Open Access Article
Open Access ArticleCo-immobilized recombinant glycosyltransferases efficiently convert rebaudioside A to M in cascade†
Zhenyang Wang‡
ae,
Wenbin Liu‡bc,
Wei Liubc,
Yuanyuan Ma*dg,
Yatong Libc,
Baoqi Wangbc,
Xiaozhen Weie,
Zhiming Liu*a and
Hao Song *bcfg
*bcfg
aCollege of Material Science and Engineering, Northeast Forestry University, Harbin 150040, China. E-mail: zhimingliu@nefu.edu.cn
bSchool of Chemical Engineering and Technology, Tianjin University, Tianjin 300072, China. E-mail: hsong@tju.edu.cn
cQingdo Institute of Ocean Engineering of Tianjin University, Qingdao 266237, China
dBiomass Conversion Laboratory, Tianjin R&D Center for Petrochemical Technology, Tianjin University, Tianjin 300072, China. E-mail: myy@tju.edu.cn
eR&D Division, Sinochem Health Company Ltd., Qingdao 266071, China
fFrontier Science Center for Synthetic Biology, Key Laboratory of Systems Bioengineering (Ministry of Education), Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China
gFrontier Technology Institute (Wuqing), Tianjin University, Tianjin 30072, China
First published on 28th April 2021
Abstract
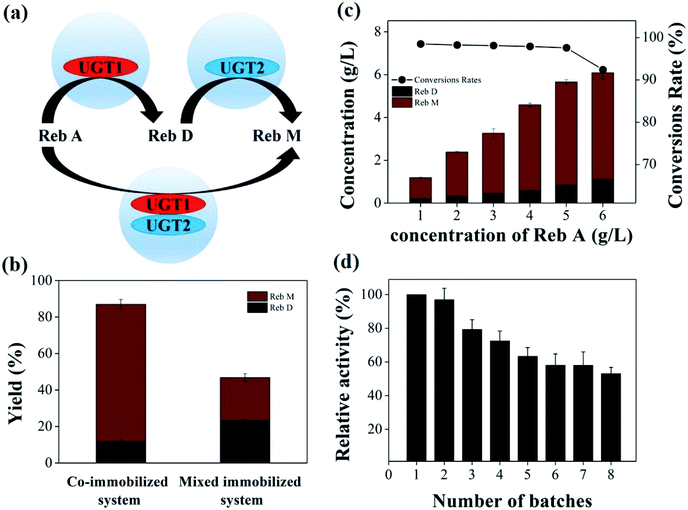
Rebaudioside M (Reb M), as a natural and healthy Stevia sweetener, is produced by two glycosyltransferases that catalyze the serial glycosylation of Rebaudioside A (Reb A) and Rebaudioside D (Reb D) in cascade. Meanwhile, it is of great importance in developing an immobilization strategy to improve the reusability of glycosyltransferases in reducing the production cost of Reb M. Here, the recombinant glycosyltransferases, i.e., OsEUGT11 (UGT1) and SrUGT76G1 (UGT2), were expressed in Escherichia coli and covalently immobilized onto chitosan beads. UGT1 and UGT2 were individually immobilized and co-immobilized onto the beads that catalyze Reb A to Reb M in one-pot. The co-immobilized enzymes system exhibited ∼3.2-fold higher activity than that of the mixed immobilized enzymes system. A fairly high Reb A conversion rate (97.3%) and a high Reb M yield of 72.2% (4.82 ± 0.11 g L−1) were obtained with a feeding Reb A concentration of 5 g L−1. Eventually, after 4 and 8 reused cycles, the co-immobilized enzymes retained 72.5% and 53.1% of their original activity, respectively, showing a high stability to minimize the total cost of enzymes and suggesting that the co-immobilized UGTs is of potentially signficant value for the production of Reb M.
1. Introduction
Steviol glycosides (SGs) in leaves of Stevia rebaudiana such as 1,2-stevioside, Rebaudioside A (Reb A), Rebaudioside D (Reb D) and Rebaudioside M (Reb M) (structures shown in Scheme 1) have attracted widespread attention for their zero-calorie energy content1 and received the Generally Recognized as Safe (GRAS) status from the United States Food and Drug Administration (USFDA).2 In S. rebaudiana, the 1,2-stevioside and Reb A are the most abundant steviol glucosides that constitute 5–10 and 2–4% w/w of the leaf dry weight, respectively.3 Reb D and Reb M have attracted extensive interest due to their high sweetness, which is 350-fold sweeter than sucrose and a much less prolonged bitter taste than most other SGs and they thus have higher prices than Reb A. Nevertheless, less abundance of Reb D and Reb M (approximately 0.4–0.5% w/w) in S. rebaudiana leads to several disadvantages in traditional extraction from the Stevia plant for industrial usage, including low efficiency, and water wasting.1,4 Recently, the biosynthesis of Reb D and Reb M in vivo or in vitro has attracted widespread interest.5,6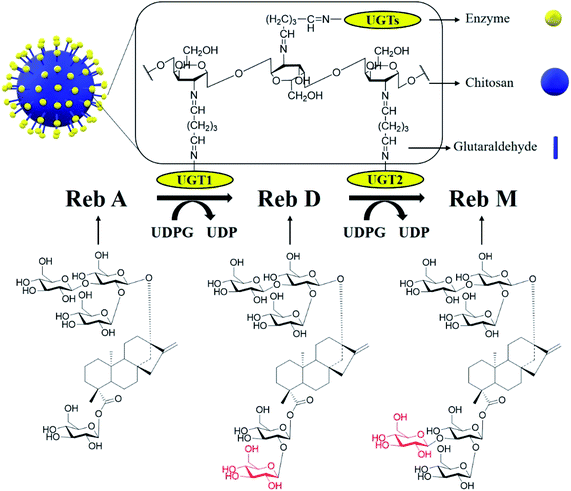 | ||
| Scheme 1 Biotransformation of Reb A to Reb D and Reb M by recombinant glycosyltransferases immobilized on chitosan beads. | ||
The biotransformation reactions of SGs in S. rebaudiana are catalyzed by a series of uridine diphosphate glucose (UDPG)-dependent glycosyltransferase (UGTs),6–8 e.g., UGT91D2 for biotransformation of Reb A to Reb D, and UGT76G1 for Reb D to Reb M.9,10 Similarly, the engineered enzymes EUGT11 from Oryza sativa L. and UGTSL2 from Solanum lycopersicum can also catalyze the biotransformation of Reb A to Reb D.9–14 Since Reb M is of much higher commercial value than that of Reb D, and its industrial production mainly comes from Reb D,1 which has a relatively high cost and low solubility. It is of great promise to develop a one-pot cascade reaction to produce Reb M directly from a cheap substrate such as Reb A to reduce the cost.
Extensive studies have shown that the expression of glycosyltransferases in bacteria typically results in rather low yields of soluble and active enzymes.6,15,16 It will lead to the increase in the enzyme cost and the high value of the final product, which seriously restricts the industrial production. Moreover, these free UGTs are not favorable to be used in industrial scale since they are not reusable and thus expensive in the enzymatic transformation process. It is of great need to develop immobilization technologies for the two cascaded enzymes to increase their reusability and reduce the production cost of these enzymes. Enzymes immobilized in insoluble supports present many advantages over bare enzymes, such as increased reusability, improved stability, sequester enzyme from product stream, and co-immobilization with other enzymes.17–19 A wide range of organic and inorganic supports, including ceramics and metal oxides, nanomaterials, and polymers have been investigated as supports for enzyme immobilization.20–23 Chitosan, as a representative of natural immobilized materials, is appropriate for immobilization of UGTs than other carriers due to its nontoxic, biocompatible nature, possessing suitable functional groups and amenable to chemical and enzymatic hydrolysis.24
Multiple free or immobilized enzymes were reported to be able to be synchronously used for the production of target products.25–28 Mixed cascade catalytic system contains two immobilized enzymes, thus the diffusion of intermediate products determines the reaction rate of the cascade reactions.18 However, the co-immobilized enzymes on a same support particle could significantly reduce the diffusion of the intermediate that mediates the cascade reactions, thus facilitating the formation of the final product. No studies had yet been conducted on the immobilization or co-immobilization of the glucosyltransferases (UGTs) for the production of Reb M from Reb A.
In this study, the recombinant enzymes, OsEUGT11 (UGT1) from Oryza sativa and SrUGT76G1 (UGT2) from S. rebaudiana, were expressed in Escherichia coli BL21(DE3). Upon purification, these enzymes were efficiently immobilized onto chitosan beads with glutaraldehyde as the cross-linking reagent. We then designed and established an effective co-immobilized system and investigated its catalytic activity. Both the co-immobilized and the mixed individually immobilized UGT1 and UGT2 can catalyze Reb A to Reb M in one-pot, while the co-immobilized enzymes system exhibited ∼3.2-fold higher activity than that of the mixed immobilized system. A high Reb A conversion rate of 97.3% and a high Reb M yield of 72.2% (4.82 g L−1) were achieved by the co-immobilized system with a Reb A feeding concentration of 5 g L−1. Finally, the co-immobilized enzyme could retain over 72.5% and 53.1% of its original activity after 4 and 8 repeated usages, showing considerable stability. Although immobilization would lead to the loss of enzyme activity, the total yield of Reb M with the successive batches exceeded that catalyzed by free enzymes, which would compensate for the increased cost of activity loss after two reuse cycles, suggesting that the co-immobilized UGTs is of significantly economic benefit for the production of Reb M. To the best of our knowledge, this is the first report on the efficient biotransformation of Reb M from Reb A with co-immobilized UGTs, establishing a basic foundation for the practical production of the sweetener Reb M.
2. Materials and methods
2.1 Materials
Reb A, Reb D and Reb M were gifted by Sinochem Health Company Ltd. UDPG disodium salt was purchased from Beijing Solarbio Science & Technology Company Ltd (Beijing, China). Chitosan, glutaraldehyde and the HPLC grade acetonitrile were purchased from Sinopharm Chemical Reagent Company Ltd (Shanghai, China). All chemicals used in this study were of analytical grade. Isopropyl-β-D-thiogalactopyranoside (IPTG) was purchased from Beijing Solarbio Science & Technology Co., Ltd. China.2.2 Production and purification of UGT1 and UGT2
Expression of recombinant UGT1 and UGT2 were referred to the method described by Kim et al.29 Briefly, the glycosyltransferase genes from Oryza sativa (UGT1, GenBank accession no. AK121682) and S. rebaudiana (UGT2, GenBank accession no. AY345974) were codon-optimized and synthesized by GenScript (Nanjing, China). Respectively, the resulting UGT1 or UGT2 was cloned in the pET28a(+) vector and transformed into the E. coli BL21(DE3) competent cells (TransGen Biotech, Beijing, China), resulting in the recombinant E. coli BL21 (pet28a-SrUGT76G1) strain or E. coli BL21 (pet28a-OsEUGT11) strain. The recombinant strain was cultivated in 400 mL Luria–Bertani (LB) medium containing kanamycin (50 mg mL−1) in 1 L flask until the OD600 reached approximately 0.6. The protein expression was induced by the addition of IPTG 0.1 mM IPTG and cells were incubated ∼14 hours at 18 °C. The cells were collected from the medium and resuspended in the Tris–HCl buffer at pH 7.0. Subsequently, the cells were lysed by ultra-sonication in an ice-water bath. After centrifugation, the His-tagged protein in the supernatant was purified through the affinity adsorption on Ni-NTA agarose column (Invitrogen). Protein mass concentration was measured by TaKaRa Bradford Protein Assay Kit with bovine serum albumin as a reference protein.2.3 Preparation of chitosan support
According to the reversed-phase suspension approach, the chitosan powder was dissolved in a 1% acetic acid aqueous solution to make a uniform transparent solution with a 4% mass volume. The resulting solution was dropped into a 4 M sodium hydroxide solution to form beads with a diameter of about 2.0 mm. The chitosan beads were washed with distilled water to neutrality and stored in distilled water at 4 °C.302.4 Enzyme immobilization
UGTs were cross-linked on chitosan beads by glutaraldehyde (Scheme 1). During the reaction, the impendent aldehyde group on the surface of chitosan beads interacted with amino group of enzymes to form imino group (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). The prepared chitosan beads were activated to a cross-linked state by adding a certain concentration of glutaraldehyde at 4 °C. Then the excess glutaraldehyde was washed off with distilled water after two hours. The activated chitosan beads were added into the enzyme solution of a certain concentration, which was oscillated at 4 °C for a certain period of time. Afterward, the beads were separated and the unbound enzyme was removed by a filter funnel with paper and washing 3 times with distilled water. The immobilized enzymes were used freshly and stored at 4 °C until use.
N–). The prepared chitosan beads were activated to a cross-linked state by adding a certain concentration of glutaraldehyde at 4 °C. Then the excess glutaraldehyde was washed off with distilled water after two hours. The activated chitosan beads were added into the enzyme solution of a certain concentration, which was oscillated at 4 °C for a certain period of time. Afterward, the beads were separated and the unbound enzyme was removed by a filter funnel with paper and washing 3 times with distilled water. The immobilized enzymes were used freshly and stored at 4 °C until use.
The effects of concentration (volume fraction) of glutaraldehyde (from 0.01% to 0.5%) and immobilized time (from 2 to 12 h) on immobilization were studied by single factor experiment. 5 mL of enzyme solution of different concentrations (from 0.01 to 0.06 mg mL−1) were added to 0.2 g of activated chitosan beads to determine the optimal amount of enzyme addition. Immobilization efficiency is reflected by the immobilization yield and the relative activity of the crosslinked enzyme in the reaction.31 The reaction was conducted at 37 °C (according to the method in Section 2.5) for 24 hours, and the maximum activity values of immobilized UGTs were regarded as 100%. The enzyme solutions before and after immobilization, and the washing solutions, were collected and their enzyme concentration was determined using bicinchoninic acid (BCA) protein assay. The immobilization yield was determined using the formula:
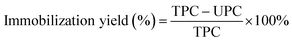 | (1) |
2.5 Assay for enzyme activity
The activity of free and immobilized UGTs refers to the method of Richman et al.32 with some modifications: a total of 0.05 mg of recombinant UGTs immobilized on 0.2 g of chitosan beads or 0.05 mg free UGTs was added to 1 mL of glucosyltransferase assay buffer (50 mM PBS buffer, pH 7.0, 3 mM MgCl2) along with corresponding substrate (1 g L−1 Reb A or 0.5 g L−1 Reb D and 1–3 mM UDPG) and incubated at 37 °C with varying reaction times toward different assays. The reaction was stopped by adding 0.4 mL of acetonitrile. Samples were filtered and detected using HPLC analysis. Under the above reaction conditions, one unit (U) of enzyme activity was defined as the amount of enzyme required to produce 1 μmol of Reb D/Reb M in 1 min under the standard assay condition. The specific activity was determined by the above method incubated for 3 hours (in the linear range of product formation, as shown in ESI Fig. 1†) and defined as the enzyme units per milligram of protein. Activity retention was defined as the ratio of the activity of the amount of the enzyme immobilized on the chitosan beads to the activity of the same amount of free enzyme.
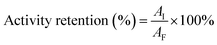 | (2) |
2.6 Co-immobilization of UGT1 and UGT2
UGT1 and UGT2 were co-immobilization onto chitosan beads. A total of 2 mL purified UGT1 (0.01 mg) and UGT2 (0.02–0.12 mg) were mixed to a scale (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 12) and incubated with 0.5 g glutaraldehyde-activated chitosan beads at one tube. The enzymes and chitosan beads were shaking at 200 rpm, 4 °C for 10 h to prepare the dual-enzyme co-immobilized system. To measure the activity of the dual-enzyme co-immobilized system, it was added to 2 mL glucosyltransferase assay buffer (50 mM PBS buffer, pH 7.0, 3 mM MgCl2) along with corresponding substrate (1 g L−1 Reb A and 4 mM UDPG). The reaction was performed at 37 °C for 6 days to detect the yield of Reb D and Reb M. The yield of Reb M catalyzed by the dual-enzyme co-immobilized system and the individually UGT1/UGT2 immobilization system were compared in this study. Furthermore, the effect of substrate concentration on the co-immobilized system was investigated under optimal conditions. The mass ratio of the substrate Reb A to the free enzyme was 1
2 to 12) and incubated with 0.5 g glutaraldehyde-activated chitosan beads at one tube. The enzymes and chitosan beads were shaking at 200 rpm, 4 °C for 10 h to prepare the dual-enzyme co-immobilized system. To measure the activity of the dual-enzyme co-immobilized system, it was added to 2 mL glucosyltransferase assay buffer (50 mM PBS buffer, pH 7.0, 3 mM MgCl2) along with corresponding substrate (1 g L−1 Reb A and 4 mM UDPG). The reaction was performed at 37 °C for 6 days to detect the yield of Reb D and Reb M. The yield of Reb M catalyzed by the dual-enzyme co-immobilized system and the individually UGT1/UGT2 immobilization system were compared in this study. Furthermore, the effect of substrate concentration on the co-immobilized system was investigated under optimal conditions. The mass ratio of the substrate Reb A to the free enzyme was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.11 in the co-immobilized system and the concentration of products was detected on day 6. Additionally, the daily volumetric productivity was calculated based on the Reb D and Reb M products concentration, which was determined by liquid chromatography and the biotransformation time, which was measured when >90% of the starting material was converted to the products.33 The conversion rate (%) of Reb A and the yield (%) of Reb D and Reb M were calculated using the following formula:
0.11 in the co-immobilized system and the concentration of products was detected on day 6. Additionally, the daily volumetric productivity was calculated based on the Reb D and Reb M products concentration, which was determined by liquid chromatography and the biotransformation time, which was measured when >90% of the starting material was converted to the products.33 The conversion rate (%) of Reb A and the yield (%) of Reb D and Reb M were calculated using the following formula:
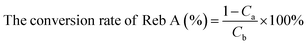 | (3) |
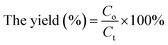 | (4) |
2.7 Determination of the optimum temperature and pH
The temperature activity profile of free and immobilized glucosyltransferase was determined in the range of 18–45 °C at pH 7.0. The effect of pH on the activity of free and immobilized glucosyltransferase was studied at 37 °C by varying the pH of the reaction mixture in the range of 6.5–8.5 using phosphate sodium buffer. The reaction was conducted for 24 hours (according to the method in Section 2.5), and the maximum activity values of immobilized UGTs were regarded as 100%.2.8 Operational stability (reusability)
To evaluate the reuse of immobilized biocatalysts, each immobilized UGT1, UGT2 or the dual-enzyme co-immobilized system was added to 1 mL of glucosyltransferase assay buffer (pH 7.0) containing substrate Reb A or Reb D. The reaction was conducted at 37 °C for 24 h and its activities was determined by the rate of product. After the reaction, the immobilized biocatalysts were filtered, washed with fresh buffer and used in a new catalysis cycle. The formula for calculating the reusable efficiency is provided below:
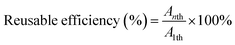 | (5) |
2.9 HPLC analysis
HPLC were used to quantitatively analyze the substrates and products in the reaction systems. Reb A, Reb D and Reb M were separated using an octadecylsilane (ODS) column (Shim-pack GIST, 4.6 × 150 mm, Shimadzu Corporation, Kyoto, Japan) under the following chromatographic conditions: UV detection wavelength 210 nm; flow rate 1.0 mL min−1; mobile phase, water and acetonitrile at 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 (v/v); and injection volume 5 μL. The retention times of Reb A, Reb D and Reb M were 6.04, 7.56 and 19.87 min, respectively.
25 (v/v); and injection volume 5 μL. The retention times of Reb A, Reb D and Reb M were 6.04, 7.56 and 19.87 min, respectively.
3. Results and discussion
3.1 Establishing one-pot cascade enzymatic transformation system from Reb A to Reb M
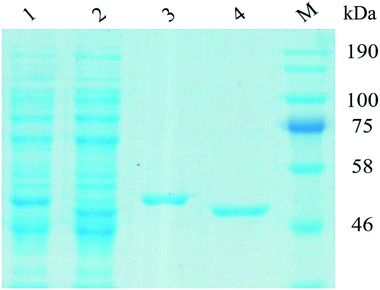
The recombinant UGT1 and UGT2 were firstly expressed in E. coli. The purified UGT1 and UGT2 showed a single band corresponding to 51.33 and 50.89 kDa on PAGE-gel (Fig. 1), demonstrating the purity level of the two enzymes.To implement the targeted production of Reb M from Reb A, we designed a one-pot cascade enzymatic transformation system. The cascade had two enzymatic reactions and two enzymes (Scheme 1). The crude extract of E. coli cells containing UGT1 and the purified UGT1 can catalyze the biotransformation from Reb A to Reb D. And the purified UGT2 and the crude extract can catalyze Reb D to produce Reb M. The specific activity of UGT1 and UGT2 were 15.8 ± 0.3 mU mg−1 for Reb A and 4.1 ± 0.1 mU mg−1 for Reb D, suggesting successful expression of the highly active UGTs. This cascade reaction route was designed with the following basic principles in mind: (1) the reaction conditions in the two reaction modules need to be compatible;34,35 (2) to reduce the precipitation of intermediate products (Reb D) for the accumulation of the final product (Reb M). Although many UGTs can catalyze the biotransformation of Reb A to Reb D and to Reb M, coordinating these two enzyme-catalyzed reactions producing Reb M directly from Reb A is challenging.6–10,36,37
3.2 Immobilization of the UGTs on chitosan beads via cross-linking with glutaraldehyde
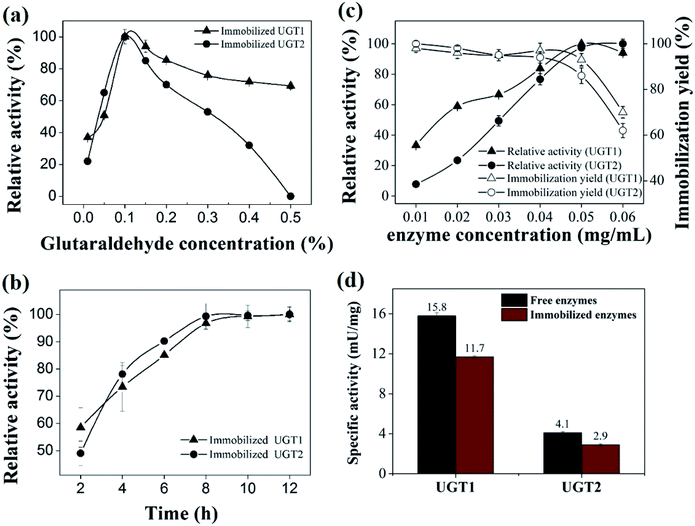
To investigate the effect of glutaraldehyde concentration on UGTs immobilization, the UGT1 and UGT2 were immobilized on chitosan beads cross-linked with different concentrations of glutaraldehyde ranging from 0.01% to 0.5% (Fig. 2a). The maximal activity was obtained when using 0.1% cross-linked beads as carrier, suggesting 0.1% glutaraldehyde is the optimal crosslinker concentration. As a versatile immobilization biomaterial, chitosan possesses a variety of functional groups that facilitate efficient binding between chitosan and enzymes.38–40 An increase in the aldehyde group on the surface of beads can provide increasing binding sites for UGTs, resulting in a higher UGTs loading and activity. A high glutaraldehyde level above 0.1% led to an extensive intramolecular or intermolecular cross-linking interaction of individual enzymes and aldehyde groups on the surface of chitosan beads would shield the active sites and limit enzyme activities.41 Therefore, 0.1% of the glutaraldehyde used to be chosen for further studies.The immobilization time has an effect on the amount of enzyme with the chitosan beads, which resulted in a deterioration of enzyme activity.42 UGTs was immobilized on chitosan beads for different times, varying from 1 h to 12 h (Fig. 2b). The immobilized enzymes exhibited the maximal activity at 8 h, an optimal crosslinking time of the enzyme with glutaraldehyde.
The effect of enzyme concentration on the activity and yield of the immobilized UGT1 and UGT2 was investigated (Fig. 2c). When the loading concentration of UGTs on chitosan beads were in the range from 0.01 to 0.05 mg mL−1, the activities of the immobilized UGTs were improved with the increase in the loading concentration. When the loading concentration of UGTs was over 0.05 mg mL−1, the activities of the immobilized UGTs were decreased significantly, suggesting the binding sites of UGTs provided by the carrier were saturated. Hence, the enzyme concentration of 0.05 mg mL−1 was selected for further investigations. Thus, the immobilized UGTs with loading level of 0.05 mg mL−1 displayed their maximum relative activities.
The purified UGTs covalently immobilized onto chitosan beads exhibited high specific activity after two cycles of reusage, which were 11.9 ± 0.1 mU mg−1 for the immobilized UGT1 and 2.9 ± 0.1 mU mg−1 for the immobilized UGT2, significantly higher than that of the original specific activity (4.4 ± 0.2 mU mg−1 of the immobilized UGT1, and 1.1 ± 0.1 mU mg−1 of the immobilized UGT2). It was likely that the active site of the enzyme was blocked due to glutaraldehyde crosslinking after immobilization and it was released continuously during the enzymatic reaction process.41 The specific activities of the optimized immobilized UGTs were comparable to those of the free enzymes (Fig. 2d). Comparative to free enzymes, the activity retention of immobilized UGT1 and UGT2 reached ∼74.1% and ∼70.7%, respectively. Chemical fixation techniques by using cross-linking agents would result in fairly stable immobilized enzymes, which could be easily sequestered from the product stream.18,43 The optimized immobilized UGTs could retain most of their activity of the free enzymes. The immobilized enzyme with regained activity at the second round of usage was then chosen for the following studies.
3.3 Effect of temperature and pH on activity and reusability of the immobilized UGTs
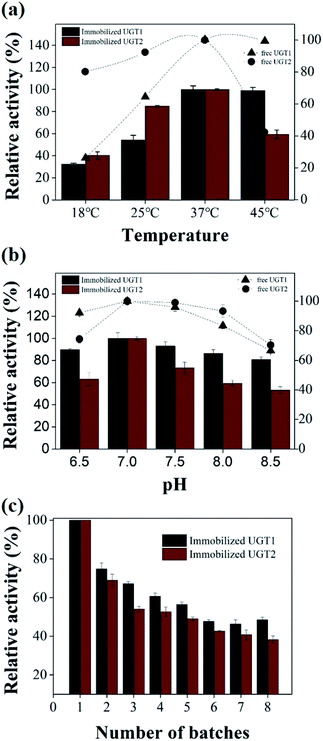
The effect of temperature on the UGTs activity of free and immobilized enzymes was studied by conducting the catalytic reaction at different temperatures between 18 and 45 °C (Fig. 3a). The optimum temperature for the immobilized UGTs was 37 °C, which was similar to free enzymes. When the temperature was 25 °C, the relative activity of immobilized UGT1 and UGT2 was 54% and 85%, indicating that the immobilized UGT2 showed a higher catalytic activity in the temperature range from 25 °C to 37 °C. When the temperature was 45 °C, the immobilized UGT1 retained 98% activity, while UGT2 only maintained 59% activity, suggesting the immobilized UGT1 showed higher activity than UGT2 in the range of 37–45 °C.The effects of pH on UGTs activities were measured at pH 6.5.0–8.5 (Fig. 3b). The optimal pH for the free and immobilized enzymes was 7, which was similar to other glycosyltransferases.14,44 The immobilized UGT2 showed 53–73% of relative activity at pH 6.5, 7.5 and 8.5, while the immobilized UGT1 retained 80% activity in the range of 6.5–8.5, suggesting that the immobilized UGT1 displayed a higher catalytic activity over a wide pH range of 6.5–8.5. Therefore, the temperature of 37 °C and pH 7.0 were chosen for the following studies.
The efficient reusability of immobilized enzymes up to several cycles is highly needed, which also plays a key role in developing a cost-effective reaction.45 The activity of the immobilized UGTs was measured for each recycle (Fig. 3c). The activity of the first batch was taken as a reference (100%). The immobilized UGT1 retained 60.6% and 48.5% relative activity after 4 and 8 repeated usages, and the immobilized UGT2 retained 52.6% and 38.2% relative activity after 4 and 8 recycles, respectively. Thus, the immobilized UGTs displayed considerable stability under operational conditions, and UGT1 has a higher stability than UGT2 after immobilization. In all, these two enzymes showed compatible properties including temperature, pH and reusability after immobilization, which were suitable for the constitution of a co-immobilization system.34,35,46
3.4 Co-immobilization of UGT1 and UGT2
Co-immobilized enzymes were established, as one of the one-pot cascade enzymatic transformation system, to improve the productivity.47–49 The UGT1 and UGT2 were co-immobilized onto the chitosan beads to form a dual-enzyme system for directly obtain Reb M from Reb A by the cascade reactions. Various ratios of UGT1 to UGT2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 1
2 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) were used for co-immobilization, the yield of Reb M was measured (Fig. 4a). When the mass ratio of UGT1 to UGT2 was 1
10) were used for co-immobilization, the yield of Reb M was measured (Fig. 4a). When the mass ratio of UGT1 to UGT2 was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, the maximum yield of the Reb M was obtained. Excess UGT2 could not improve the yield of Reb M catalyzed by the co-immobilized UGT1 and UGT2. The appropriate mass ratio of enzymes in the co-immobilized system would reduce the precipitation of intermediate products and accumulate the final product.50,51 Thus, in this study, 1
10, the maximum yield of the Reb M was obtained. Excess UGT2 could not improve the yield of Reb M catalyzed by the co-immobilized UGT1 and UGT2. The appropriate mass ratio of enzymes in the co-immobilized system would reduce the precipitation of intermediate products and accumulate the final product.50,51 Thus, in this study, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (UGT1 to UGT2) was selected as an optimum mass ratio of the two enzymes to further study the effects of the other process variables.
10 (UGT1 to UGT2) was selected as an optimum mass ratio of the two enzymes to further study the effects of the other process variables.
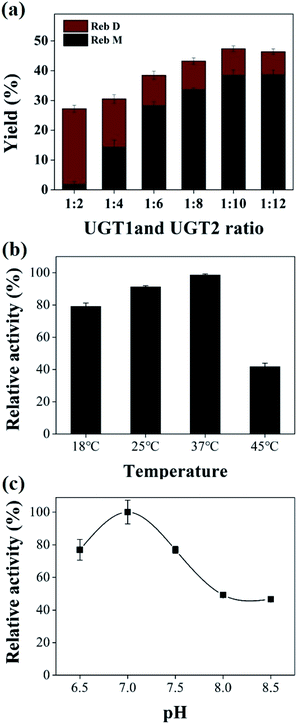
The co-immobilized UGTs showed the highest catalytic activity at 37 °C (Fig. 4b). 80.0–91.2% of the maximum activity could be retained in the range of 18 to 25 °C. The relative activity of co-immobilized UGTs reached 41% of the maximum at 45 °C. The co-immobilized enzyme system had a similar activity trend to the free UGT2 at various temperature conditions. The optimal pH of the co-immobilized UGTs was 7.0 for Reb M production (Fig. 4c). The co-immobilized UGTs showed 46–76% of relative activity at pH 6.5, 7.5 and 8.5, which has a similar trend to that of the immobilized UGT2. Therefore, 37 °C and pH of 7.0 were selected for the subsequent studies.
3.5 Comparison of the co-immobilized and the mixed immobilized UGTs
UGT1 and UGT2 were individually immobilized on two carriers to form a mixed immobilized system. The yield of Reb M catalyzed by the co-immobilized and the mixed immobilized UGTs were compared (Fig. 5a). The co-immobilized UGTs can convert 87.1% of Reb A, which is 1.7-fold higher than that by using mixed UGTs (Fig. 5b). The two systems with various ratios of UGT1 to UGT2 were also compared (ESI Fig. 2†). Similarly, the co-immobilized enzymes exhibited higher activity than that of the mixed immobilized enzymes. The yield of Reb M was ∼75.4% by the dual-enzyme co-immobilized system, ∼3.2-fold higher than that by the mixed immobilized UGTs (∼23.8%). These data illustrated that the co-immobilized enzymes had higher efficiency for the Reb M production than the mixed immobilized system. When the two enzymes were immobilized on different carrier beads, the substrate transfer had to overcome double diffusion barriers, restricting the cascade catalytic reactions.26 In contrast, the two enzymes were fixed in the same carrier bead in the dual-enzyme co-immobilization system would reduce the transport time of the substrate and intermediate loss due to diffusion,52 the substrate was instantly converted to the final product due to that two closely enzymes constituted a efficiently cascade biocatalyst.28,53–55 Therefore, this co-immobilization system enabled a fast transfer of substrate, thus enhancing the overall catalytic efficiency of the cascade reactions.Product with high concentration can yield relatively low separation costs, which is valuable for the large-scale production. Therefore, the effect of substrate concentration on the co-immobilized system was investigated under optimized mass ratio of UGTs, temperature and pH conditions (Fig. 5c). The ratio of substrate to enzyme was 1 mg Reb A to 0.55 mg of UGTs immobilized onto chitosan beads. The conversion rate of Reb A changed from 98.1% to 92.4% at the 6th day of incubation, with the feeding concentration of Reb A from 1 to 6 g L−1 (Table 1). When the feeding substrate concentration was 5–6 g L−1, the products reached 5.66 ± 0.12 g L−1 (including 0.84 ± 0.01 g L−1 Reb D, 4.82 ± 0.11 g L−1 Reb M) and 6.09 ± 0.32 g L−1 (including 1.09 ± 0.04 g L−1 Reb D, 4.99 ± 0.28 g L−1 Reb M), and the Reb M yield of 72.2% and 62.3% were obtained via the formula (4), respectively. Additionally, the daily volumetric productivity reached 0.94 ± 0.02 g L−1 and 1.02 ± 0.04 g L−1, respectively. Thus, a high conversion rate (97.3%) and Reb M yield (72.2%) with 5 g L−1 Reb A feeding demonstrated a great potential of the co-immobilized UGTs for Reb M production from Reb A in one-pot. The trace residue of Reb D was caused by the relatively low UGT2 activity and stability, which may be solved by enhanced stability and activity of UGT2 with the glycosylated C19-carboxylic acid functional group.9,56
| The feeding substrate Reb A concentration (g L−1) | 1 | 2 | 3 | 4 | 5 | 6 |
| Conversion rate (%) | 98.5 | 98.2 | 98.1 | 97.9 | 97.6 | 92.4 |
| Reb D concentration (g L−1) | 0.20 ± 0.01 | 0.32 ± 0.02 | 0.44 ± 0.02 | 0.57 ± 0.03 | 0.83 ± 0.03 | 1.09 ± 0.04 |
| Reb M concentration (g L−1) | 0.99 ± 0.02 | 2.06 ± 0.05 | 2.81 ± 0.19 | 4.02 ± 0.09 | 4.83 ± 0.11 | 4.99 ± 0.28 |
| Daily volumetric productivity (g L−1) | 0.2 ± 0.01 | 0.4 ± 0.01 | 0.54 ± 0.03 | 0.77 ± 0.04 | 0.94 ± 0.02 | 1.02 ± 0.04 |
The operational stability of the dual-enzyme co-immobilization system was investigated by measuring the activity of the optimized co-immobilized enzyme after cycling 8 repeated usages (Fig. 5d). The highest activity (100%) was considered as the first cycle. As the number of repeated cycles increased, the activity of the co-immobilized UGTs decreased. Additionally, the reusability of the mixed immobilized UGTs same with that of the immobilized UGT2 showed a downtrend (ESI Fig. 3†). The washing step of the immobilization process would lead to a loss of enzymes, and the inactivation of UGTs molecules in the repeatedly adjusted solution have influenced the active site of enzyme during the cycling process.57–59 After 4 and 8 repeated uses, the co-immobilized enzyme retained above 72.5% and 53.1% of its original activity, respectively. The stability of the enzyme was greatly improved upon co-immobilization onto the beads.60–62
The costs of each component for Reb M production catalyzed by free and co-immobilized enzyme at the laboratory level was calculated to assess the cost difference for producing 1.0 g Reb M from Reb A. The total costs for the former case have ∼4-fold higher than that of the latter case. The cost difference is mainly due to the consumption of free enzymes. In addition, the immobilization costs include the carrier and crosslinker that are neglectable due to their low costs.63,64 Therefore, according to the yield of Reb M for eight cycles of reusage by the co-immobilized enzymes, the costs for the two case would be estimated and compared. Although immobilization may lead to a loss of enzyme activity, the total yield of Reb M with the successive batches of reusage has exceeded that catalyzed by free enzymes, which would compensate for the increased cost of activity loss. In addition, the yield of Reb M was ∼75.4% by the dual-enzyme co-immobilized system, and it retained above 97% of its original activity after two repeats. Thus, the co-immobilized enzymes showed a significant economic advantage after two reusing cycles.
4. Conclusions
In summary, the purified recombinant glycosyltransferase UGT1 and UGT2 from E. coli BL21(DE3) were successfully produced, purified, and immobilized into the chitosan beads to improve the reusability of these glycosyltransferases. Enzymes would be sequestered from product stream and the production cost was thus minimized. To reduce the transport time of the substrate and intermediate loss due to diffusion in the multi-enzyme cascade reactions, we developed a co-immobilization strategy that could efficiently convert Reb A to Reb M in cascade in one-pot. The co-immobilized system of UGT1 and UGT 2 exhibited ∼3.2-fold higher activity than that of the individually immobilized system. A high substrate (Reb A) conversion rate of 97.3% and a Reb M yield of 72.2% (4.82 ± 0.11 g L−1) were obtained with the Reb A feeding concentration of 5 g L−1. Although a loss of enzyme activity upon immobilization, the co-immobilized enzymes retained 72.5% and 53.1% of its original activity after 4 and 8 rounds of reusages, respectively. The successive batches of reusage would compensate for the loss of enzyme activity. The co-immobilized UGT1 and UGT2 is thus a viable way to efficiently produce Reb M from Reb A in one-pot.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We acknowledge the financial support from the National Key Research and Development Program of China (2017YFD0601004).References
- K. Olsson, S. Carlsen, A. Semmler, E. Simon, M. D. Mikkelsen and B. L. Moller, Microb. Cell Fact., 2016, 15, 207 CrossRef PubMed.
- M. Younes, G. Aquilina, K. H. Engel, P. Fowler, M. J. Frutos Fernandez, P. Furst, R. Gurtler, U. Gundert-Remy, T. Husoy, W. Mennes, P. Moldeus, A. Oskarsson, R. Shah, I. Waalkens-Berendsen, D. Wolfle, G. Degen, D. Gott, J. C. Leblanc, L. Herman, J. Aguilera, A. Giarola, A. M. Rincon and L. Castle, EFSA J., 2019, 17, e05867 Search PubMed.
- V. Chatsudthipong and C. Muanprasat, Pharmacol. Ther., 2009, 121, 41–54 CrossRef CAS PubMed.
- A. U. Jackson, A. Tata, C. Wu, R. H. Perry, G. Haas, L. West and R. G. Cooks, Analyst, 2009, 134, 867–874 RSC.
- Y. H. Zhang, Biotechnol. Bioeng., 2010, 105, 663–677 CAS.
- A. A. Mohamed, S. Ceunen, J. M. Geuns, W. Van den Ende and M. De Ley, J. Plant Physiol., 2011, 168, 1136–1141 CrossRef CAS PubMed.
- R. R. Zheng, Z. Y. Zhu, Y. L. Wang, S. Y. Hu, W. Xi, W. Xiao, X. L. Qu, L. L. Zhong, Q. Fu and C. Y. Wang, Front. Plant Sci., 2019, 10, 12 CrossRef PubMed.
- B. P. Wu, X. M. Cao, H. R. Liu, C. Q. Zhu, H. Klee, B. Zhang and K. S. Chen, J. Exp. Bot., 2019, 70, 925–936 CrossRef CAS PubMed.
- Z. Liu, J. Li, Y. Sun, P. Zhang and Y. Wang, Plant Commun., 2020, 1, 100004 CrossRef PubMed.
- P. Indra, M. Avetik and B. Cynthia, Foods, 2014, 3, 162–175 CrossRef PubMed.
- M. Lin, F. Wang and Y. Zhu, Biochem. Eng. J., 2020, 159, 107626 CrossRef CAS.
- L. Chen, P. Sun, F. Zhou, Y. Li, K. Chen, H. Jia, M. Yan, D. Gong and P. Ouyang, Food Chem., 2018, 259, 286–291 CrossRef CAS PubMed.
- J. Tao, G. Li, X. Liang, A. Tao and S. Diego, Enzymatic method for preparing rebaudioside M, US Pat., US20190233866A1, 2019 Search PubMed.
- L. Chen, R. Cai, J. Weng, Y. Li, H. Jia, K. Chen, M. Yan and P. Ouyang, Microb. Biotechnol., 2020, 13, 974–983 CrossRef CAS PubMed.
- S. M. Forget, J. Na, N. E. McCormick and D. L. Jakeman, Org. Biomol. Chem., 2017, 15, 2725–2729 RSC.
- D. M. Liang, J. H. Liu, H. Wu, B. B. Wang, H. J. Zhu and J. J. Qiao, Chem. Soc. Rev., 2015, 44, 8350–8374 RSC.
- V. L. Sirisha, A. Jain and A. Jain, Adv. Food Nutr. Res., 2016, 79, 179–211 CAS.
- R. DiCosimo, J. McAuliffe, A. J. Poulose and G. Bohlmann, Chem. Soc. Rev., 2013, 42, 6437–6474 RSC.
- M. C. R. Franssen, P. Steunenberg, E. L. Scott, H. Zuilhof and J. P. M. Sanders, Chem. Soc. Rev., 2013, 42, 6491–6533 RSC.
- E. D. Yushkova, E. A. Nazarova, A. V. Matyuhina, A. O. Noskova, D. O. Shavronskaya, V. V. Vinogradov, N. N. Skvortsova and E. F. Krivoshapkina, J. Agric. Food Chem., 2019, 67, 11553–11567 CrossRef CAS PubMed.
- R. J. White, R. Luque, V. L. Budarin, J. H. Clark and D. J. Macquarrie, Chem. Soc. Rev., 2009, 38, 481–494 RSC.
- C. Sanchez, P. Belleville, M. Popall and L. Nicole, Chem. Soc. Rev., 2011, 40, 696–753 RSC.
- A. Carné, C. Carbonell, I. Imaz and D. Maspoch, Chem. Soc. Rev., 2011, 40, 291–305 RSC.
- B. Krajewska, Enzyme Microb. Technol., 2004, 35, 126–139 CrossRef CAS.
- E. T. Hwang and S. Lee, ACS Catal., 2019, 9, 4402–4425 CrossRef CAS.
- S. Ren, C. Li, X. Jiao, S. Jia, Y. Jiang, M. Bilal and J. Cui, Chem. Eng. J., 2019, 373, 1254–1278 CrossRef CAS.
- Y. Zhang and H. Hess, ACS Catal., 2017, 7, 6018–6027 CrossRef CAS.
- J. F. Kornecki, D. Carballares, P. W. Tardioli, R. C. Rodrigues, Á. Berenguer-Murcia, A. R. Alcántara and R. Fernandez-Lafuente, Catal. Sci. Technol., 2020, 10, 5740–5771 RSC.
- M. J. Kim, J. Zheng, M. H. Liao and I. C. Jang, Plant Biotechnol. J., 2019, 17, 1037–1047 CrossRef CAS PubMed.
- S. Lei, H. Yu, C. Huang and X. Zheng, Food Sci., 2013, 34, 233–238 CAS.
- I. Kaleem, A. Rasool, B. Lv, N. Riaz, J. U. Hassan, R. Manzoor and C. Li, Chem. Eng. Sci., 2017, 162, 332–340 CrossRef CAS.
- A. Richman, A. Swanson, T. Humphrey, R. Chapman, B. McGarvey, R. Pocs and J. Brandle, Plant J., 2005, 41, 56–67 CrossRef CAS PubMed.
- J. H. Seo, H. H. Kim, E. Y. Jeon, Y. H. Song, C. S. Shin and J. B. Park, Sci. Rep., 2016, 6, 28223 CrossRef CAS PubMed.
- D. Zhang, X. Jing, W. Zhang, Y. Nie and Y. Xu, RSC Adv., 2019, 9, 29927–29935 RSC.
- M. Zhao, X. Hong, Abdullah, R. Yao and Y. Xiao, Green Chem., 2021, 23, 838–847 RSC.
- T. Yang, J. Zhang, D. Ke, W. Yang, M. Tang, J. Jiang, G. Cheng, J. Li, W. Cheng, Y. Wei, Q. Li, J. H. Naismith and X. Zhu, Nat. Commun., 2019, 10, 3214 CrossRef PubMed.
- Z. Wang, J. Hong, S. Ma, T. Huang, Y. Ma, W. Liu, W. Liu, Z. Liu and H. Song, Int. J. Biol. Macromol., 2020, 163, 1669–1676 CrossRef CAS PubMed.
- M. L. Verma, S. Kumar, A. Das, J. S. Randhawa and M. Chamundeeswari, Environ. Chem. Lett., 2020, 18, 315–323 CrossRef CAS.
- R. C. F. Cheung, T. B. Ng, J. H. Wong and W. Y. Chan, Mar. Drugs, 2015, 13, 5156–5186 CrossRef CAS PubMed.
- G. Hojnik Podrepšek, Ž. Knez and M. Leitgeb, Nanomaterials, 2020, 10, 1913 CrossRef PubMed.
- I. Migneault, C. Dartiguenave, M. J. Bertrand and K. C. Waldron, BioTechniques, 2004, 37, 798–802 CrossRef PubMed.
- D. S. Jiang, S. Y. Long, J. Huang, H. Y. Xiao and J. Y. Zhou, Biochem. Eng. J., 2005, 25, 15–23 CrossRef CAS.
- M. L. Verma, Environ. Chem. Lett., 2017, 15, 555–560 CrossRef CAS.
- L. Bungaruang, A. Gutmann and B. Nidetzky, Adv. Synth. Catal., 2013, 355, 2757–2763 CrossRef CAS PubMed.
- Z. Lei and Q. Jiang, J. Agric. Food Chem., 2011, 59, 2592–2599 CrossRef CAS PubMed.
- D. L. Morris, J. Campbell and W. E. Hornby, Biochem. J., 1975, 147, 593–603 CrossRef CAS PubMed.
- S. Talekar, S. Desai, M. Pillai, N. Nagavekar, S. Ambarkar, S. Surnis, M. Ladole, S. Nadar and M. Mulla, RSC Adv., 2013, 3, 2265–2271 RSC.
- E. J. Cho, S. Jung, H. J. Kim, Y. G. Lee, K. C. Nam, H.-J. Lee and H.-J. Bae, Chem. Commun., 2012, 48, 886–888 RSC.
- N. Ghéczy, K. Sasaki, M. Yoshimoto, S. Pour-Esmaeil, M. Kröger, P. Stano and P. Walde, RSC Adv., 2020, 10, 18655–18676 RSC.
- H. Torabizadeh and E. Montazeri, Carbohydr. Res., 2020, 488, 107904 CrossRef CAS PubMed.
- D. Y. Wu, M. Z. Zhuang, G. X. Su, P. F. Gao, Y. Z. Jin and H. Y. Fu, IOP Conf. Ser. Earth Environ. Sci., 2020, 435, 012006 CrossRef.
- X. Yu, Z. Zhang, J. Li, Y. Su, M. Gao, T. Jin and G. Chen, Bioresour. Technol., 2021, 321, 124509 CrossRef CAS PubMed.
- F. Kazenwadel, M. Franzreb and B. E. Rapp, Anal. Methods, 2015, 7, 4030–4037 RSC.
- J. Zdarta, K. Bachosz, O. Degórska, A. Zdarta, E. Kaczorek, M. Pinelo, A. S. Meyer and T. Jesionowski, Materials, 2019, 12, 3167 CrossRef CAS PubMed.
- P. Han, X. Zhou and C. You, Front. Bioeng. Biotechnol., 2020, 8, 380 CrossRef PubMed.
- Y. Yang, S. Huang, Y. Han, H. Yuan, C. Gu and Y. Zhao, Plant Physiol. Biochem., 2014, 80, 220–225 CrossRef CAS PubMed.
- Z. Gao, Y. Yi, J. Zhao, Y. Xia, M. Jiang, F. Cao, H. Zhou, P. Wei, H. Jia and X. Yong, Bioresour. Bioprocess., 2018, 5, 27 CrossRef.
- V. C. Badgujar, K. C. Badgujar, P. M. Yeole and B. M. Bhanage, Enzyme Microb. Technol., 2021, 144, 109738 CrossRef CAS PubMed.
- X. Han, G. Liu, W. Song and Y. Qu, Bioresour. Technol., 2018, 248, 248–257 CrossRef CAS PubMed.
- J. J. Virgen Ortíz, S. G. Pedrero, L. Fernandez Lopez, N. Lopez Carrobles, B. C. Gorines, C. Otero and R. Fernandez Lafuente, Molecules, 2017, 22, 91 CrossRef PubMed.
- Y. Chou, C. Ko, L. O. Chen, C. Yen and J. Shaw, Molecules, 2015, 20, 3744–3757 CrossRef CAS PubMed.
- T. S. Kim, S. K. S. Patel, C. Selvaraj, W. S. Jung, C. H. Pan, Y. C. Kang and J. K. Lee, Sci. Rep., 2016, 6, 33438 CrossRef CAS PubMed.
- K. Pospiskova and I. Safarik, Carbohydr. Polym., 2013, 96, 545–548 CrossRef CAS PubMed.
- Y. Zhang, D. Wang, X. Gao, W. Xu, P. Liang and Y. Xu, Am. J. Analyt. Chem., 2013, 4, 48–53 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10574k |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2021 |