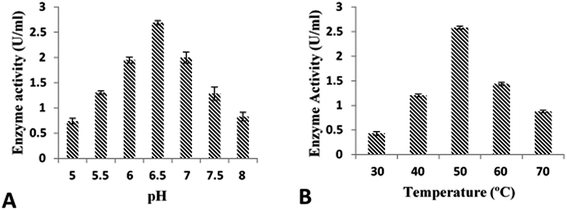
Utilization of various IPTG concentrations (0.1–0.6 mM) revealed that maximum cellobiohydrolase expression (2.16 ± 0.045 U ml−1) was achieved when recombinant E. coli BL21(DE3) bacterial cells were induced with 0.5 mM IPTG. However, reduction in cellobiohydrolase expression was observed at lower concentrations (Fig. 3A). Effect of time of induction was also monitored to obtain maximum production of cellobiohydrolase enzyme. Various time scale (1 to 6 h) was used for incubation of recombinant culture. The maximum production (2.35 ± 0.085 U ml−1) of the cellobiohydrolase gene with 1.04 ± 0.012 mg ml−1 total protein contents was determined after 4 h of incubation at 37 °C and 0.5 mM IPTG induction (Fig. 3B). Reduced production of enzyme (1.38 ± 0.036 U ml−1, 1.72 ± 0.104 U ml−1 and 1.76 ± 0.124 U ml−1) was obtained after 2 h, 3 h and 5 h incubation, respectively at 37 °C. Effect of temperature was also investigated to get the most favorable temperature for high production of cellobiohydrolase. Recombinant E. coli cells were cultured at different temperatures (16 °C to 42 °C) after induction with 0.5 mM IPTG for 4 h. Highest expression (2.57 ± 0.041 U ml−1) with 1.56 ± 0.014 mg ml−1 total protein contents was obtained at 37 °C for 4 h. At elevated temperatures, enzyme activity was decreased and dropped to 1.81 ± 0.121 U ml−1 at 42 °C (Fig. 3C). pH of the medium has obvious effect on the growth of microorganisms. Maximum production (2.78 ± 0.145 U ml−1) of cellobiohydrolase enzyme was observed at pH 6.0. At pH 4, 5, 7, 8 and 9 less enzyme activity (1.25 ± 0.987 U ml−1, 1.68 ± 0.72 U ml−1, 2.11 ± 0.921 U ml−1, 1.56 ± 0.998 U ml−1 and 0.98 ± 0.958 U ml−1) was obtained (Fig. 3D). The best culture conditions for cellobiohydrolase production was obtained at 37 °C, at initial pH 6.0, with addition of 0.5 mM IPTG in the medium for 4 h incubation.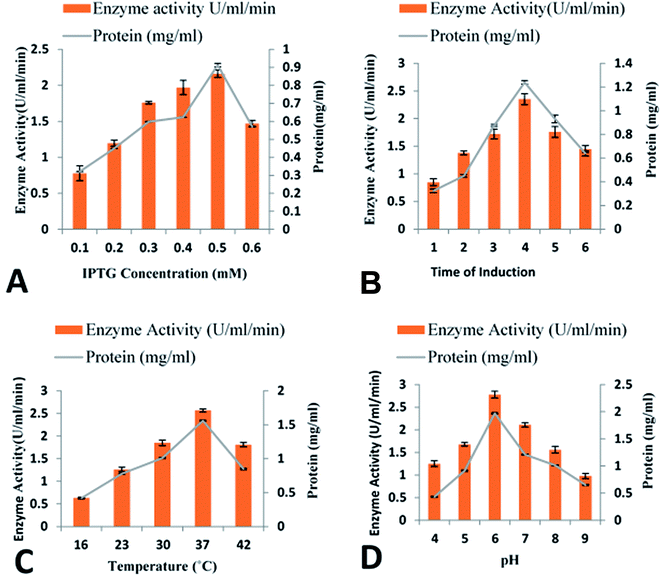 | ||
| Fig. 3 Optimization of cloned cellobiohydrolase gene expression: (A) effect of IPTG concentration (B) effect of time of induction (C) effect of incubation temperature (D) effect of pH. | ||
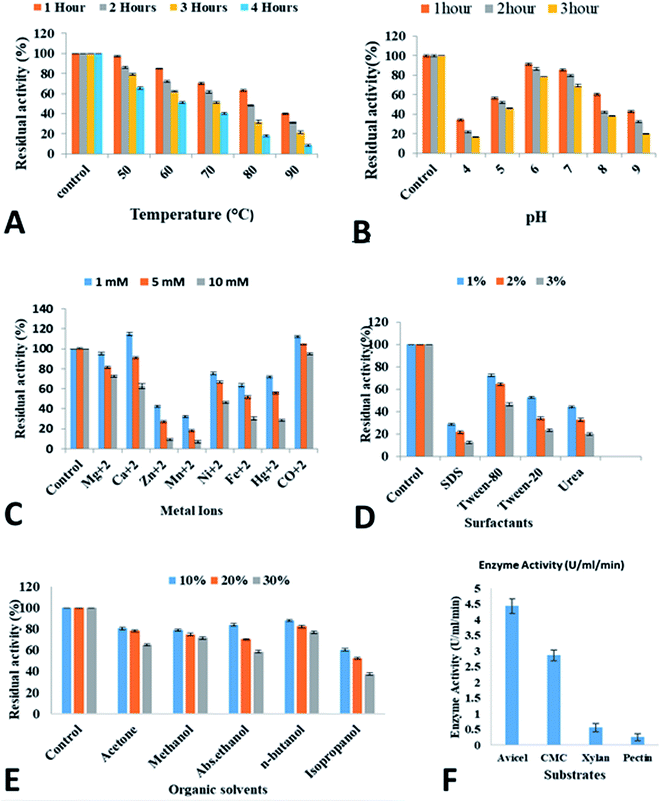
Characterization of recombinant cellobiohydrolase
Purified recombinant cellobiohydrolase enzyme was characterized for the various parameters below:| Pretreated biomass type | Reducing sugars released (mg ml−1) | Saccharification (%) | Cellulose contents in biomass (%) |
|---|---|---|---|
| Sugarcane bagasse | 4.79 | 21.57 | 62 (Guilherme et al., 2015)107 |
| Saccharum munja (hard) | 1.77 | 7.96 | 35 (Kumar et al., 2019)108 |
| Saccharum munja (soft) | 3.86 | 17.4 | 70 (Singh et al., 2017)109 |
| Sawdust | 3.11 | 13.9 | 45 (Klash et al., 2010)110 |
| Hazelnut cob | 3.5 | 15.75 | 52 (Demirbraş, 2005)111 |
| Hazelnut shell | 2.5 | 11.5 | 42 (Demirbraş, 1999)112 |
| Rhododendron | 2.88 | 12.96 | 47 (Shrestha & Budhathoki, 2012)113 |
Discussion
Biofuel production from lignocellulosic plant biomass is attracting the intense attention of researchers in recent decades in response to the search for alternative fuels. The major portion of lignocellulosic plant biomass is comprised of complex cellulose polysaccharides and the degradation of this complex cellulose requires a complete set of cellulases. Exoglucanase is one of the most important cellulases which acts upon the ends of cellulose chains and results in the production of cellobiose and glucose units. In this study, the cellobiohydrolase gene from C. clariflavum was cloned into E. coli BL21(DE3) using the expression vector pET 21a(+). The recombinant cellobiohydrolase enzyme activity (2.78 ± 0.145 U ml−1) was obtained with 4 h of IPTG (0.5 mM) induction at 37 °C. Higher concentrations of IPTG resulted in reduced cellobiohydrolase enzyme production due to the toxicity of IPTG that reduced the rate of ribosomal RNA synthesis.The present study showed much higher purifications when compared to many recent reports. The specific activity of the purified enzyme reached 57.4 U mg−1 after IMAC processing. Similarly, the percentage recovery was 35.17% and a purification fold of 3.90 was attained. In comparison to this current study, much less purification fold of cellobiohydrolases has been recently reported. There, a purification fold of 1.07 and 1.11 was reported for glucanases from Trichoderma viride and C. thermocellum respectively.41,42 Similarly, reduced recovery yields of 10.26% and 1.06% was achieved after the final purification of cellobiohydrolases from T. harzianum and Bacillus stearothermophilus.43,44 In another study, a similar low recovery of cellobiohydrolase from Aspergillus oryzae was reported after its purification which was in-compatible with recovery obtained in present study.45
In this study, the enzyme retained maximum activity when incubated at acidic pH (6.0). Many studies involving cellobiohydrolase (cellobiohydrolase) from thermophiles showed similar results. Cellobiohydrolase from C. thermocellum showed maximum activity at pH 5.7.46 Similarly, the cellobiohydrolases from Thermoascus aurantiacus47 and Actinomyces sp.48 remained stable at pH 6.0 and the cellobiohydrolase from Thermotoga petrophila retained 79% activity at pH 4.49 On the other hand, T. maritima showed stability at varied pH range between 6.0–7.5.50 However, various cellulases from T. reesei and Rasamsonia emersonii showed stability between pH values 4–6.51
One study makes use of the presence of all nine tryptophan residues in the catalytic domain of cellobiohydrolase from mutant T. reesei to demonstrate the effect of pH on the enzyme activity utilizing fluorescence measurement.105 It was found that drastic changes occur in the catalytic site of the enzyme at lower and higher pH values that resulted in less binding with substrate and showed less fluorescence intensity due to change in polarity of some of all tryptophans in the active site tunnel of Cel7A when substrate is bound. Similar results indicated high alkaline pH value caused radical changes in secondary structure of cellobiohydrolase from T. harzianum and resulted in drastic decrease in enzyme activity.106
An important feature of cellobiohydrolase is its stability at different ranges of temperatures from 50–90 °C for 1–4 h. Specifically, the cellobiohydrolase retained 63% activity at 80 °C for 1 h. Jeong et al.52 demonstrated that cellobiohydrolase from Fervidobacterium islandicum was stable at 65 °C for 1 h. In the present study, cellobiohydrolase retained reasonable stability of 52% for 4 h at 60 °C. In one study, cellobiohydrolase from T. petrophila showed 81% residual activity at 70 °C for 4 h.49 Recently, around 50% cellobiohydrolase activity from Lentinula edodes was demonstrated at 60 °C.53 Several reports have reported less residual activities of thermostable cellobiohydrolases at pre-incubation at high temperature. Cellobiohydrolase obtained from C. thermocellum,42 T. maritima,50 hot spring isolate,54 Thermoanaerobacterium thermosaccharolyticum,55 Thermoascus aurantiacus,47 Fervidobacterium islandicum52 all showed highly reduced enzyme activity when incubated between 70 and 90 °C. However, two reports demonstrated that cellobiohydrolase from T. maritima50 and T. neapolitana56 maintained their catalytic activity during pre-incubation at 90 °C.
It has been proposed that the presence of rigid protein structures at high temperatures establish fluidity and flexibility in their structural characteristics.57,58 Helix stabilization is one of the key factors that is governed by the low level presence of C-β-branched amino acids like valine, isoleucine and threonine.59 Presence of specific amino acids like proline at helical ends also contribute in helix stabilization and leads to enzyme stability.60 Similarly, there are many interactions between domain/subunits that can stabilize the structure of thermostable enzyme. These interactions include hydrophobic interactions, hydrogen bonds, disulphide bridges aromatic interactions, docking loose ends and ion-pair networks.61–65 Sometimes oligomer formation helps in the prolonged stability of enzyme at high temperature.60,63,66 Apart from this, dense packing in enzyme structures due to increased core hydrophobicity63 and the presence of stable surface-exposed amino acids like glutamine that cause deamidation60 or oxidative degradation due to presence of cysteine and methionine67 contribute significantly to enzyme stability.
It is well established that thermostable enzymes possess rigid structure that could be converted into flexible and fluidity structures at high temperature.42,56 In another interesting study, the enzymatic activity of thermophilic α-amylase obtained from Bacillus licheniformis has been reported at high temperature, however, loss of enzyme activity has been described at high temperature in mesophilic B. amyloliquefaciens.68 On the other hand, the main contributors that sustain the molecular integrity of enzymes from psychrophilic microorganisms near zero celsius are decreased rigidity in enzyme core and reduced inter domain interactions but their enzyme structures resemble with mesophilic and thermophilic enzymes.69 Decreased rigidity in protein core and decreased inter domain interactions contribute in the molecular structure of psychrophilic microorganisms at sub-zero temperature.70 Further, it is also generally assumed that the increase in stability from psychrophilic to thermophilic proteins arises from a decrease in conformational flexibility.71,72
In industrial applications, generally thermostable enzymes are preferred that can survive high temperature (>55 °C) needed in industrial processes. Additional advantages of thermostable enzymes usage include high tolerance for organic solvents,73 reduced risk of contamination and increased stability specially when used in raw material pre-treatments. The stability of cellobiohydrolase from C. clariflavum make it the enzyme of choice for industrial use especially in saccharification of biomass. The wide range of industrial uses of thermostable cellobiohydrolase including paper and pulp industry, food industry, textile industry as well as in biofuel industry has been reported.74
Cellobiohydrolase from C. clariflavum was studied in the presence of various metal ions. The cellobiohydrolase showed tolerance and increased enzymatic activity in the presence of Ca2+, Ni2+, Co2+ and Mg2+. However, Mn2+, Fe2+ and Zn2+ and Hg2+ showed adverse effect on the activity of the enzyme. Many previous reports on cellobiohydrolase have shown similar results. It has been shown that Rhizopus oryzae cellobiohydrolase demonstrated low activity in the presence of Zn2+ and Hg2+.75 Recently, it has been illustrated that cellobiohydrolase from Fervidobacterium islandicum remained stable in the presence of Co2+, Na2+, Ca2+, Mg2+ and Mn2+ ions while showed reduced enzymatic activity in the presence of Zn2+, Fe2+ and Cd2+ metal ions.52 Other studies by Ghori et al.76 and Nasir et al.41 reported that cellobiohydrolase from A. niger and T. viride respectively were inhibited in the presence of Hg2+ and Fe2+ while enzymes retained high residual activity in the presence of other metal ions. Most of the glycoside hydrolases are inhibited by Hg2+ that oxidizes indole rings of tryptophan residues of the enzyme.77 This phenomenon has been observed in cellulases from Bacillus sp. BG-CS10 and B. sphaericus.78,79 On the other hand, damage of active site due to disturbance in thiol groups in the cellobiohydrolase enzyme could also be the main cause of the decreased activity of cellobiohydrolase.80 Some studies have demonstrated the increased residual activity of cellobiohydrolase in the presence of Ca2+.81,82 The possible effect of metal ions on cellobiohydrolase has not been clearly explained in the literature. However, it is assumed that it could probably occur due to redox effect on amino acids, thus, enhancing or reducing the enzyme activities. Binding of metal ions with sulfhydryl (–SH) groups of cysteine result in denaturing of thiol group and consequently change in conformational structure of enzyme disallow binding with the substrate.83,84
The effect of divalent ions on cellobiohydrolase is not clearly understood and possibly occurs by redox effects on the amino acids, increasing or decreasing their activities. The reduced activity of the enzyme is probably due to the damage of the active site by denaturation of the existing thiol groups.83 It is assumed that metal ions generally bind to sulfhydryl (–SH) groups of the cysteine present in an enzyme and by denaturing of the thiol group, thus resulting in the conformational change of an enzyme which prohibits binding with the substrate.82,84 Previously, stereochemical aspect of α-amylase in hyperthermophilic archaebacterium Pyrococcus furious showed that higher activation energies are required for optimum function of enzyme at high temperature.71
One of the significant feature of cellobiohydrolase from C. clariflavum is its tolerance against various organic compounds and surfactants. Azadian et al.85 has shown that catalytic activity of cellobiohydrolase from Bacillus was increased to 161% and 152% in the presence of methanol and chloroform respectively while it was inhibited to 61% by DMF. Thomas et al.86 has explained the organic solvent, n-hexane and isooctane, tolerant cellobiohydrolase enzyme from Promicromonospora sp. Gaur and Tiwari87 showed increased residual activity of cellobiohydrolase from B. vallismortis to 242.5, 178.7, 117, 170.3, 180.2, 143, 160.3, 115.9, 131.9 and 133% after incubation for 24 h to 48 h with 30%, n-decane, n-dodecane, iso-octane, toluene, xylene, n-hexane, acetone, methanol, n-butanol and cyclohexane. Similarly, other studies have reported that cellobiohydrolase from hot spring isolate and Hermetia illucens remained stable in the presence of isopropanol, propanol and methanol.54,88 However, some studies reported decreased residual activity of cellobiohydrolase enzyme from soil metagenome library.89 It has been shown previously that β-glucosidases from T. aurantiacus, A. oryzae and Fusarium oxysporum were activated by short chain alcohols.90–92 It has been proposed that alcohols alter the polarity of the medium that help to stabilize the enzyme conformation.93 Less cellobiohydrolase activity has been demonstrated in the presence of 1% various surfactants like Tween 80, Tween 20, Triton X-100, SDS and H2O2. Generally, it is suggested that stability of cellulases in presence of Tween 80 corresponds to the hydrophobicity and secondary structure propensity of these proteins.94
Some groups have studied the cellobiohydrolase obtained from commercial company, microbes isolated from agricultural wastes and bacteria isolated from hot spring.41,54 It is assumed that a several complex mechanisms confers adaptations and allow bacteria to survive against organic solvents.95 The development of reflux pumps in bacteria due to periplasmic space helps them tolerate against the organic solvents.96 Structurally and functionally unrelated compounds are removed from the cytoplasm of the bacteria due to these efflux pumps.97 On the other hand, organic solvents can penetrate the active sites of the enzyme thus making these enzymes ineffective.98,99 Water molecules interactions with enzymes help retain the three dimensional structure and its activity.100
For the complete degradation of cellulose to simple sugars, cellobiohydrolases play a very important role. These enzymes hydrolyze the cellulose at the polysaccharide chain ends and results in production of cellobiose as well as glucose units.101 In this study recombinant purified cellobiohydrolase from C. clariflavum was found to be very active for the hydrolysis of pretreated plant biomass. Maximum saccharification (21.57%) was observed against pretreated sugarcane bagasse while all other tested biomass (sawdust, hazelnut cob, hazelnut shell, rhododendron, Saccharum munja hard, Saccharum munja soft) were also significantly hydrolyzed by the action of recombinant enzyme (Table 2). Very few literatures on the exclusive use of cellobiohydrolase for the saccharification of plant biomass has been previously reported although the saccharification of plant biomass by the combined effect of cellulases is reported in previous reports. In a previous study, 9.03% saccharification was obtained against sugarcane bagasse and 5.92% saccharification of sawdust utilizing the cellulases of A. oryzae102 while 5.12% saccharification of wheat straw and 7.31% saccharification of sugarcane bagasse was reported for the endoglucanase of C. thermocellum in another study.103 A 24% saccharification of sugarcane bagasse was observed by the action of endoglucanase from B. licheniformis.104
Conclusion
The present study describes the cloning of cellobiohydrolase gene from a thermophilic bacterium C. clariflavum in E. coli BL21(DE3) and its expression in pET21a(+) expression system. This study presented the optimized conditions for the maximum production of cellobiohydrolase enzyme in E. coli BL21(DE3) using pET21a(+) expression system. This study is innovative for the industrial scale production of recombinant cellobiohydrolase enzyme.Funding
This research was funded by the Higher Education Commission of Pakistan for Biofuel project no. 5535 and the Pak-Turk Researchers Mobility Program.Conflicts of interest
We have no competing interests.Acknowledgements
The manuscript is thoroughly read and corrected for English language by Prof. Dr Stephen J. Perkins, Professor Emeritus, Department of Structural and Molecular Biology, Darwin Building, Gover Street, University College of London, United Kingdom.References
- A. A. Rafindadi and I. Ozturk, Impacts of renewable energy consumption on the German economic growth: evidence from combined cointegration test, Renewable Sustainable Energy Rev., 2017, 75, 1130–1141 CrossRef.
- J. Xu, J. J. Cheng, R. R. Sharma-Shivappa and J. C. Burns, Sodium hydroxide pretreatment of switchgrass for ethanol production, Energy Fuels, 2010, 24, 2113–2119 CrossRef CAS.
- Z. Wang, G. Cao, C. Jiang, J. Song, J. Zheng and Q. Yang, Butanol production from wheat straw by combining crude enzymatic hydrolysis and anaerobic fermentation using Clostridium acetobutylicum ATCC824, Energy Fuels, 2013, 27, 5900–5906 CrossRef CAS.
- F. J. Ríos-Fránquez, E. González-Bautista, T. Ponce-Noyola, A. C. Ramos-Valdivia, H. M. Poggi-Varaldo, J. García-Mena and A. Martinez, Expression of a codon-optimized β-glucosidase from Cellulomonas flavigena PR-22 in Saccharomyces cerevisiae for bioethanol production from cellobiose, Arch. Microbiol., 2017, 199, 605–611 CrossRef.
- V. Astolfi, A. L. Astolfi, M. A. Mazutti, E. Rigo, M. Di Luccio, A. F. Camargo, C. Dalastra, S. Kubeneck, G. Fongaro and H. Treichel, Cellulolytic enzyme production from agricultural residues for biofuel purpose on circular economy approach, Bioprocess Biosyst. Eng., 2019, 42, 677–685 CrossRef CAS.
- F. Shah, B. Ranawat and S. Mishra, An Approach Toward Cellulase Production, Bioconversion, and Utilization in Adv. Bioproc. Alter. Fuels, Biobased Chemicals, and Bioproduct., Elsevier, 2019, pp. 207–223 Search PubMed.
- G. Henriksson, A. Nutt, H. Henriksson, B. Pettersson, J. Ståhlberg, G. Johansson and G. Pettersson, Endoglucanase 28 (Cel12A), a new Phanerochaete chrysosporium cellulase, Eur. J. Biochem., 1999, 259, 88–95 CrossRef CAS.
- A. Shrotri, H. Kobayashi and A. Fukuoka, Catalytic conversion of structural carbohydrates and lignin to chemicals, in Adv. Catalys., Elsevier, 2017, vol. 60, pp. 59–123 Search PubMed.
- Y. H. P. Zhang, M. E. Himmel and J. R. Mielenz, Outlook for cellulase improvement: screening and selection strategies, Biotechnol. Adv., 2006, 24, 452–481 CrossRef CAS.
- M. Himmel, M. Tucker, S. Lastick, K. Oh, J. Fox, D. Spindler and K. Grohmann, Isolation and characterization of a 1, 4-beta-D-glucan glucohydrolase from the yeast, Torulopsis wickerhamii, J. Biol. Chem., 1986, 261, 2948–12955 CrossRef.
- C. J. Yeoman, Y. Han, D. Dodd, C. M. Schroeder, R. I. Mackie and I. K. Cann, Thermostable enzymes as biocatalysts in the biofuel industry, in Advances in applied microbiology, Elsevier, 2010, pp. 1–55 Search PubMed.
- P. K. GhattyVenkataKrishna, E. M. Alekozai, G. T. Beckham, R. Schulz, M. F. Crowley, E. C. Uberbacher and X. Cheng, Initial recognition of a cellodextrin chain in the cellulose-binding tunnel may affect cellobiohydrolase directional specificity, Biophys. J., 2013, 104, 904–912 CrossRef CAS.
- E. A. Bayer, L. J. Shimon, Y. Shoham and R. Lamed, Cellulosomes—structure and ultrastructure, J. Struct. Biol., 1998, 124, 221–234 CrossRef CAS.
- A. L. Demain, M. Newcomb and J. D. Wu, Cellulase, clostridia, and ethanol, Microbiol. Mol. Biol. Rev., 2005, 69, 124–154 CrossRef CAS.
- R. H. Doi, A. Kosugi, K. Murashima, Y. Tamaru and S. O. Han, Cellulosomes from mesophilic bacteria, J. Bacteriol., 2003, 185, 5907–5914 CrossRef CAS.
- C. Moreau, S. Tapin-Lingua, S. Grisel, I. Gimbert, S. Le Gall, V. Meyer, M. Petit-Conil, J.-G. Berrin, B. Cathala and A. Villares, Lytic polysaccharide monooxygenases (LPMOs) facilitate cellulose nanofibrils production, Biotechnol. Biofuels, 2019, 12, 1–13 CrossRef CAS.
- A. Levasseur, E. Drula, V. Lombard, P. M. Coutinho and B. Henrissat, Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes, Biotechnol. Biofuels, 2013, 6, 41 CrossRef CAS.
- P. F. de Gouvêa, L. E. Gerolamo, A. V. Bernardi, L. Pereira, S. A. Uyemura and T. M. Dinamarco, Lytic polysaccharide monooxygenase from Aspergillus fumigatus can improve enzymatic cocktail activity during sugarcane bagasse hydrolysis, Protein Pept. Lett., 2019, 26, 377–385 CrossRef.
- D. Kracher, Z. Forsberg, B. Bissaro, S. Gangl, M. Preims, C. Sygmund, V. G. Eijsink and R. Ludwig, Polysaccharide oxidation by lytic polysaccharide monooxygenase is enhanced by engineered cellobiose dehydrogenase, FEBS J., 2020, 287, 897–908 CrossRef CAS.
- S. Fritsche, C. Hopson, J. Gorman, R. Gabriel and S. W. Singer, Purification and characterization of a native lytic polysaccharide monooxygenase from Thermoascus aurantiacus, Biotechnol. Lett., 2020, 42, 1897–1905 CrossRef CAS.
- H. Shiratori, H. Ikeno, S. Ayame, N. Kataoka, A. Miya, K. Hosono, T. Beppu and K. Ueda, Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor, Appl. Environ. Microbiol., 2006, 72, 3702–3709 CrossRef CAS.
- H. Shiratori, K. Sasaya, H. Ohiwa, H. Ikeno, S. Ayame, N. Kataoka, A. Miya, T. Beppu and K. Ueda, Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge, Int. J. Syst. Evol. Microbiol., 2009, 59, 1764–1770 CrossRef CAS.
- J. A. Izquierdo, L. Goodwin, K. W. Davenport, H. Teshima, D. Bruce, C. Detter, R. Tapia, S. Han, M. Land and L. Hauser, Complete genome sequence of Clostridium clariflavum DSM 19732, Stand. Genomic Sci., 2012, 6, 104–115 CrossRef CAS.
- C. M. Fontes and H. J. Gilbert, Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates, Annu. Rev. Biochem., 2010, 79, 655–681 CrossRef CAS.
- A. Mohajeri, Y. Pilehvar-Soltanahmadi, J. Abdolalizadeh, P. Karimi and N. Zarghami, Effect of culture condition variables on human endostatin gene expression in Escherichia coli using response surface methodology, Jundishapur J. Microbiol., 2016, 9, e34091 Search PubMed.
- M. R. Couto, J. L. Rodrigues and L. R. Rodrigues, Optimization of fermentation conditions for the production of curcumin by engineered Escherichia coli, J. R. Soc., Interface, 2017, 14, 20170470 CrossRef.
- L. P. Meleiro, S. Carli, R. Fonseca-Maldonado, M. da Silva Torricillas, A. L. R. L. Zimbardi, R. J. Ward, J. A. Jorge and R. P. M. Furriel, Overexpression of a cellobiose-glucose-halotolerant endoglucanase from Scytalidium thermophilum, Appl. Biochem. Biotechnol., 2018, 185, 316–333 CrossRef CAS.
- J. Xu, B. He, B. Wu, B. Wang, C. Wang and L. Hu, An ionic liquid tolerant cellulase derived from chemically polluted microhabitats and its application in in situ saccharification of rice straw, Bioresour. Technol., 2014, 157, 166–173 CrossRef CAS.
- I. B. Hmad, M. Boudabbous, H. Belghith and A. Gargouri, A novel ionic liquid-stable halophilic endoglucanase from Stachybotrys microspora, Process Biochem., 2017, 54, 59–66 CrossRef.
- L. W. Yoon, G. C. Ngoh, A. S. M. Chua, M. F. A. Patah and W. H. Teoh, Process intensification of cellulase and bioethanol production from sugarcane bagasse via an integrated saccharification and fermentation process, Chem. Eng. Process., 2019, 142, 107528 CrossRef CAS.
- R. I. Ladeira-Ázar, T. Morgan, G. P. Maitan-Alfenas and V. M. Guimarães, Inhibitors compounds on sugarcane bagasse saccharification: effects of pretreatment methods and alternatives to decrease inhibition, Appl. Biochem. Biotechnol., 2019, 188, 29–42 CrossRef.
- P. V. Volkov, A. M. Rozhkova, A. V. Gusakov and A. P. Sinitsyn, Homologous cloning, purification and characterization of highly active cellobiohydrolase I (Cel7A) from Penicillium canescens, Protein Expression Purif., 2014, 103, 1–7 CrossRef CAS.
- J.-H. Lim, C.-R. Lee, V. Dhakshnamoorthy, J. S. Park and S.-K. Hong, Molecular characterization of Streptomyces coelicolor A (3) SCO6548 as a cellulose 1, 4-β-cellobiosidase, FEMS Microbiol. Lett., 2016, 363, fnv245 CrossRef.
- X. Guan, P. Chen, Q. Xu, L. Qian, J. Huang and B. Lin, Expression, purification and molecular characterization of a novel endoglucanase protein from Bacillus subtilis SB13, Protein Expression Purif., 2017, 134, 125–131 CrossRef CAS.
- M. M. Bradford, A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- G. L. Miller, Use of dinitrosalicylic acid reagent for determination of reducing sugar, Anal. Chem., 1959, 31, 426–428 CrossRef CAS.
- S. S. Ahmed, M. Akhter, M. Sajjad, R. Gul and S. Khurshid, Soluble Production, Characterization, and Structural Aesthetics of an Industrially Important Thermostable β-Glucosidase from Clostridium thermocellum in Escherichia coli, BioMed Res. Int., 2019, 2019, 1–8 Search PubMed.
- D. Sahoo, J. Andersson and B. Mattiasson, Immobilized metal affinity chromatography in open-loop simulated moving bed technology: purification of a heat stable histidine tagged β-glucosidase, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 1651–1656 CrossRef CAS.
- U. K. Laemmli, Cleavage of structural proteins during the assembly of the head of bacteriophage T4, Nature, 1970, 227, 680–685 CrossRef CAS.
- E. Z. Hoşgün, D. Berikten, M. Kıvanç and B. Bozan, Ethanol production from hazelnut shells through enzymatic saccharification and fermentation by low-temperature alkali pretreatment, Fuel, 2017, 196, 280–287 CrossRef.
- I. Hafiz Muhammad Nasir, A. Ishtiaq, Z. Muhammad Anjum and I. Muhammad, Purification and characterization of the kinetic parameters of cellulase produced from wheat straw by Trichoderma viride under SSF and its detergent compatibility, Adv. Biosci. Biotechnol., 2011, 149–156 Search PubMed.
- V. V. Zverlov, G. A. Velikodvorskaya and W. H. Schwarz, A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose, Microbiology, 2002, 148, 247–255 CrossRef CAS.
- S. Ahmed, A. Bashir, H. Saleem, M. Saadia and A. Jamil, Production and purification of cellulose-degrading enzymes from a filamentous fungus Trichoderma harzianum, Pak. J. Bot., 2009, 41, 1411–1419 CAS.
- A. Bahobil, R. Bayoumi, H. Atta and M. El-Sehrawey, Production of thermoalkalistable microbial cellulases from some agricultural wastes under solid state fermentation conditions, Int. J. Curr. Microbiol. Appl. Sci., 2014, 3, 99–117 Search PubMed.
- M. F. Begum and N. Absar, Purification and characterization of intracellular cellulase from Aspergillus oryzae ITCC-4857.01, Mycobiology, 2009, 37, 121–127 CrossRef CAS.
- Y. J. Liu, S. Liu, S. Dong, R. Li, Y. Feng and Q. Cui, Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum, Biotechnol. Biofuels, 2018, 11, 6 CrossRef.
- J. Hong, H. Tamaki, K. Yamamoto and H. Kumagai, Cloning of a gene encoding thermostable cellobiohydrolase from Thermoascus aurantiacus and its expression in yeast, Appl. Microbiol. Biotechnol., 2003, 63, 42–50 CrossRef CAS.
- S. C. Kim, S. H. Kang, E. Y. Choi, Y. H. Hong, J. D. Bok, J. Y. Kim, S. S. Lee, Y. J. Choi, I. S. Choi and K. K. Cho, Cloning and characterization of an endoglucanase gene from Actinomyces sp. Korean native goat 40, Asian-Australas. J. Anim. Sci., 2016, 29, 126 CrossRef CAS.
- S. F. Tahir, M. N. Aftab, F. Akram, A. Nawaz and H. Mukhtar, Purification and characterization of a thermostable cellobiohydrolase from Thermotoga petrophila, Protein Pept. Lett., 2018, 25, 1003–1014 CrossRef.
- K. Bronnenmeier, A. Kern, W. Liebl and W. L. Staudenbauer, Purification of Thermotoga maritima enzymes for the degradation of cellulosic materials, Appl. Environ. Microbiol., 1995, 61, 1399–1407 CrossRef CAS.
- C. Schiano-di-Cola, N. Røjel, K. Jensen, J. Kari, T. H. Sørensen, K. Borch and P. Westh, Systematic deletions in the cellobiohydrolase (CBH) Cel7A from the fungus Trichoderma reesei reveal flexible loops critical for CBH activity, J. Biol. Chem., 2019, 294, 1807–1815 CrossRef CAS.
- W. S. Jeong, D. H. Seo, J. H. Jung, D. H. Jung, D.-W. Lee, Y. S. Park and C. S. Park, Enzymatic Characteristics of a Highly Thermostable β-(1-4)-Glucanase from Fervidobacterium islandicum AW-1 (KCTC 4680), J. Microbiol. Biotechnol., 2017, 27, 271–276 CrossRef CAS.
- L. Li, M. Qu, C. Liu, K. Pan, L. Xu, K. OuYang, X. Song, Y. Li and X. Zhao, Expression of a recombinant Lentinula edodes cellobiohydrolase by Pichia pastoris and its effects on in vitro ruminal fermentation of agricultural straws, Int. J. Biol. Macromol., 2019, 134, 146–155 CrossRef CAS.
- D. Zarafeta, D. Kissas, C. Sayer, S. R. Gudbergsdottir, E. Ladoukakis, M. N. Isupov, A. Chatziioannou, X. Peng, J. A. Littlechild and G. Skretas, Discovery and characterization of a thermostable and highly halotolerant GH5 cellulase from an icelandic hot spring isolate, PLoS One, 2016, 11, e0146454 CrossRef.
- L. Zhao, Q. Pang, J. Xie, J. Pei, F. Wang and S. Fan, Enzymatic properties of Thermoanaerobacterium thermosaccharolyticum β-glucosidase fused to Clostridium cellulovorans cellulose binding domain and its application in hydrolysis of microcrystalline cellulose, BMC Biotechnol., 2013, 13, 1–8 CrossRef.
- J. D. Bok, D. A. Yernool and D. E. Eveleigh, Purification, characterization, and molecular analysis of thermostable cellulases CelA and CelB from Thermotoga neapolitana, Appl. Environ. Microbiol., 1998, 64, 4774–4781 CrossRef CAS.
- K. Khajeh, S. Khezre-Barati and M. Nemat-Gorgani, Proteolysis of mesophilic and thermophilic α-amylases, Appl. Biochem. Biotechnol., 2001, 94, 97–109 CrossRef CAS.
- T. Sundaram, I. Wright and A. Wilkinson, Malate dehydrogenase from thermophilic and mesophilic bacteria. Molecular size, subunit structure, amino acid composition, immunochemical homology, and catalytic activity, Biochemistry, 1980, 19, 2017–2022 CrossRef CAS.
- M. E. Bruins, A. E. Janssen and R. M. Boom, Thermozymes and their applications, Appl. Biochem. Biotechnol., 2001, 90, 155 CrossRef CAS.
- C. Vieille and G. J. Zeikus, Hyperthermophilic enzymes: sources, uses, and molecular mechanisms for thermostability, Microbiol. Mol. Biol. Rev., 2001, 65, 1–43 CrossRef CAS.
- R. Ladenstein and B. Ren, Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles, FEBS J., 2006, 273, 4170–4185 CrossRef CAS.
- A. Karshikoff and R. Ladenstein, Ion pairs and the thermotolerance of proteins from hyperthermophiles: a ‘traffic rule’ for hot roads, Trends Biochem. Sci., 2001, 26, 550–557 CrossRef CAS.
- M. J. Danson and D. W. Hough, Structure, function and stability of enzymes from the Archaea, Trends Microbiol., 1998, 6, 307–314 CrossRef CAS.
- M. A. Arnott, R. A. Michael, C. R. Thompson, D. W. Hough and M. J. Danson, Thermostability and thermoactivity of citrate synthases from the thermophilic and hyperthermophilic archaea, Thermoplasma acidophilum and Pyrococcus furiosus, J. Mol. Biol., 2000, 304, 657–668 CrossRef CAS.
- S. J. Crennell, G. O. Hreggvidsson and E. N. Karlsson, The structure of Rhodothermus marinus Cel12A, a highly thermostable family 12 endoglucanase, at 1.8 Å resolution, J. Mol. Biol., 2002, 320, 883–897 CrossRef CAS.
- S. J. Crennell, D. Cook, A. Minns, D. Svergun, R. L. Andersen and E. N. Karlsson, Dimerisation and an increase in active site aromatic groups as adaptations to high temperatures: X-ray solution scattering and substrate-bound crystal structures of Rhodothermus marinus endoglucanase Cel12A, J. Mol. Biol., 2006, 356, 57–71 CrossRef CAS.
- J. K. Yano and T. L. Poulos, New understandings of thermostable and peizostable enzymes, Curr. Opin. Biotechnol., 2003, 14, 360–365 CrossRef CAS.
- J. Fitter, R. Herrmann, N. Dencher, A. Blume and T. Hauss, Activity and stability of a thermostable α-amylase compared to its mesophilic homologue: mechanisms of thermal adaptation, Biochemistry, 2001, 40, 10723–10731 CrossRef CAS.
- S. Violot, N. Aghajari, M. Czjzek, G. Feller, G. K. Sonan, P. Gouet, C. Gerday, R. Haser and V. Receveur-Brechot, Structure of a full length psychrophilic cellulase from Pseudoalteromonas haloplanktis revealed by X-ray diffraction and small angle X-ray scattering, J. Mol. Biol., 2005, 348, 1211–1224 CrossRef CAS.
- N. Aghajari, G. Feller, C. Gerday and R. Haser, Structures of the psychrophilic Alteromonas haloplanctis α-amylase give insights into cold adaptation at a molecular level, Structure, 1998, 6, 1503–1516 CrossRef CAS.
- K. A. Laderman, B. R. Davis, H. C. Krutzsch, M. S. Lewis, Y. Griko, P. L. Privalov and C. B. Anfinsen, The purification and characterization of an extremely thermostable alpha-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus, J. Biol. Chem., 1993, 268, 24394–24401 CrossRef CAS.
- P. ZAvodszky, J. Kardos, Á. Svingor and G. A. Petsko, Adjustment of conformational flexibility is a key event in the thermal adaptation of proteins, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 7406–7411 CrossRef CAS.
- J. K. Kristjansson, Thermophilic organisms as sources of thermostable enzymes, Trends Biotechnol., 1989, 7, 349–353 CrossRef CAS.
- H. D. Jang and K. S. Chang, Thermostable cellulases from Streptomyces sp.: scale-up production in a 50-l fermenter, Biotechnol. Lett., 2005, 27, 239–242 CrossRef CAS.
- S. Mukherjee, M. Karmakar and R. R. Ray, Production of extra cellular exoglucanase by Rhizopus oryzae from submerged fermentation of agro wastes, Recent Res. Sci. Technol., 2011, 69–75 CAS.
- M. I. Ghori, S. Ahmed, M. A. Malana and A. Jamil, Kinetics of exoglucanase and endoglucanase produced by Aspergillus niger NRRL 567, Afr. J. Biotechnol., 2012, 11, 7227–7231 CAS.
- Y. Hu, G. Zhang, A. Li, J. Chen and L. Ma, Cloning and enzymatic characterization of a xylanase gene from a soil-derived metagenomic library with an efficient approach, Appl. Microbiol. Biotechnol., 2008, 80, 823 CrossRef CAS.
- G. Zhang, S. Li, Y. Xue, L. Mao and Y. Ma, Effects of salts on activity of halophilic cellulase with glucomannanase activity isolated from alkaliphilic and halophilic Bacillus sp. BG-CS10, Extremophiles, 2012, 16, 35–43 CrossRef CAS.
- J. Singh, N. Batra and R. C. Sobti, Purification and characterisation of alkaline cellulase produced by a novel isolate, Bacillus sphaericus JS1, J. Ind. Microbiol. Biotechnol., 2004, 31, 51–56 CrossRef CAS.
- M. Hussain, M. Asghar, M. Yaqoob and Z. Iqbal, A study of optimum conditions for exoglucanase production by Arachniotus sp, Int. J. Agric. Biol., 1999, 4, 342–344 Search PubMed.
- D.-s. Xue, L.-y. Liang, D.-q. Lin and S. J. Yao, Thermal inactivation kinetics and secondary structure change of a low molecular weight halostable exoglucanase from a marine Aspergillus niger at high salinities, Appl. Biochem. Biotechnol., 2017, 183, 1111–1125 CrossRef CAS.
- S. M. Marino and V. N. Gladyshev, Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces, J. Mol. Biol., 2010, 404, 902–916 CrossRef CAS.
- L. B. Poole, The basics of thiols and cysteines in redox biology and chemistry, Free Radical Biol. Med., 2015, 80, 148–157 CrossRef CAS.
- A. Tejirian and F. Xu, Inhibition of cellulase-catalyzed lignocellulosic hydrolysis by iron and oxidative metal ions and complexes, Appl. Environ. Microbiol., 2010, 76, 7673–7682 CrossRef CAS.
- F. Azadian, A. Badoei-dalfard, A. Namaki-Shoushtari, Z. Karami and M. Hassanshahian, Production and characterization of an acido-thermophilic, organic solvent stable cellulase from Bacillus sonorensis HSC7 by conversion of lignocellulosic wastes, J. Genet. Eng. Biotechnol., 2017, 15, 187–196 CrossRef.
- L. Thomas, H. Ram, A. Kumar and V. P. Singh, Production, optimization, and characterization of organic solvent tolerant cellulases from a lignocellulosic waste-degrading actinobacterium, Promicromonospora sp. VP111, Appl. Biochem. Biotechnol., 2016, 179, 863–879 CrossRef CAS.
- R. Gaur and S. Tiwari, Isolation, production, purification and characterization of an organic-solvent-thermostable alkalophilic cellulase from Bacillus vallismortis RG-07, BMC Biotechnol., 2015, 15, 19 CrossRef.
- C. M. Lee, Y. S. Lee, S. H. Seo, S. H. Yoon, S. J. Kim, B. S. Hahn, J. S. Sim and B. S. Koo, Screening and characterization of a novel cellulase gene from the gut microflora of Hermetia illucens using metagenomic library, J. Microbiol. Biotechnol., 2014, 24, 1196–1206 CrossRef CAS.
- R. Garg, R. Srivastava, V. Brahma, L. Verma, S. Karthikeyan and G. Sahni, Biochemical and structural characterization of a novel halotolerant cellulase from soil metagenome, Sci. Rep., 2016, 6, 39634 CrossRef CAS.
- N. J. Parry, D. E. Beever, E. Owen, I. Vandenberghe, J. V. Beeumen and M. K. Bhat, Biochemical characterization and mechanism of action of a thermostable β-glucosidase purified from Thermoascus aurantiacus, Biochem. J., 2001, 353, 117–127 CrossRef CAS.
- C. Riou, J.-M. Salmon, M.-J. Vallier, Z. Günata and P. Barre, Purification, characterization, and substrate specificity of a novel highly glucose-tolerant β-glucosidase from Aspergillus oryzae, Appl. Environ. Microbiol., 1998, 64, 3607–3614 CrossRef CAS.
- P. Christakopoulos, P. W. Goodenough, D. Kekos, B. J. Macris, M. Claeyssens and M. K. Bhat, Purification and Characterisation of an Extracellular β-Glucosidase with Transglycosylation and Exo-glucosidase Activities from Fusarium oxysporum, Eur. J. Biochem., 1994, 224, 379–385 CrossRef CAS.
- J. J. Mateo and R. Di Stefano, Description of the β-glucosidase activity of wine yeasts, Food Microbiol., 1997, 14, 583–591 CrossRef CAS.
- F. Chiti, M. Stefani, N. Taddei, G. Ramponi and C. M. Dobson, Rationalization of the effects of mutations on peptide and protein aggregation rates, Nature, 2003, 424, 805–808 CrossRef CAS.
- H. J. Heipieper, G. Neumann, S. Cornelissen and F. Meinhardt, Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems, Appl. Microbiol. Biotechnol., 2007, 74, 961–973 CrossRef CAS.
- H. Nikaido, Multidrug efflux pumps of gram-negative bacteria, J. Bacteriol., 1996, 178, 5853 CrossRef CAS.
- O. Lomovskaya, M. Warren, J. Galazzo, R. Fronko, M. Lee and S. Chamberland, Targeting efflux pumps in Pseudomonas aeruginosa, 39th Interscience Conference for Antimicrobial Agents and Chemotherapy, San Francisco, 1999 Search PubMed.
- O. Lomovskaya, M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland and T. Renau, Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy, Antimicrob. Agents Chemother., 2001, 45, 105–116 CrossRef CAS.
- A. M. Klibanov, Why are enzymes less active in organic solvents than in water?, Trends Biotechnol., 1997, 15, 97–101 CrossRef CAS.
- R. M. Daniel, R. V. Dunn, J. L. Finney and J. C. Smith, The role of dynamics in enzyme activity, Annu. Rev. Biophys. Biomol. Struct., 2003, 32, 69–92 CrossRef CAS.
- C. J. Yeoman, Y. Han, D. Dodd, C. M. Schroeder, R. I. Mackie and I. K. Cann, Thermostable enzymes as biocatalysts in the biofuel industry, in Adv. Appl. Microbiol., Elsevier, 2010, vol. 70, pp. 1–55 Search PubMed.
- A. R. Alimon, Bioconversion and saccharification of some lignocellulosic wastes by Aspergillus oryzae ITCC-4857.01 for fermentable sugar production, Electron. J. Biotechnol., 2011, 14, 3 Search PubMed.
- I. ul Haq, F. Akram, M. A. Khan, Z. Hussain, A. Nawaz, K. Iqbal and A. J. Shah, CenC, a multidomain thermostable GH9 processive endoglucanase from Clostridium thermocellum: cloning, characterization and saccharification studies, World J. Microbiol. Biotechnol., 2015, 31, 1699–1710 CrossRef CAS.
- A. Zafar, S. Aftab, M. N. Aftab, H. Mukhtar and M. A. Saleem, Purification and characterization of cloned endo-1,4-β-glucanase gene of Bacillus licheniformis for saccharification of plant biomass, Pak. J. Bot., 2019, 51, 957–966 CAS.
- H. Boer, T. T. Teeri and A. Koivula, Characterization of Trichoderma reesei cellobiohydrolase Cel7A secreted from Pichia pastoris using two different promoters, Biotechnol. Bioeng., 2000, 69, 486–494 CrossRef CAS.
- F. Colussi, V. Serpa, P. d. S. Delabona, L. R. Manzine, M. L. Voltatodio, R. Alves, B. L. Mello, N. Pereira, C. S. Farinas and A. M. Golubev, Purification, and biochemical and biophysical characterization of cellobiohydrolase I from Trichoderma harzianum IOC 3844, J. Microbiol. Biotechnol., 2011, 21, 808–817 CrossRef CAS.
- A. Guilherme, P. Dantas, E. Santos, F. Fernandes and G. Macedo, Evaluation of composition, characterization and enzymatic hydrolysis of pretreated sugar cane bagasse, Braz. J. Chem. Eng., 2015, 32, 23–33 CrossRef.
- M. Kumar, P. Mishra and S. Upadhyay, Pyrolysis of Saccharum munja: optimization of process parameters using response surface methodology (RSM) and evaluation of kinetic parameters, Bioresour. Technol. Rep., 2019, 8, 100332 CrossRef.
- G. P. Singh, P. V. Madiwale and R. Adivarekar, Investigation of properties of cellulosic fibres extracted from Saccharum Munja grass and its application potential, International Journal of Fiber and Textile Research, 2017, 7, 30–37 Search PubMed.
- A. Klash, E. Ncube, B. Du Toit and M. Meincken, Determination of the cellulose and lignin content on wood fibre surfaces of eucalypts as a function of genotype and site, Eur. J. For. Res., 2010, 129, 741–748 CrossRef CAS.
- A. Demirbaş, Estimating of structural composition of wood and non-wood biomass samples, Energy Sources, 2005, 27, 761–767 CrossRef.
- A. Demirbaş, Properties of charcoal derived from hazelnut shell and the production of briquettes using pyrolytic oil, Energy, 1999, 24, 141–150 CrossRef.
- R. M. Shrestha and N. pratapsingh Budhathoki, The chemical compositions of Rhododendron arboreum, “Laligunras”, J. Nepal Chem. Soc., 2012, 30, 97–106 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra00545f |
| ‡ Both authors have equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |